Abstract
Aggregation of prion proteins is the cause of various prion related diseases. The infectious form of prions, amyloid fibrils, exist as multiple strains. The strains are thought to represent structurally different prion protein molecules packed into amyloid fibrils, but the knowledge on the structure of different types of aggregates is limited. Here we report on the use of AFM (Atomic Force Microscopy) and TERS (Tip-Enhanced Raman Scattering) to study morphological heterogeneity and access underlying conformational features of individual amyloid aggregates. Using AFM we identified morphology of amyloid fibrils formed by the peptide (CGNNQQNY) from the yeast prion protein Sup35 that is critically involved in the aggregation of the full protein. TERS results demonstrate that morphologically different amyloid fibrils are composed of a distinct set of conformations. Fibrils formed at pH 5.6 are composed of a mixture of peptide conformations (β-sheets, random coil and α-helix) while fibrils formed in pH~2 solution primarily have β-sheets. Additionally, peak positions in the amide III region of the TERS spectra suggested that peptides have parallel arrangement of β-sheets for pH~2 fibrils and antiparallel arrangement for fibrils formed at pH 5.6. We also developed a methodology for detailed analysis of the peptide secondary structure by correlating intensity changes of Raman bands in different regions of TERS spectra. Such correlation established that structural composition of peptides is highly localized with large contribution of unordered secondary structures on a fibrillar surface.
1. Introduction
Self-assembly of proteins and peptides into amyloid fibrils is a ubiquitous phenomenon characteristic to the majority if not all polypeptide sequences.[1, 2] The first critical step in the self-assembly process is the acquisition of an aggregation prone conformation followed by self-association events resulting in the formation of aggregated species.[3] The formation of aberrant protein aggregates has been long associated with the development of many neurodegenerative diseases. A vivid example of aberrant aggregation is the formation of prions. Prions are infectious species that are “misfolded” proteins with abnormal conformation. They propagate by imposing their conformation onto normal cellular proteins leading to the formation of aggregates and ultimately to neurodegeneration.[4] One of the most intriguing phenomena is the formation of various strains by the same prion protein with the same primary sequence. These strains are associated with amyloids that can adopt distinct conformations and produce distinct phenotypes.[5] Fibrils of the same polypeptide sequence can form distinct morphologies exhibiting a great wealth of polymorphism.[6] Structural characteristics of a polypeptide sequence forming aggregates dictate amyloid morphology and strength of the phenotype.[7]
We have reported recently that aggregation patterns of an octapeptide (CGNNQQNY) from the yeast prion protein Sup35 is significantly affected by the environmental conditions.[8] Subtle changes in the environment influence the morphology of aggregated structures. In particular we showed that the electrostatic interactions define the kinetics of peptide's aggregation as well as the final morphology of the aggregated species. Both, neutralization of positive charges of the peptide sequence by high ionic strength and the compensation of the overall peptide charge by pH lead to accelerated kinetics of the aggregate assembly. These studies suggested that the peptide may adopt different conformations that lead to different morphologies of aggregates. Testing this hypothesis was the major goal of this paper. In this study we compare two types of aggregates formed in pH ~2 solution and at pH near the peptide's pI value (pH 5.6). These conditions produce distinct aggregation kinetics but similar intensities of ThT fluorescence at the end of the aggregation process. We also employed a combination of AFM and Tip-Enhanced Raman Scattering (TERS) to characterize these two types of aggregates.
TERS allows for structural characterization of amyloids without bulk averaging.[9] As a combination of AFM and Raman spectroscopy this method exceeds the diffraction limit and sensitivity of conventional Raman technique while preserving its ability to provide spectroscopic fingerprint. This spectroscopic signature recognizes specific Raman bands that report on the secondary structure of proteins and peptides composing the fibrils.[9, 10] Recently, TERS has been utilized to study secondary structure and amino acid composition of twisted insulin fibrils[11] as well as insulin fibril polymorphs with flat topology.[12] Furthermore, TERS detects a particular aminoacid distribution and corresponding secondary structure at any measured point on a fibrillar surface with a resolution of only a few nanometers.[10, 13] These studies demonstrated the potential of TERS to assess structural characteristics of amyloid fibril with high lateral resolution.[14]
In the present study we employed TERS method to characterize two types of fibrils formed by a short peptide CGNNQQNY from yeast prion protein Sup35. First, using AFM we found dramatic differences in fibril length and height for the fibrils formed at pH 5.6 and pH~2. pH 5.6 generally favored shorter fibrillar structures compared to pH~2 conditions. Second, TERS identified differences in the secondary structures of peptides composing morphologically different fibrils. Analysis of TERS spectra revealed that the fibrils formed under each condition have different structural properties. Fibrils formed in pH~2 solution are primarily composed of parallel β-sheet while fibrils formed at pH 5.6 are composed of a mixture of conformations with β-sheet structure arranged in antiparallel manner. We showed the possibility of in depth analysis of amide III region to delineate contributions from α-helix, β-sheet and unordered secondary structures. Also, a correlation between Raman peak intensities corresponding to α-helix, β-sheet and unordered secondary structures in amide III region and amide I region indicated that both regions provide adequate representation of structural characteristics of amyloid surfaces. Furthermore, using 2D analysis we correlated amide III intensity changes with the intensities of the bands corresponding to Cα-H in plane bending. This correlation suggested high localization of structural properties and large contribution of unordered non-helical secondary structures present in the peptides on a fibrillar surface.
2. Experimental Section
2.1 Materials
The peptide CGNNQQNY was synthesized by Peptide 2.0 (Chantilly, VA). Synthesized peptide was purified by C18 reverse-phase HPLC, and its molecular weight was confirmed by MALDI-TOF mass spectroscopy. Acetate buffer (pH 5.6) was prepared from sodium acetate/acetic acid with total ionic strength 11 mM. Mica was purchased from Asheville-Schoonmaker Mica Co., Newport News, VA.
2.2 Aggregation of Peptides
Lyophilized peptide was dissolved in either deionized water or an acetate buffer to the final concentration of 0.5 mg/mL. Dissolving peptide in water resulted in low pH value (~2) of the solution. The low pH value is due to residual presence of trifluoroacetic acid (TFA) that remained after HPLC purification of the peptide. The amount of TFA was the same for both DI water solution and acetate buffer. Peptide concentration was estimated from the absorbance at 274 nm using an extinction coefficient of 1405 M−1 cm−1. After the peptide was dissolved we have analyzed the solution using mass-spectrometry (4800 MALDI TOF analyzer from Applied Biosystems, Foster City, CA). One major peak was present at m/z ~ 940 corresponding to molecular weight of CGNNQQNY peptide (MWpep). No peaks at m/z ~ 2×MWpep were observed suggesting that no dimers due to disulfide bridge formation were present in solution. Approximately 100 μL of freshly prepared peptide solution was incubated at room temperature in 500 μL Eppendorf tubes while stirring on a magnetic stirrer (Model 190M, Hanna Instruments) for 15 min every 2 hours with micro stir bars (Fisher Scientific). The extent of peptide aggregation was followed by ThT fluorescence (Figure S1). 2 μL aliquots from the samples were withdrawn periodically and added to 118 μL of 5 μM ThT solution. Fluorescence intensity was detected on a Cary Eclipse spectrofluorimeter (Varian Inc., Palo Alto, CA) at an emission wavelength of 485 nm while exciting at wavelength 450 nm using Cary sub-micro fluorometer cell (60 μL total volume). Samples for AFM imaging analysis and TERS measurements were taken after the aggregation was complete as indicated by the maximum ThT fluorescence reaching a plateau.
2.3 Atomic Force Microscopy
Samples of aggregated peptides were filtered through Microcon centrifugal filter device (10,000 MWCO) to remove buffer components from the solution. For that 5 μL of sample was loaded onto the filter diluted with 95 μL of water and spun at 14,000 g for 10 minutes. 3 μL of the supernatant was deposited from a pipette, with the end of the tip cut off, onto mica disks for 2 min followed by a gentle rinse with deionized water and drying with argon.
Images were acquired in air using a MultiMode SPM NanoScope IV system (Bruker, Santa Barbara, CA) operating in tapping mode. Silicon-etched tapping mode probes (OTESPA) with nominal spring constants of ~42 N/m and a resonant frequency of ~330 Hz were used. Analyses of images were done using Femtoscan Online (Advanced Technologies Center, Moscow, Russia).
2.4 TERS measurements
Both types of fibrils were deposited on mica substrates as described above in Sample preparation section. The TERS setup consists of an AFM (Nanowizard, JPK, Berlin, Germany) mounted on an inverted microscope (Olympus IX70, Hamburg, Germany) equipped with a 60x (N.A. 1.45) oil immersion objective (Olympus, Hamburg, Germany). For excitation a krypton ion laser (Innova 300C Coherent, St. Clara, California, USA) at λ = 568 nm, P = 1.4 mW was used. Spectra were recorded with a confocal Raman spectrometer (LabRam HR, Horiba Jobin Yvon, Villeneuve d'Ascq, France) with a liquid nitrogen cooled CCD camera (ISA Spectrum One, Horiba Jobin Yvon, Villeneuve d'Ascq, France). The z-focus of the laser was maintained by mounting the objective on a piezo device (PIFOC, Physik Instrumente, Karlsruhe Germany). TERS tips were prepared by evaporating 20 nm silver on commercial AFM silicon tips (NSG 10, NT-MDT, Moscow, Russia). Tip-sample distance was kept constant by working in closed-loop feedback. The sample was mounted on a 100×100 μm scanning stage (P-734, Physik Instrumente, Karlsruhe, Germany). After defining a grid on a single fibril TERS spectra were recorded on 7 nm distant points with an acquisition time of 10 s. Tip contamination was excluded by a reference measurement next to the fibril after the grid measurement. The measured reference spectrum was subtracted from all the measured spectra of the grid. Figure S3 in the supplemental information shows a comparison of such a reference spectrum with a Raman spectrum taken on a fibril. Also, additional Raman spectra taken with TERS probe engaged/disengaged are shown in Figure S3. Raman spectra with subtracted reference were used for 2D analysis without any further treatment or normalization. Generalized Two-Dimensional Correlation Spectroscopy (2DCOS) was performed using 2DShige software (developed by Shigeaki Morita, Osaka Electro-Communication University, Japan).
3. RESULTS AND DISCUSSION
3.1 Aggregation pattern and morphology of aggregated CGNNQQNY peptide
CGNNQQNY peptide readily aggregates upon dissolving of lyophilized powder in aqueous solution. This octapeptide forms a number of polymorphs depending on aggregation conditions.[8] Here, we characterized two types of fibrils formed at pH~2 and pH 5.6. Distinct aggregation kinetics is observed for these two conditions, faster at pH 5.6 with lag time τlag=10.8 h and slower at pH~2 with τlag=40.8 h (Figure S1 in Supporting Information).
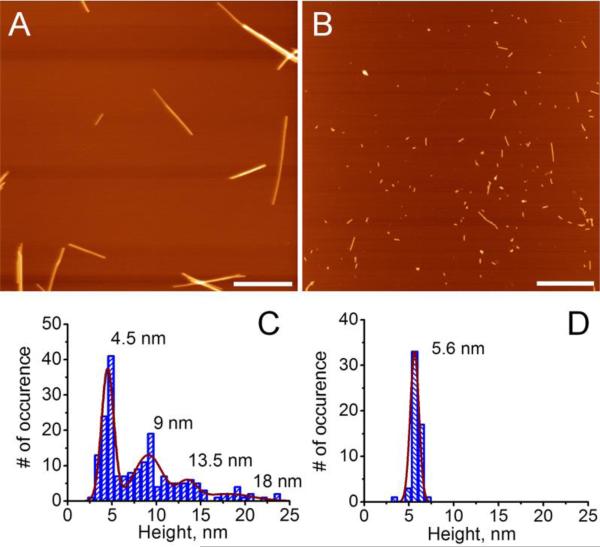
We analyzed morphology of the aggregated species for CGNNQQNY formed at these two conditions using AFM. Figure 1 illustrates that morphology of the aggregates is dominated by fibrillar structures regardless of environmental conditions. However, the two conditions, pH~2 (Figure 1, A) and pH 5.6 (Figure 1, B) produce fibrils that are morphologically distinct. Fibrils obtained at pH~2 appear longer and brighter on the image than those formed at pH 5.6.
Figure 1.
AFM images of amyloid fibrils formed by CGNNQQNY peptide in A) pH~2 and B) pH 5.6 buffer solution. Scale bar is 500 nm. C) and D) corresponding heights of the fibrils.
Detailed analysis and comparison of the heights of the fibrils formed in pH~2 and pH 5.6 buffer conditions is presented in Figure 1, C and D. At pH 5.6, the majority of all aggregates had heights around 5.6 nm (Figure 1, D). Fibrils at pH~2 had a larger range of heights from 4 nm to 25 nm (Figure 1, C). Additionally, pH~2 grown fibrils associate with one another forming bundles. The histogram for the heights shows multiples of single fibril height and this is evident from the histogram showing several peaks at 4.5, 9, 13.5 and 18 nm. On the other hand, fibrils grown in pH 5.6 buffer are homogeneous in height around 5.6 nm.
3.2 Tip-Enhanced Raman Spectroscopy (TERS) of amyloid fibrils
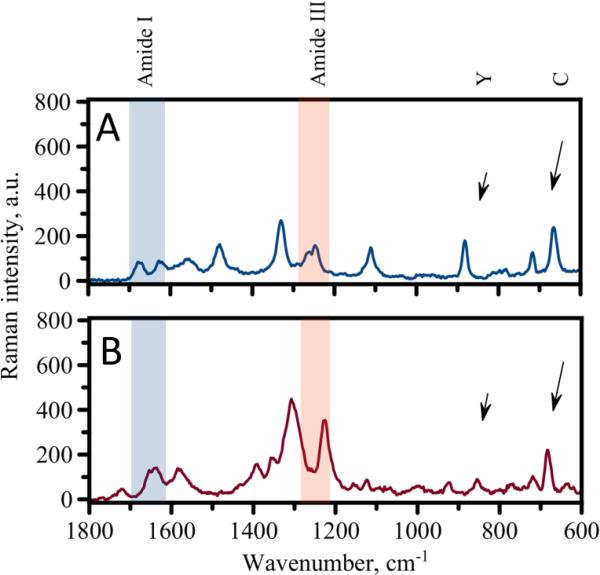
Next, we used TERS to assess conformational differences/similarities of peptides in these two forms of fibrils. Figure 2 shows TERS spectra obtained for the probing of the fibrils formed in acetate buffer - pH 5.6 (Figure 2, A) at pH ~2 and (Figure 2, B). Both spectra are similar with well pronounced peaks in several regions. Several characteristic Raman bands can be identified and assigned in the spectra. One group of bands corresponds to signals from amino acids. It was possible to assign Raman bands corresponding to cysteine (C) and tyrosine (Y) amino acids (marked in the spectra with arrows) as those amino acids have specific spectral features known from previous TERS studies.[15] Another group of strong signals was observed in the regions specific for amide bands of peptide backbone (amide I and III in Fig. 2). These amide bands are considered to be signatures which characterize peptide conformation.[16] Despite the general similarities, the spectra for the two types of fibrils exhibit some differences. The observed differences are mostly manifested in the positions of the characteristic amide bands. The positions of the amide I and amide III bands are most often used for determination of peptide backbone conformations. Variations in these regions, therefore, suggest that the secondary structure of peptides composing these fibrils is different. Depending on the nature of the amino acid residues in a peptide sequence it might be possible to use either amide I or amide III regions for secondary structure evaluation.[10, 11] Recent reports indicated that the amide I band is sensitive to the nature of amino acid residues and can be effectively suppressed in TERS and SERS, especially for bulky amino acids such as tyrosine.[17, 18]. Thus we focused first on the analysis of the amide III band region for secondary structure assessment.
Figure 2.
TERS spectra for fibrils formed in acetate buffer (A) and pH~2 solution (B). Arrows indicate positions of Raman bands assigned to tyrosine (Y) and cytosine (C).
The spectrum shown in Figure 2, B of the pH~2 fibrils features one peak at 1223 cm−1 (beta-sheet) in the amide III region. On the other hand the amide III region for the fibrils formed at pH 5.6 is more complex with several clear peaks (Figure 2, A). Namely, we were able to distinguish β-sheet (1237 cm−1), random coil (1252 cm−1) and α-helix (1273 cm−1). Spectra measured for pH 5.6 fibrils always showed significant contribution from non beta-sheet structures. Thus, we hypothesize that the surfaces of the fibrils formed at pH~2 primarily have beta-sheet structure while fibrils formed at pH 5.6 are composed of a mixture of conformations. Interestingly, our results suggest that despite uniform morphology of pH 5.6 fibrils they are composed of peptides that have adopted various secondary structures on a fibrillar surface.
3.3 Amide III band position and structure of amyloid fibril
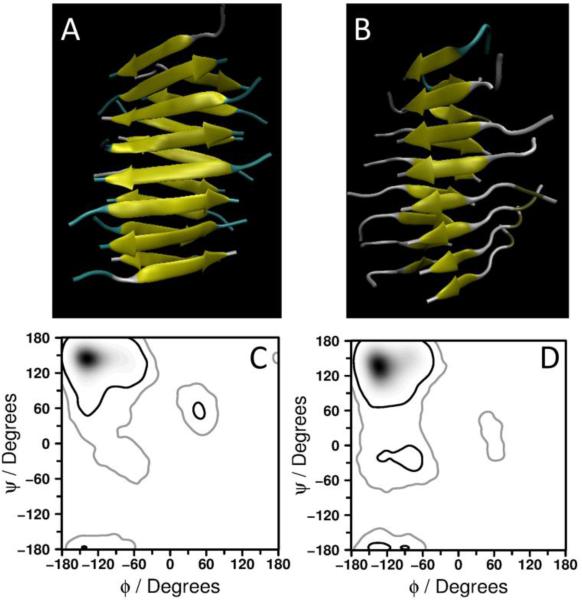
Comparison of the band position of beta-sheet structure for two fibrillar forms shows quite significant difference. Low wavenumber (1223 cm−1) is observed for pH~2 fibrils while fibrils formed in acetate buffer exhibit a higher wavenumber (1237 cm−1) in the spectra shown in Figure 2. We interpret these differences in the band positions for β-sheet of the two types of fibrils as following. Parallel β-sheets arrangement in pH~2 fibrils causes smaller Raman shift in the spectrum while antiparallel arrangement of the peptides in the fibrils formed in acetate buffer results in a larger shift. It has been recently reported that the amide III band position strongly depends on Ψ dihedral angle resulting from coupling between the peptide N-H and Cα-H in-plane bending vibrations while the dependence on Φ dihedral angle was reported to be only marginal.[19] Indeed, different arrangement of the peptides within the fibril would result in different Ψ dihedral angle as typically presented by Ramachandran plot. We used Molecular Dynamics (MD) simulations to build fibrillar structures of two sheets composed of 16 monomers with parallel and antiparallel orientation of the peptide. The models are shown in Figures 3A and B. From these simulations we have estimated the values of Ψ angle corresponding to parallel or antiparallel configurations. The Ramachandran plots generated for the structures of the two different types of aggregates show preferred angles accommodated by the peptides in these structures (Figures 3C and D). Based on the plots obtained from MD simulations most favorable Ψ angle of 140-155° can be found in an aggregate with peptides arranged in antiparallel configuration, while for parallel arrangement this angle spans values 125-140°. We computed expected values of Ψ dihedral angle for both types of fibrils using previously proposed relationship between Ψ and the amide III band frequency (Equation 1).[19] According to the structural model for the peptide GNNQQNY,[1, 13, 20] extended β-sheets are formed by individual molecules stabilized with hydrogen bonds of both backbone and side chains of the peptide. Further structural organization involves the formation of tight interface completely excluding water between β-sheets.[13] Thus, we adopted eq. (1) for anhydrous case[19] to estimate the differences in Ψ dihedral angle for beta conformation in the two types of fibrils:
| (1) |
Figure 3.
(A) and (B): structures of 16-mers of CGNNQQNY peptide arranged in antiparallel (A) and parallel (B) configuration. (C) and (D): Ramachandran plots for antiparallel (C) and parallel (D) structures of 16-mer of CGNNQQNY peptide.
Although the correlation of the band frequency and the values of the dihedral angle (Ψ) was originally proposed for the deep UV resonance Raman (DUVRR) conditions, recent studies[21, 22] suggested that data obtained under non-resonance conditions also seem to show similar correlations with available empirical relations developed in [19]. The estimate of the Ψ angle using eq. (1) suggests that the low wavenumber 1223 cm−1 observed for pH~2 corresponds to an average Ψ angle of 135°, while 1237 cm−1 observed for pH=5.6 corresponds to a Ψ angle of 155° (Table 1). Thus, we conclude that the two types of fibrils studied here have different arrangement of β-strands: pH~2 fibrils have parallel arrangement and pH 5.6 fibrils have antiparallel arrangement. The possibility of forming both parallel and antiparallel configurations for this short peptide has been recently reported.[23] Among the two possible structures, the parallel β-sheet is more stable than antiparallel configuration contributing to a larger stability of the amyloid core.[23] Theoretical and experimental observations also concluded that antiparallel arrangement is mostly stabilized by van der Waals interactions between complementary side chains, while parallel arrangement is stabilized by a dense network of backbone H-bonds.[24-26]
Table 1.
Amide III band position, computed angle and predicted beta-strand arrangement.
| Ψ | β-strand arrangement | ||
|---|---|---|---|
| pH~2 solution | 1223 cm−1 | 135° | parallel |
| Acetate buffer (pH 5.6) | 1237 cm−1 | 155° | antiparallel |
3.4 Mapping secondary structure of the fibril with TERS
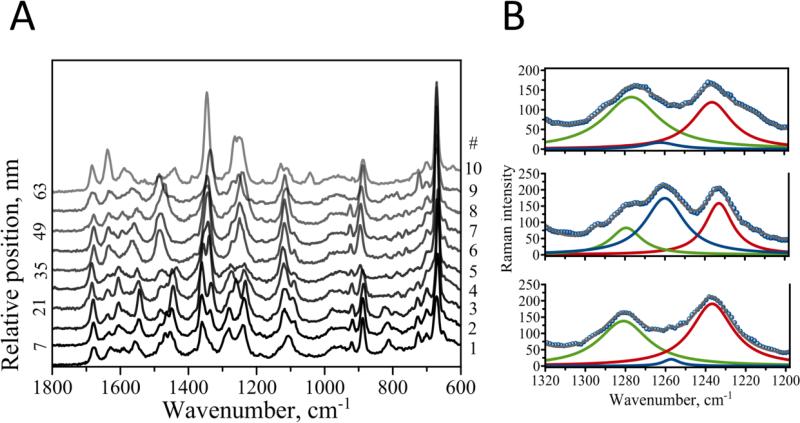
Structural mapping is one of the advantages of the TERS method. Typically, the sample topography is first acquired with the AFM module of the TERS setup. Next, the tip is positioned in the area of interest, and TERS spectra are point-wise collected along the selected area. The spatial resolution of TERS has been recently demonstrated to reach a few nanometers for fibrillar biological samples,[10, 14] subnanometer range and even single molecules under certain experimental conditions.[27, 28] This allows for highly resolved structural maps to be obtained with TERS for biological samples including amyloid fibrils.[10] For fibrils with varying secondary structures, TERS enables precise localization and assignment of different conformations, which is not amenable to other methods. We took advantage of this capability of TERS to map conformational characteristics in fibril formed in acetate buffer (pH 5.6). A map with a step size of 7 nm was measured along a fibril. Figure 4 shows 10 consecutive Raman spectra taken 7 nm apart. Although, all ten spectra exhibit a great degree of similarity, positions of the bands and their relative intensities change. Major changes occur in the amide regions of the spectra.
Figure 4.
A) Ten consecutive TERS spectra measured along the fibril formed at pH 5.6. B) Detailed analysis of amide III bands for the spectra at relative position 7 (bottom, spectrum #2), 14 (middle, spectrum #3) and 21 (top, spectrum #4) with the following color coding: α-helix (green), random coil/unordered (blue) and β-sheet (red).
Figure 4 B shows an example of such detailed analysis of the amide III performed on spectra #2, #3 and #4 (at 7, 14 and 21 nm). The amide III obviously consists of several bands with apparent Lorentzian profile. Fitting these bands resulted in 3 distinct peaks, which we assign to α-helix (green), random coil/unordered (blue) and β-sheet (red). Sections below discuss a thorough analysis of the amide III band, its variation with measured position and correlation with the bands in amide I region.
Figure 4, A indicates that a mixture of β-sheet, α-helix and random coil conformations are present at any given measuring position. Tip Enhanced Raman Scattering allowed us to characterize the secondary structure of the peptide along the fibril. For example, the band intensity of random coil/disordered structure changes from 20 to 173 (in spectrum # 2 to spectrum # 3 in Figure 4B) and then steeply declines to 14 (spectrum # 4). Similar variations were found for α-sheet (spectra # 5 through # 9 shown in Figure S2, A) and α-helical (spectra # 3 through # 5 in Figure S2, A) conformations. Such a strong variation may be due to the fact that secondary structures analyzed with TERS preferentially represent contributions from fibrillar surface rather than fibrillar core where β-sheet is expected to dominate. Although, band intensity corresponding to each particular secondary structure changes drastically (Figure S2, A), the bands’ positions stay the same with only minor variation within the following ranges for alpha-helical: 1271-1283 cm−1, random coil: 1248-1261 cm−1, and beta-sheet: 1233-1242 cm−1 (Figure S2, B).
We interpret these changes as the existence of heterogeneity of peptide secondary structure on a fibrillar surface. Note that the intensity variation in the spectra was measured with 7 nm steps, suggesting that the structural TERS analysis can be obtained with at least 7 nm lateral resolution. For example, the intensity of the marker band at 723 cm−1 varies throughout the investigated region (Figure 4, A). Spectrum #9 features this peak with relatively high intensity while spectra #8 and #10 only show residual intensity of this peak. A similar intensity change can be found for the band at 1040 cm−1. The signal in this case appears only in the spectrum #10. This supports our conclusion that the lateral resolution of TERS measurements is high enough to obtain local structural characteristics of amyloid fibrils. This is also in a good agreement with recently reported TERS study on insulin fibrils indicating that the resolution can be only a few nanometers.[10, 11]
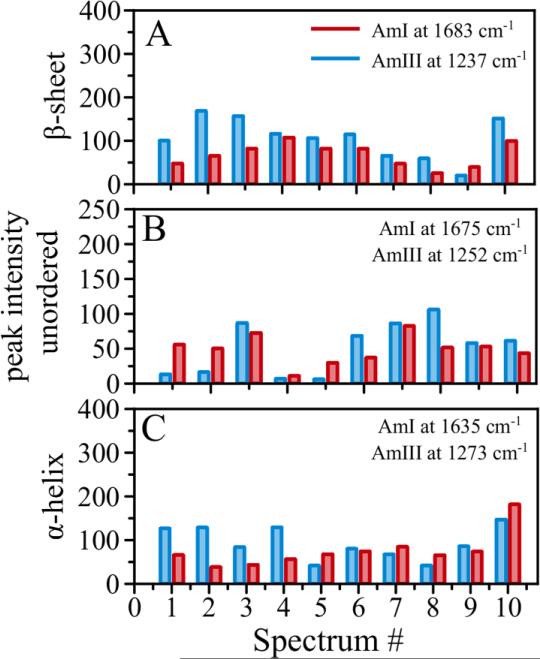
From our spectral analysis it seems that amide III bands are very sensitive to peptide conformational changes. Another region commonly used to evaluate peptide secondary structure is amide I. Intensity of amide I bands (beta, alpha, unordered structures) was compared with the intensities of corresponding bands in amide III region for the 10 spectra from Figure 4. The analysis is shown in Figure 5. Amide I peaks at 1683 cm−1 was correlated with amide III at 1237 cm−1 (β-sheet, Figure 5, A); AmI@1675 cm−1 with AmIII@1252 cm−1 (unordered, Figure 5, B); and AmI@1635 cm−1 was correlated with AmIII@1273 cm−1 peak (α-helix, Figure 5, C). A nice correlation is observed between the amide III bands and corresponding bands in amide I region as indicated by the overall trend in intensity changes. Despite very nice correlation for this set of spectra we found it more challenging to differentiate β-sheet from unordered structure in the amide I region compare to amide III due to the fact that the bands of these structures in the amide I region highly overlap and their maxima are very close to each other (1683 cm−1/ 1675 cm−1). This suggests that the Raman bands in amide III region do provide adequate representation of secondary structure analysis in TERS spectra and it might be more practical to use amide III bands for structural characterization in amyloid fibrils.
Figure 5.
Comparative analysis of band intensities for A) beta-sheet, B) unordered, and C) alpha-helical structure. Blue bars represent intensities in amide III region, red bars represent intensities in amide I region.
3.5 Amide III/Cα-H (bending) Correlation
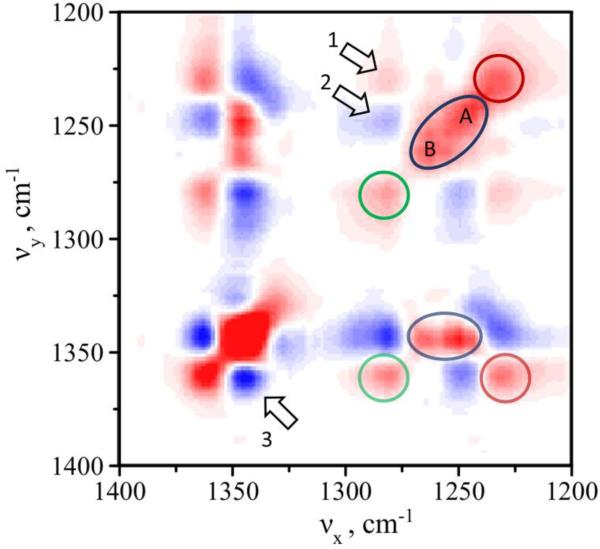
Next we correlated changes in secondary structure observed in amide III region with the intensity of the bands other than amide peaks throughout the spectral window by calculating the 2D synchronous spectrum. Particularly, we were interested in correlating changes in amide III with other intense peaks such as Cα–H (bending) Raman bands. The correlated 2D spectrum was constructed using the data set shown in Figure 4A. The results of spectral intensity correlation of amide III and Cα–H bending peaks are presented in Figure 6. The 2D synchronous map correlates changes in the spectra showing how intensity change at a given frequency is coordinated with the intensity changes at other frequencies. There are two types of peaks in the 2 D maps: 1) diagonal autopeaks due to intensity changes at the same frequency and 2) off-diagonal cross-peaks due to coincidental changes of spectral intensities at two different Raman shifts. Figure 6 shows a range from 1400 to 1200 cm−1 covering both amide III and Cα–H bending regions. The amide III region spans between 1220 and 1300 with 1235 cm−1 (red circle) normally assigned to β-sheet secondary structure, 1280 cm−1 (green circle) is α-helix and ~1248-1265 cm−1 (blue oval) was assigned to unordered/random coil conformation.
Figure 6.
2D synchronous map for amide III and Cα-H regions. Positive and negative regions are shown in red and blue, respectively. Diagonal autopeaks in the amide III region are indicated with the red circle for β-sheet secondary structure, green circle for α-helix and blue oval for unordered/random coil conformation. Similar representation was used for off-diagonal cross-peaks showing correlation between amide III and Cα-H. Arrows indicate correlation observed between peaks within the same region: 1) 1280/1235 cm−1, 2) 1280/1249 cm−1, and 3) 1360/1340 cm−1.
Two noticeable observations can be made from the 2D map of amide III. 1) β-sheet (red) and α-helix (green) are localized, while peak corresponding to unordered/random coil conformation seems to have a complex structure with two apparent maxima at 1249 cm−1 - (A) and 1262 cm−1 - (B). 2) α-helix and β-sheet are positively correlated as indicated by cross peak 1. There is no obvious correlation between unordered structure and β-sheet as no cross peaks are present for these two bands. Cross peak 2 indicates negative correlation between α-helical and unordered structure A. At the same time, no correlation (no cross peaks) between α-helix and unordered structure B is observed.
The 2D map shows that large changes in intensity are also observed in the Cα-H region, which is dominated by two major peaks: 1360 cm−1 and 1340 cm−1. The presence of negative cross peak 3 (1360/1340 cm−1) indicates that these two peaks substitute each other when TERS tip moves along the fibril. The correlation analysis between amide III and Cα-H (bending) suggests that the peak 1340 cm−1 is associated with unordered structure (positive 1340/1249 and positive 1340/1262). The peak at 1360 cm−1 is associated with ordered structures (positive 1360/1280 and positive 1360/1235). Cα-H (bending) peaks are expected to be largely affected by the secondary structure (primarily non-helical conformations) due to strong coupling of Cα-H bending motion with N-H bending and C-N stretching.[29] It is enhanced by coupling to the amide III mode via C-H bending and C-C stretching.[30] Our data suggest a strong and direct correlation of the intensity of these peaks with changes in amide III region in the TERS spectra. Such correlation further corroborates our interpretation of structural heterogeneity of the amyloid fibril. Additionally, large intensity of the peak at 1340 cm−1 also suggests high content of unordered structure. Cross peaks of Cα-H (bending) with amide III bands indicate coincidental changes in intensity for unordered/ordered structures in opposite directions. The plot also suggests that there are two types of unordered structure A and B and only A directly correlates with intensity gain of alpha-helical structure but does not have any correlation with beta-sheet. Thus, only type A unordered structure seems to give rise to ordered alpha-helical, while beta-sheet structure develops from ordered alpha-helical and not from unordered structure as suggested by positive cross peak 1 (Figure 6, arrow 1).
4. CONCLUSIONS
We have found dramatic differences in morphology (length and height) for the fibrils formed at two distinct conditions – pH 5.6 and pH~2. Tip-enhanced Raman Scattering identified differences in secondary structures of peptides composing these morphologically different fibrils. A mixture of peptide secondary structures for fibrils assembled at pH 5.6 was observed in contrast to primarily β-sheet structure for the fibrils assembled at pH~2. By correlating the Raman peak positions corresponding to β-sheet structure in the amide III region with Ψ dihedral angle obtained from Molecular Dynamics simulations of 16-mer fibrils we hypothesized that TERS can provide information about packing arrangement of peptides within the fibrillar structures. Our data suggested an antiparallel arrangement for pH 5.6 fibrils and parallel arrangement for the fibrils formed at pH~2. High spatial resolution of TERS allowed us to laterally resolve conformational features of peptides composing amyloid fibrils within only a few nanometers. New ways to perform detailed analysis of secondary structure of peptides with the use of 2D correlation spectra and construct conformational maps were proposed. Overall, we demonstrated the potential of TERS as a sensitive tool to correlate structural changes in single protein aggregate which can be done with high spatial resolution.
Supplementary Material
Paper Highlights.
This paper reports on the study of morphological and structural heterogeneity of amyloid aggregates. Distinct amyloid polymorphs with dramatic differences in morphology (length and height) formed at two conditions – pH 5.6 and pH ~2 were characterized using AFM (Atomic Force Microscopy) and TERS (Tip-Enhanced Raman Scattering). Tip-enhanced Raman Scattering identified a mixture of peptide secondary structures for fibrils assembled at pH 5.6 in contrast to primarily beta-sheet structure for the fibrils assembled at pH 2. We also established that TERS can provide information about packing arrangement (parallel versus antiparallel) of peptides within the fibrillar structures. This was done by correlating the Raman peak positions corresponding to β-sheet structure in the amide III region with Ψ dihedral angle obtained from Molecular Dynamics simulations. Furthermore, we developed a methodology for detailed analysis of the peptide secondary structure by correlating intensity changes of amide III peaks with other regions in TERS spectra. Such correlation established that structural composition is highly localized with large contribution of unordered secondary structures. Overall, this study demonstrates the potential of TERS as a sensitive tool to analyze structural changes within single protein aggregate.
ACKNOWLEDGEMENTS
The work was supported by grants 5R01 GM096039-04 (NIH) and EPS – 1004094 (NSF) to YLL.
SUPPORTING INFORMATION. SI describes aggregation pattern of CGNNQQNY peptide, provides details of Molecular Dynamics (MD) simulations, shows bands’ intensity and position changes in the amide III region, and compares Raman spectra measured with TERS probe engaged/disengaged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Budich C, Neugebauer U, Popp J, Deckert V. Cell wall investigations utilizing tip-enhanced Raman scattering. J Microsc. 2008;229:533–539. doi: 10.1111/j.1365-2818.2008.01939.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang P, Zhang W, Liang O, Pantoja M, Katzer J, Schroeder T, Xie Y-H. Giant Optical Response from Graphene–Plasmonic System. ACS Nano. 2012;6:6244–6249. doi: 10.1021/nn301694m. [DOI] [PubMed] [Google Scholar]
- 3.Stadler J, Schmid T, Zenobi R. Nanoscale chemical imaging using top-illumination tip-enhanced Raman spectroscopy. Nano Lett. 2010;10:4514–4520. doi: 10.1021/nl102423m. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner SB. Cell biology. A unifying role for prions in neurodegenerative diseases. Science. 2012;336:1511–1513. doi: 10.1126/science.1222951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frederick KK, Debelouchina GT, Kayatekin C, Dorminy T, Jacavone AC, Griffin RG, Lindquist S. Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem Biol. 2014;21:295–305. doi: 10.1016/j.chembiol.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tycko R. Amyloid Polymorphism: Structural Basis and Neurobiological Relevance. Neuron. 2015;86:632–645. doi: 10.1016/j.neuron.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 8.Portillo AM, Krasnoslobodtsev AV, Lyubchenko YL. Effect of electrostatics on aggregation of prion protein Sup35 peptide. Journal of physics. Condensed matter : an Institute of Physics journal. 2012;24:164205. doi: 10.1088/0953-8984/24/16/164205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krasnoslobodtsev AV, Portillo AM, Deckert-Gaudig T, Deckert V, Lyubchenko YL. Nanoimaging for prion related diseases. Prion. 2010;4:265–274. doi: 10.4161/pri.4.4.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deckert-Gaudig T, Kammer E, Deckert V. Tracking of nanoscale structural variations on a single amyloid fibril with tip-enhanced Raman scattering. J Biophotonics. 2012;5:215–219. doi: 10.1002/jbio.201100142. [DOI] [PubMed] [Google Scholar]
- 11.Kurouski D, Deckert-Gaudig T, Deckert V, Lednev IK. Structure and Composition of Insulin Fibril Surfaces Probed by TERS. Journal of the American Chemical Society. 2012;134:13323–13329. doi: 10.1021/ja303263y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurouski D, Deckert-Gaudig T, Deckert V, Lednev Igor K. Surface Characterization of Insulin Protofilaments and Fibril Polymorphs Using Tip-Enhanced Raman Spectroscopy (TERS) Biophys J. 2014;106:263–271. doi: 10.1016/j.bpj.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Treffer R, Bohme R, Deckert-Gaudig T, Lau K, Tiede S, Lin X, Deckert V. Advances in TERS (tip-enhanced Raman scattering) for biochemical applications. Biochem Soc Trans. 2012;40:609–614. doi: 10.1042/BST20120033. [DOI] [PubMed] [Google Scholar]
- 14.Deckert V, Deckert-Gaudig T, Diegel M, Gotz I, Langeluddecke L, Schneidewind H, Sharma G, Singh P, Trautmann S, Zeisberger M, Zhang Z. Spatial resolution in Raman spectroscopy. Faraday Discuss. 2015;177:9–20. doi: 10.1039/c5fd90014j. [DOI] [PubMed] [Google Scholar]
- 15.Deckert-Gaudig T, Bailo E, Deckert V. Tip-enhanced Raman scattering (TERS) of oxidised glutathione on an ultraflat gold nanoplate. Physical chemistry chemical physics : PCCP. 2009;11:7360–7362. doi: 10.1039/b904735b. [DOI] [PubMed] [Google Scholar]
- 16.Jakobsen RJ, Brown LL, Hutson TB, Fink DJ, Veis A. Intermolecular interactions in collagen self-assembly as revealed by Fourier transform infrared spectroscopy. Science. 1983;220:1288–1290. doi: 10.1126/science.6857249. [DOI] [PubMed] [Google Scholar]
- 17.Kurouski D, Postiglione T, Deckert-Gaudig T, Deckert V, Lednev IK. Amide I vibrational mode suppression in surface (SERS) and tip (TERS) enhanced Raman spectra of protein specimens. Analyst. 2013;138:1665–1673. doi: 10.1039/c2an36478f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blum C, Schmid T, Opilik L, Weidmann S, Fagerer SR, Zenobi R. Understanding tip-enhanced Raman spectra of biological molecules: a combined Raman, SERS and TERS study. Journal of Raman Spectroscopy. 2012;43:1895–1904. [Google Scholar]
- 19.Mikhonin AV, Bykov SV, Myshakina NS, Asher SA. Peptide secondary structure folding reaction coordinate: correlation between uv raman amide III frequency, Psi Ramachandran angle, and hydrogen bonding. J Phys Chem B. 2006;110:1928–1943. doi: 10.1021/jp054593h. [DOI] [PubMed] [Google Scholar]
- 20.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharya S, Ghosh S, Pandey NK, Chaudhury S, Dasgupta S, Roy A. Distribution of Protein Ramachandran Psi (ψ) Angle Using Non-Resonance Visible Raman Scattering Measurements. The Journal of Physical Chemistry B. 2013;117:13993–14000. doi: 10.1021/jp408009y. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Wang Y, Lu HP. Revealing the secondary structural changes of amyloid β peptide by probing the spectral fingerprint characters. Journal of Raman Spectroscopy. 2013;44:670–674. [Google Scholar]
- 23.Qi X, Hong L, Zhang Y. A variational model for oligomer-formation process of GNNQQNY peptide from yeast prion protein Sup35. Biophys J. 2012;102:597–605. doi: 10.1016/j.bpj.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng J, Ma B, Tsai CJ, Nussinov R. Structural stability and dynamics of an amyloid-forming peptide GNNQQNY from the yeast prion sup-35. Biophys J. 2006;91:824–833. doi: 10.1529/biophysj.106.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitagliano L, Esposito L, Pedone C, De Simone A. Stability of single sheet GNNQQNY aggregates analyzed by replica exchange molecular dynamics: antiparallel versus parallel association. Biochem Biophys Res Commun. 2008;377:1036–1041. doi: 10.1016/j.bbrc.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 26.Nelson R, Eisenberg D. Recent atomic models of amyloid fibril structure. Curr Opin Struct Biol. 2006;16:260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Zhang R, Zhang Y, Dong ZC, Jiang S, Zhang C, Chen LG, Zhang L, Liao Y, Aizpurua J, Luo Y, Yang JL, Hou JG. Chemical mapping of a single molecule by plasmon-enhanced Raman scattering. Nature. 2013;498:82–86. doi: 10.1038/nature12151. [DOI] [PubMed] [Google Scholar]
- 28.Jiang S, Zhang Y, Zhang R, Hu C, Liao M, Luo Y, Yang J, Dong Z, Hou JG. Distinguishing adjacent molecules on a surface using plasmon-enhanced Raman scattering. Nat Nano. 2015;10:865–869. doi: 10.1038/nnano.2015.170. [DOI] [PubMed] [Google Scholar]
- 29.Ianoul A, Boyden MN, Asher SA. Dependence of the Peptide Amide III Vibration on the Φ Dihedral Angle. Journal of the American Chemical Society. 2001;123:7433–7434. doi: 10.1021/ja0023128. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Purrello R, Jordan T, Spiro TG. UVRR spectroscopy of the peptide bond. 1. Amide S, a nonhelical structure marker, is a C.alpha.H bending mode. Journal of the American Chemical Society. 1991;113:6359–6368. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.