Abstract
Murine sex chromosome aneuploidies (SCAs) provide powerful models for charting sex chromosome influences on mammalian brain development. Here, building on prior work in X-monosomic (XO) mice, we use spatially non-biased high-resolution imaging to compare and contrast neuroanatomical alterations in XXY and XO mice relative to their wild-type XX and XY littermates. First, we show that carriage of a supernumerary X chromosome in XXY males (1) does not prevent normative volumetric masculinization of the bed nucleus of the stria terminalis (BNST) and medial amygdala, but (2) causes distributed anatomical alterations relative to XY males, which show a statistically unexpected tendency to be colocalized with and reciprocal to XO-XX differences in anatomy. These overlaps identify the lateral septum, BNST, ventral group thalamic nuclei and periaqueductal gray matter as regions with replicable sensitivity to X chromosome dose across two SCAs. We then harness anatomical variation across all four karyotype groups in our study—XO, XX, XY and XXY—to create an agnostic data-driven segmentation of the mouse brain into five distributed clusters which (1) recover fundamental properties of brain organization with high spatial precision, (2) define two previously uncharacterized systems of relative volume excess in females vs. males (“forebrain cholinergic” and “cerebelo-pontine-thalamo-cortical”), and (3) adopt stereotyped spatial motifs which delineate ordered gradients of sex chromosome and gonadal influences on volumetric brain development. Taken together, these data provide a new framework for the study of sexually dimorphic influences on brain development in health and disrupted brain development in SCA.
Keywords: rain anatomy, Sexual dimorphism, XO, XXY
Introduction
Sex chromosome aneuploidies (SCAs) provide powerful models to study sex chromosome influences on brain development (Lenroot et al. 2009). Better understanding these influences is important for (1) specifying biological contributors to sex differences in brain structure and function (Rutter et al. 2003; Giedd et al. 2012), and (2) clarifying brain mechanisms for the elevated rates of neurodevelopmental disorder that is seen in human SCA syndromes (Lee et al. 2012; Visootsak et al. 2007; Simpson et al. 2003).
Neuroimaging studies have identified widespread structural and functional brain alterations in humans SCAs suggesting that variations in sex chromosome gene dosage may have direct effects on mammalian brain development (Lenroot et al. 2009; Raznahan et al. 2010; Lepage et al. 2012; Bryant et al. 2011; Skakkebaek et al. 2013; Itti et al. 2006). However, a number of factors complicate the use of human SCA syndromes as a model for examination of sex chromosome gene-dosage effects, including background genetic and environmental diversity, karyotypic mosaicism (Wolff et al. 2010; Garcia-Quevedo et al. 2011), and X chromosome parent of origin (Lepage et al. 2013). These considerations make murine SCA models an attractive complement to the study of SCA in humans. Murine SCAs not only provide greater control over genetic background and environmental variation than is achievable in human populations, but they are also free of karyotypic mosaicism and allow X chromosome parent of origin to be predetermined (Davies et al. 2005; Swerdloff et al. 2011). The best-characterized murine SCAs are female X-monosomy (Probst et al. 2008) (XO), and presence of a supernumerary X chromosome in males (Swerdloff et al. 2011) (XXY). XO mice have been studied as a potential model for Turner syndrome (TS, 45, X karyotype)—with an emphasis on X haploinsufficiency effects on gene expression (Lopes et al. 2010), somatic growth (Burgoyne et al. 2002), fertility (Burgoyne and Baker 1985) and behavior (Lynn and Davies 2007). XXY mice have primarily been studied as a model for the cognitive-behavioral alterations and evolving postnatal gonadal dysfunction seen in Klinefelter syndrome (KS, 47, XXY karyotype) (Liu et al. 2010; Lue et al. 2005).
To date, murine SCAs have remained largely unexploited as a means for studying sex chromosome influences on brain development, primarily due to the labor-intensive nature of classical histochemical methods for the study of murine anatomy. However, advances in structural magnetic resonance image (sMRI) acquisition and processing have now made it possible to conduct a highly efficient and technically homogenous survey of anatomy across the entire mouse brain in a spatially unbiased manner (Lerch et al. 2011). We recently applied these methods in XO mice, and found evidence for X chromosome gene-dosage effects at classical sites of androgen-dependent sexual dimorphism within the brain (Raznahan et al. 2013). Here, we extend this earlier work in three new directions. First, we chart neuroanatomical alterations in the XXY SCA mouse model for the first time. We were specifically interested in determining (1) if foci of sexually dimorphic murine brain volume that are detectible by sMRI, such as relative male volume excess in BNST and amygdala relative to females (Raznahan et al. 2013), are preserved in XXY males, and (2) if XXY mice recapitulate any of the better-replicated anatomical alterations in studies of XXY humans [e.g., amygdala and hippocampal volume reductions relative to typically developing males (Bryant et al. 2011; Steinman et al. 2009)]. Second, we use reciprocal anatomical abnormalities in XXY and XO mice to detect replicable X-dosage effects on the brain that are independent of gonadal or Y chromosome status. Our prior work in XO mice identified multiple regions where murine brain volume may be influenced by X gene dosage, including the olfactory bulb, BNST, amygdala, nucleus accumbens, thalamus, hypothalamus, periaqueductal gray matter and parieto-temporal cortex (Raznahan et al. 2013). Finally, we apply data triangulation methods across XO, XX, XY and XXY mice to fractionate the brain based on its sensitivity to differences in gonad and sex chromosome complement. These data-mining analyses are wholly agnostic and yield empirically determined clusters of brain regions defined by distinct patterns of anatomical variation across the four karyotype groups represented in our study.
Methods
Mice
We studied 88 mice from four karyotype groups (10 XO, 26 XX, 30 XY and 22 XXY), which came from one of two cohorts. XXY cohort. This cohort was generated using a four-generation breeding scheme involving mice with a structurally rearranged Y chromosome (XY*). In the first generation, XY* mice (C57BL/6JEi-Chr YA/HeJ/EiJ, JAX, Bar Harbour, ME: JAX stock number 002602) were crossed with purebred C57BL/6J (B6) females (JAX stock number 000664) to derive XY*X females. These mice were then crossed with wild-type B6 males, and errors in female meiosis arising from the structurally abnormal X chromosome led to multiple different sex chromosome aneuploidies in offspring. Chromosomal analyses in this third generation identified XYY*X males, who were bred with wild-type B6 females, generating XXY karyotype in 50 % of male offspring. XX (n = 14), XY (n = 19) and XXY (n = 22) mice from this fourth generation constituted the XXY cohort. All XXY mice in the study bore a supernumerary X chromosome of paternal origin. Karyotype was assessed by counting-stained metaphase chromosomes in cultured fibroblasts obtained from ear clips in adult mice. XO cohort. These data were previously described in Raznahan et al. (2013); briefly, in the grandparental generation of this cohort, female XO mice carrying the tabby (EdaTa) coat-color marker on an agouti background (JAX, Bar Harbor, ME: JAX stock number 000314) (Probst et al. 2008) was crossed with B6CBACaF1/J-Aw-J/A males (JAX stock number 001201) to derive XO mice without the tabby coat-color marker. The XO females arising from this cross (the parental generation) were then bred with purebred C57BL/6J males (JAX stock number 000664) to generate XO (n = 10), XX (n = 12), and XY (n = 11) mice in this cohort. All XO mice in this study bore an X chromosome of paternal origin. X chromosome copy number was determined by quantitative PCR (qPCR) (Raznahan et al. 2013).
All mice were housed in a specific pathogen free (SPF), Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC)-accredited animal facilities at either LA Biomedical Research Institute at Harbor-UCLA (XXY cohort) or Baylor College of Medicine (XO cohort). Food and water were provided ad libitum. This work was approved by the Institutional Animal Care and Use Committees (IACUC) of both institutions.
Brain sample preparation
Mice were killed in adulthood (Mean age in postnatal days: XO cohort—80, XXY cohort—548) following deep anesthesia by intraperitoneal injection with a combination of ketamine (Fort Dodge Animal Health, Fort Dodge, IA, 150 mg/kg) and xylazine (MP Biomedicals, LLC, Solon, OH, 10 mg/kg) (see (Raznahan et al. 2013) for manufacturers for the XO cohort. Thoracic cavities were opened, and animals were perfused through the left ventricle with 30 mL of phosphate-buffered saline (PBS) (pH 7.4) plus heparin with 2 mM ProHance® (gadoteridol, Bracco Diagnostics Inc., Princeton, NJ) at room temperature (25 °C) at a rate of approximately 100 mL/h. This was followed by infusion with 30 mL of 4 % paraformaldehyde (PFA) in PBS with 2 mM ProHance® at the same rate. Following perfusion, the heads were removed along with the skin, lower jaw, ears and the cartilaginous nose tip. The remaining skull structures were allowed to postfix in 4 % PFA at 4 °C with 2 mM ProHance® for 12 h. Following an incubation period of 5 days in PBS plus 0.02 % sodium azide with 2 mM ProHance® at 4 °C, the skulls were transferred to a PBS and 2 mM ProHance® solution for at least 7 days at 4 °C.
Image acquisition and preprocessing
A multi-channel 7.0 Tesla MRI scanner (Varian Inc., Palo Alto, CA) with a 6-cm inner bore diameter insert gradient was used to acquire anatomical images of brains within skulls. Prior to imaging, the samples were removed from the contrast agent solution, blotted and placed into plastic tubes (13-mm diam) filled with a proton-free susceptibility-matching fluid (Fluorinert FC-77, 3M Corp., St. Paul, MN). Three custom-built, solenoid coils (14 mm diam, 18.3 cm in length) with over wound ends were used to image three brains in parallel. Parameters used in the scans were optimized for gray/white matter contrast: a T2-weighted, 3D fast spin-echo sequence with six echoes, with TR/TE = 325/32 ms, four averages, field-of-view 14 × 14 × 25 mm3 and matrix size = 432 × 432 × 780 giving an image with 32-µm isotropic voxels. Geometric distortion due to the position of the three coils inside the magnet was calibrated using a precision-machined MR phantom. Total imaging time was 11.3 h (Cahill et al. 2012). All 88 acquired anatomical MRIs in the XXY cohort were analyzed using an image registration pipeline described previously (Lerch et al. 2011). Briefly, after an initial affine registration to correct for postural differences, pairwise 12-parameter registrations were computed to define a consensus space for all scans. Three generations of non-linear registrations using a diffeomorphic registration algorithm (Avants et al. 2008) then brought all scans into precise alignment. The Jacobian determinant of the deformation fields was used as the metric of local volume change.
Statistical analysis
Anatomical variation across XX, XY and XXY mice
Significant anatomical differences across XXY mice and their XX and XY littermates were defined using an F test for the omnibus effect of group at each voxel within the XXY cohort, and thresholding the resulting statistical map using False Discovery Rate (FDR) correction with q (the expected proportion of falsely rejected null hypotheses) set at 0.05 (Genovese et al. 2002).
Mapping anatomical differences between XXY and XY mice, and their conjunction with XO-XX differences
Anatomical differences between XXY and XY littermates did not survive FDR correction for multiple comparisons. Given this, and the need to calculate conjunctions between XXY-XY and XO-XX contrasts with unequal power to detect statistically significant group differences, we present XXY-XY findings, and calculate XXY-XY and XO-XX contrast conjunctions using a nominal p < 0.05 threshold. Thus, the conjunction between XXY-XY and XO-XX maps identified regions where XXY and XO karyotypes both conferred nominally significant anatomical differences from their respective gonadal control littermates—either in opposing (“reciprocal X dose effects”) or similar directions. Because a relaxed statistical threshold in each contrast increases the risk of spurious conjunctions between XXY-XY and XO-XX differences, we created analogous conjunction maps for 1,000 permutations of group membership within each contrast, and compared the observed proportion of “reciprocal X-effect” voxels to this “null” distribution. None of the aforementioned contrasts combined mice from different cohorts.
Clustering the brain based on anatomical variability across XO, XX, XY and XXY karyotypes
To integrate data across all four karyotype groups, we first removed aneuploidy-independent cohort differences from our anatomical data. To do this, we first calculated the deformation fields required to bring brains of non-aneuploidic mice from the XXY cohort into alignment their counterparts in the XO cohort. This step provided voxel-specific estimates of any cohort effects on brain anatomy, which were then subtracted from the deformation maps bringing all XXY cohort brains into consensus space. As a result, we could assess karyotype effects on brain anatomy in the absence of any potentially confounding cohort effects on brain anatomy (e.g., secondary to genetic, environmental or developmental differences between XXY and XO cohorts). Next, an F-test for the omnibus effect of group at each voxel was thresholded using FDR correction for multiple comparisons (with q < 0.05) to define regions of the brain that differed significantly across XO, XX, XY and XXY groups. By averaging deformation maps within each karyotype group, we derived estimates of mean volume for each karyotype group at each of the ~200,000 voxels within the F test mask. This step resulted in a 200,000 by 4 matrix with rows representing voxels and columns karyotype groups. We used the “kmeans” function in the R language and environment for statistical computing (with iter.max and nstart set at 100) to cluster voxels based on karyotype group mean volumes. Visual inspection of the resulting “scree-plot” showing within cluster residuals vs. cluster number for 2 through 15 clusters indicated an optimal 5-cluster solution. Voxel-wise cluster designations were then visualized within the F test mask using color codes. Mean voxel volume for each cluster was calculated in each brain to examine karyotype group differences in cluster volume.
Anatomical localization
Regional findings were attributed to underlying structures by first referring to anatomical labels provided by a fully-automated multi-atlas segmentation algorithm as previously described and validated (Mallar Chakravarty et al. 2012). However, as our statistical maps frequently displayed highly organized spatial structure within single atlas labels, finer-scale anatomical attributions were made through visual comparison with Allen Atlas coronal and sagittal reference postnatal day 56 mouse brain atlases (http://mouse.brain-map.org/static/atlas). To facilitate reader assessment of attributions so made, we provide corresponding Allen Atlas coronal (C#) and sagittal (S#) position numbers for selected statistical maps in our study.
Results
Anatomical variation in XX, XY and XXY littermates
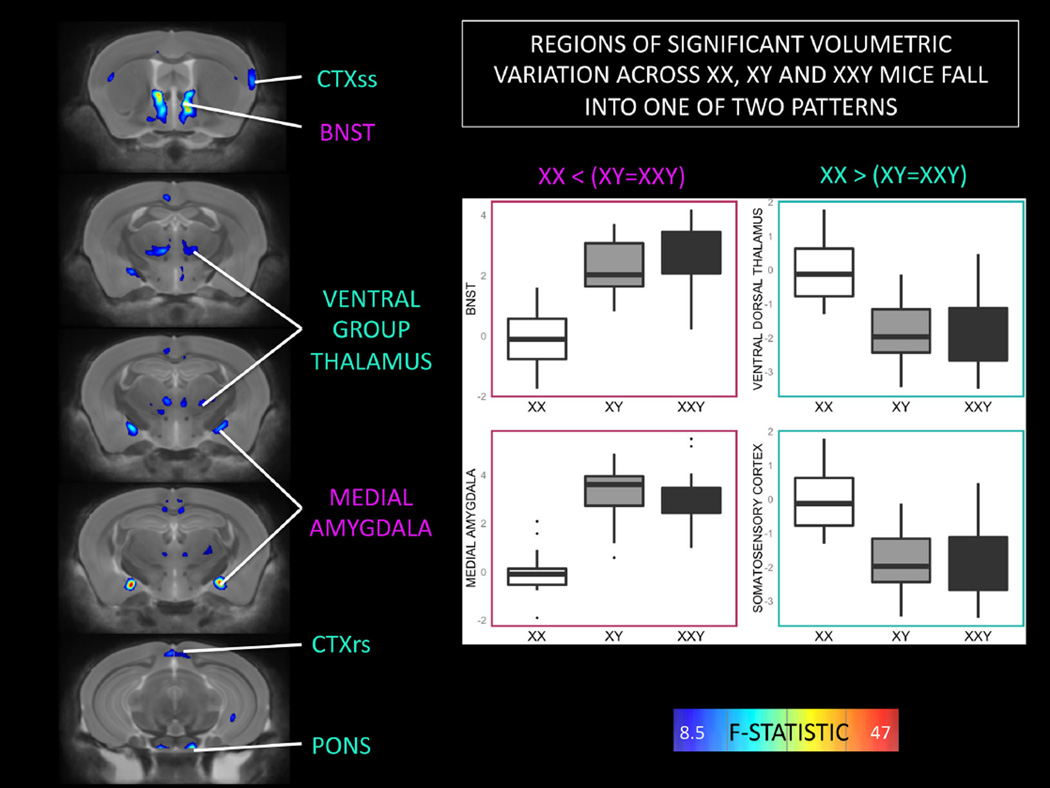
An omnibus F test across XX, XY and XXY mice identified multiple bilaterally symmetric foci of significant anatomical variation (surviving FDR correction) within the amygdala, bed nucleus of the stria terminalis (BNST), thalamus, somatosensory cortex and pons (Fig. 1). Posthoc group comparisons established that brain volume within these regions followed one of two patterns based on the direction of volume difference between XX and XY mice (with XXY mice resembling their XY littermates) (Fig. 1 inset boxplots).
Fig. 1.
Coronal views of the brain showing regions of statistically significant anatomical variation across XX, XY and XXY groups. Brain images show a thresholded F-statistic map after (after False Discovery Rate correction for multiple comparisons) for the omnibus effect of “Group” on brain volume (color scale as per F-statistic bar). Two patterns of significant group effect are detected. Both XY and XXY mice show volume increases relative to XX females within the bed nucleus of the stria terminalis (BNST) and medial amygdala. Conversely, XY and XXY mice both show volume decreases relative to XX females within the somatosensory and retrosplenial cortices (CTXss and CTXrs respectively), and the ventral group nuclei of the dorsal thalamus
Reciprocal anatomical alterations in XXY and XO mice
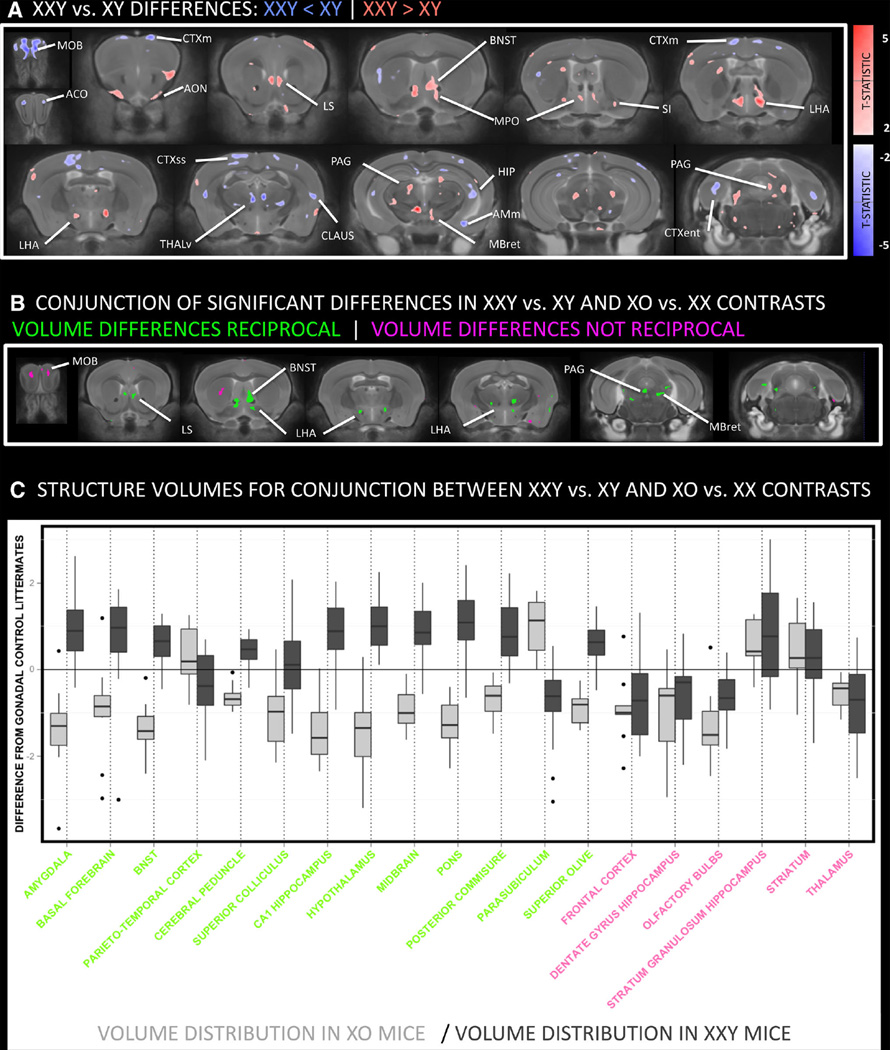
Brain regions where volume differences between XXY mice and their XY littermates survived a relaxed threshold for statistical significance (p < 0.05) were largely bilaterally symmetric. Relative volume reduction was seen in XXY mice within the main olfactory bulb (MOB), olfactory limb of the anterior commissure (ACO), thalamus (THAL), hippocampus (HIP), amygdala (AMY), sensorimotor cortex (CTXsm), entorhinal cortex (CTXent) and claustrum (CLAUS)]. Conversely, XXY mice showed a relative volume increase within anterior olfactory nucleus (AON), lateral septal region (LS), bed nucleus of the stria terminalis (BNST), medial preoptic hypothalamus (MPO), and lateral hypothalamic area (LHA), substantia innominata (SI), periaqueductal gray matter (PAG) and midbrain reticular nucleus (MBret) (Fig. 2a). By deriving an analogous contrast map for the comparison between XO mice and their XX littermates, we established that 70 % of all voxels with overlapping XO-XX and XXY-XY differences showed reciprocal X-dose effects (i.e., XXY > XY where XO < XX or vice versa) (Fig. 2b). The observed proportion of voxels showing such effects exceeded that seen in 81 % of 1,000 overlap maps generated after permutation of genotype group membership amongst XXY and XY mice (Fig. 2c).
Fig. 2.
Coronal views of the brain showing anatomical differences between XXY and XY mice and the conjunction of this contrast with XO-XX differences. a Thresholded T-statistic map (at nominal p < 0.05) for volume increases (red) and decreases (blue) in XXY mice relative to their XY littermates. XXY mice show greater volume than XY mice within the anterior olfactory nucleus (AON), lateral septal region (LS), bed nucleus of the stria terminalis (BNST), hypothalamus, substantia innominata (SI), periaqueductal gray matter (PAG) and midbrain reticular nucleus (MBret). Relative volume decreases in XXY mice are seen within the main olfactory bulb (MOB), olfactory limb of the anterior commissure (AC), thalamus, hippocampus (HIP), motor (CTXm), entorhinal cortex (CTXent) and claustrum (CLAUS). b Regions where XXY and XO mice both show anatomical differences relative to their respective gonadal control littermates (i.e., XY and XX). The presence of color indicates overlapping XXY-XY and XO-XX differences in brain volume (when each contrast is thresholded at p < 0.05). These volume differences are reciprocal in green regions [such that XXY > XY where XO < XX (and vice versa)], and not reciprocal in pink regions. c Effect size box and whisker plots for main regions of overlapping XXY-XY and XO-XX differences. Regional color codes correspond to those in b. For each structure, XO (light gray) and XXY (dark gray) volume distribution within voxels of overlapping XXY-XY and XO-XX differences are shown relative to littermate XX and XY controls (respectively). AMc amygdala, central, AMm amygdala, medial, AON anterior olfactory nucleus, BNSTa anterior division of bed nucleus stria terminalis, BNSTp posterior division of bed nucleus stria terminalis, CP caudate putamen, CBctx cerebellar cortex, CLAUS claustrum, CTXent cerebral cortex: entorhinal, CTXm cerebral cortex: motor, CTXrs cerebral cortex: retrosplenial, CTXss cerebral cortex: somatosensory, HIP hippocampus, LHA lateral hypothalamic area, LS lateral septum, MBret midbrain reticular nucleus, MOB main olfactory bulb, MPO medial preoptic area of hypothalamus, MS medial septal nucleus, NA nucleus accumbens, OT olfactory tubercle, PAG periaqueductal gray, SI substantia innominata, THr Reuniens nucleus of thalamus, THv ventral group nuclei of thalamus
Clustering the brain by patterns of significant anatomical variation across XO, XX, XY and XXY mice
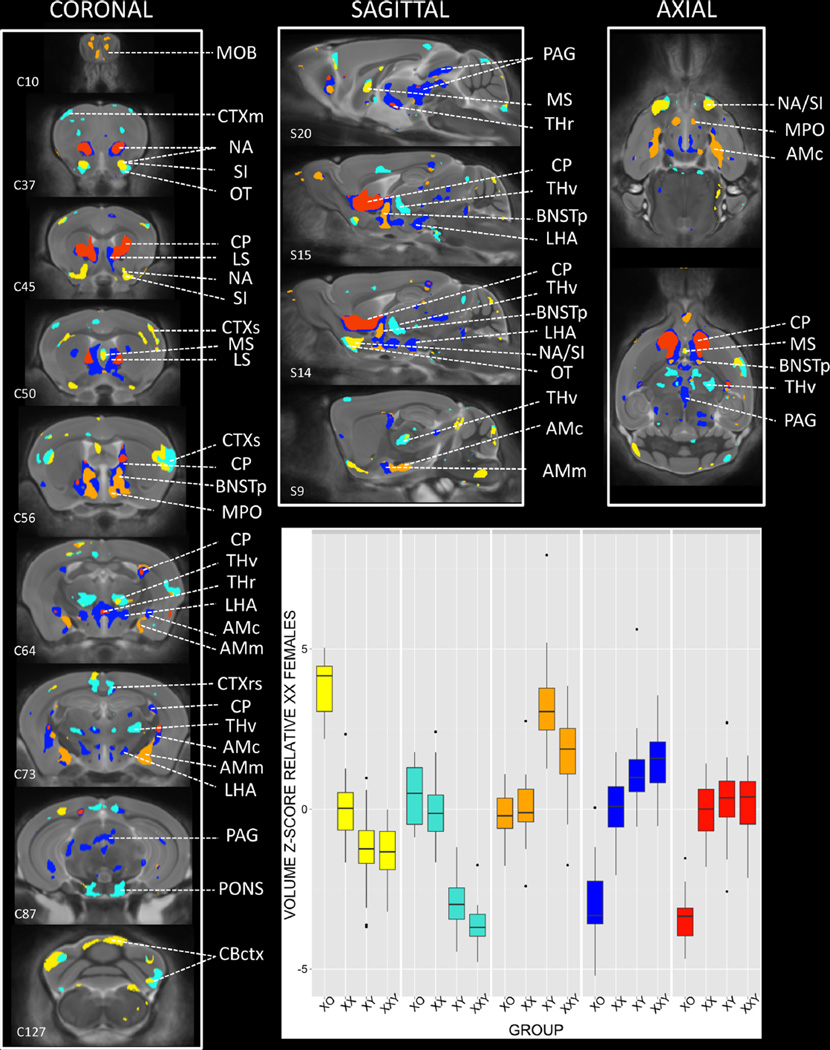
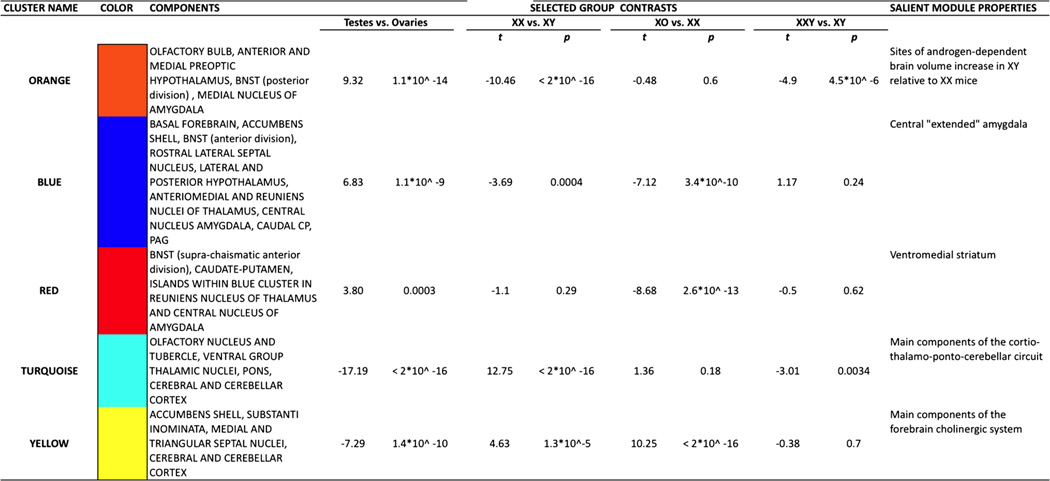
Brain regions of significant anatomical variation across all four karyotype groups were attributed to one of five clusters [Orange, Blue, Red, Turquoise, Yellow] based on K-means clustering of voxels using group mean voxel volume for XO, XX, XY and XXY mice. These clusters showed pronounced bilateral symmetry, and were organized such that cluster boundaries either adjoined Orange, Blue and Red clusters, or Turquoise and Yellow clusters (Fig. 3; Movie 1). Clusters generally defined distinct sets of brain regions, although certain structures—such as the BNST, amygdala, lateral septal region, hypothalamus and cerebral cortex—encompassed more than one cluster (Table 1).
Fig. 3.
Regions of significant anatomical variation across XO, XX, XY, and XXY groups, clustered by observed pattern of anatomical differences between groups. Coronal, sagittal and axial views of the brain show colored regions where an omnibus F test for the effect of group on brain volume survived False Discovery Rate Correction for multiple comparisons with q (expected proportion falsely rejected nulls) set at 0.05. Mean volume for each karyotype group was calculated at every such voxel, and the resulting 200,000 × 4 matrix was subjected to K-means clustering. Visual inspection of the K-means scree plot indicated the optimal number of clusters was 5. Each of these clusters is shown in a different color, with inset box plots displaying the distribution of volume for each cluster across XO, XX, XY, and XXY groups. C# and S# provided for selected slices relate to corresponding Coronal and Sagittal sections within the Allen Institute Reference Mouse Brain Atlas. AMc amygdala, central, AMm amygdala, medial, AON anterior olfactory nucleus, BNSTa anterior division of bed nucleus stria terminalis, BNSTp posterior division of bed nucleus stria terminalis, CP caudate putamen, CBctx cerebellar cortex, CLAUS claustrum, CTXent cerebral cortex: entorhinal, CTXm cerebral cortex: motor, CTXrs cerebral cortex: retrosplenial, CTXss cerebral cortex: somatosensory, HIP hippocampus, LHA lateral hypothalamic area, LS lateral septum, MBret midbrain reticular nucleus, MOB main olfactory bulb, MPO medial preoptic area of hypothalamus, MS medial septal nucleus, NA nucleus accumbens, OT olfactory tubercle, PAG periaqueductal gray, SI substantia innominata, THr Reuniens nucleus of thalamus, THv ventral group nuclei of thalamus
Table 1.
Five clusters in the mouse brain distinguished by observed anatomical variation across XO, XX, XY and XXY groups
This table highlights cluster color, content, selected group contrast in cluster volume and functional/structural themes represented by each cluster
Each cluster showed a distinct pattern of volumetric group differences (Fig. 3; Table 1). Orange and Blue clusters were both larger in gonadal males vs. gonadal females, but differed in the magnitude of X aneuploidy effect observed in each gonadal group (X chromosome aneuploidy effect within males in Orange, but within females in Blue). The closely associated Red cluster was characterized by volume reductions in XO mice relative to all three other groups—which were in turn statistically indistinguishable from each other. Turquoise and Yellow clusters were both larger in gonadal females than gonadal males, and—echoing the Orange–Blue distinction—differed from each other in the magnitude of X aneuploidy effect observed in each gonadal group (karyotype effects within males in Turquoise, but within females in Yellow).
Discussion
Our study provides the first neuroanatomical characterization of the XXY murine SCA model, specifies strong candidate foci for X chromosome gene dose effects on brain volume through conjunction analysis, and provides a data-driven segmentation of the mouse brain based on its volumetric sensitivity to differences in sex chromosome and gonadal complement.
Anatomical phenotype of XXY mice
Anatomical variation within the XXY mouse cohort indicates that presence of a supernumerary X chromosome in males does not prevent penetrance of normative sexual dimorphisms: XY and XXY males both show greater volume than XX females at classic sites of sexual dimorphism within the BNST and amygdala (Hines et al. 1992). XXY mice also group with their XY littermates at lesser-known but previously reported foci of relative brain volume excess in XX females (e.g., thalamus and pons) (Raznahan et al. 2013; Spring et al. 2007). However, direct anatomical comparison between XXY mice and their XY littermates indicates that presence of a supernumerary X chromosome still has an impact on regional brain volumes in males (albeit subtle and at a nominal level of statistical significance). Some of these XXY-XY differences fall within known sites of androgen-sensitive sexual dimorphism such as the BNST and amygdala (Zuloaga et al. 2008; Wu et al. 2009)—suggesting that although the XXY karyotype does not ablate normative volumetric masculinization of the mouse brain, it can interfere with the processes that establish or maintain such sex differences.
Our finding of altered amygdalar structure in XXY mice relative to XY littermates is notable in light of the known importance of amygdalar circuitry in appetitive conditioning and social behavior (Fernando et al. 2013; Felix-Ortiz and Tye 2014), and documented abnormalities in both these domains amongst XXY mice (Lue et al. 2005; Liu et al. 2010). However, the most pronounced anatomical alterations detected by our study impact non-amygdala structures such as the lateral septum, BNST and hypothalamus—indicating the need for broader behavioral characterization of XXY mice, especially through tests of fear conditioning, stress reactivity, aggression and defensive behavior (Koolhaas et al. 1998; Risold and Swanson 1997; Calandreau et al. 2007).
Neuroanatomical alterations in XXY mice recapitulate some of the more robust findings in structural MRI studies of XXY humans: volume reductions relative to XY karyotype in the amygdala and hippocampus (Bryant et al. 2011; Steinman et al. 2009). However, several of the structures that we find to be volumetrically altered in XXY mice are technically difficult to assay using conventional structural neuroimaging methods humans including the BNST, substantia innominata, hypothalamus and PAG. Given that similar brain regions were also highlighted in our earlier neuroanatomical study of XO mice (Raznahan et al. 2013), closer examination of these structures in human SCAs using specially tailored imaging methods is warranted.
Localizing X chromosome aneuploidy effects on brain volume
Our study design allowed us to identify voxels of significant volume difference in both the XO vs XX and XXY vs XY contrast. The proportion of these conjunction voxels where X chromosome haploinsufficiency in females and X chromosome supernumeracy in males produces reciprocal anatomical effects (70 %) is significantly greater than that expected by chance alone. Finding that inverse alterations of X chromosome dosage—in contrasting Y chromosome and gonadal contexts—confer inverse alterations of the same phenotype constitutes strong evidence for direct X chromosome gene-dosage effects. Therefore, our study identifies specific regions within the lateral septum (rostral part), BNST (anterior division), hypothalamus (lateral preoptic and lateral hypothalamic area), thalamus (ventral group nuclei) and midbrain (periaqueductal gray and reticular formation) as strong candidate foci for direct X chromosome gene effects on brain morphometry. Little is known regarding the impact of sex chromosomes and gonadal steroids on the anatomy of these brain regions, although many have been reported to show sexually dimorphic neurochemistry, functioning or connectivity (Krzanowska et al. 2002; Loyd and Murphy 2006; De Vries et al. 2002; Dumais et al. 2013).
The X chromosome genes most likely to show reciprocally altered expression in XO and XXY karyotypes are those that are predicted to undergo step-wide changes in expression with altering X chromosome count by virtue of (1) being located in the “pseudoautosomal region” (PAR) of shared sequence and obligate meiotic pairing between X and Y chromosomes [e.g., Sts (Palmer et al. 1997)] (Otto et al. 2011), or (2) escaping X chromosome inactivation (XCI)—the random “silencing” of one X chromosome in female XX cells (Lyon 1961) that is hypothesized to compensate for male–female differences in X chromosome count [e.g., Ddx3x, Eif2s3x, Itm2a, Kdm5c, Kdm6a, Mid1 (Wu et al. 2014; Lopes et al. 2010; Werler et al. 2011)].
Data-driven brain segmentation across XO, XX, XY and XXY mice recovers known aspects of brain organization
By clustering voxels based on the profile of their volume differences across XO, XX, XY and XXY mice, we identified five distinct “modes” of volumetric sensitivity to the alterations in gonadal profile and X chromosome dose that is represented in our study. The spatial distribution of these five clusters recapitulates multiple aspects of brain organization that were not encoded in the data used to generate our clustering scheme. Specifically, these clusters (1) show striking bilateral symmetry, (2) commonly recover known anatomical boundaries, and (3) divide a large number of spatially dispersed and functionally distinct brain structures into five subsets that share common connections, functions or neurotransmitter profiles.
The sexually dimorphic Orange cluster brings together classic sites of sexual dimorphism in the rodent brain, which are all larger in males than females [BNST, medial amygdala, hypothalamus (overlying anterior hypothalamic and medial preoptic subregions) (Hines et al. 1992; Morris et al. 2004; Shah et al. 2004; Wu et al. 2009)], as well as interconnected sexually dimorphic regions of the olfactory bulb (Segovia and Guillamon 1993). The Blue cluster adjoins the Orange, and is also larger in XY males as compared to XX females. Orange–Blue cluster boundaries recapitulate the intra-amygdala distinction between medial and central subnuclei (Fig. 3, C73), the intra-hypothalamic distinction between medial preoptic and lateral subnuclei (Fig. 3, C56/64), and the distinction between anterior and posterior divisions of the BNST (Fig. 3, S15). These distinctions, and Orange and Blue cluster designations outside the amygdala, hypothalamus and BNST subcomponents, recover the topography of known projections in the rodent brain [e.g., Orange: Medial amygdala—posterior division BNST (Dong et al. 2001), medial preoptic area of the hypothalamus (Pardo-Bellver et al. 2012); Blue: Central amygdala—anterior division BNST (Dong et al. 2001), lateral hypothalamic area (Ono et al. 1985), PAG (Rizvi et al. 1991)]. The Red cluster adjoins the Blue, and defines large bilaterally symmetric ventromedial regions of the striatum, along with smaller thalamic (overlying the Reuniens nucleus) and amygdalar (overlying the central nucleus) subregions. The prominent parasagittal Blue–Red cluster boundary separates lateral septal structures from the striatum.
The two remaining clusters identified by karyotype group differences in brain volume frequently co-localize and both show greater volume in females as compared to males. The Yellow cluster brings together the key components of the forebrain cholinergic system (Mesulam et al. 1983) [medial and triangular nuclei of the lateral septum, substantial innominata] and closely associated regions of the accumbens (Heimer et al. 1997), while the Turquoise cluster encompasses major way stations in the cerebello-thalamo-cortical system (Shinoda et al. 1993).
Data-driven brain segmentation across XO, XX, XY and XXY mice suggests spatial patterning of chromosomal and gonadal influences on brain development
The spatial distribution of clusters within the brain, and differences between clusters in the profile of volumetric variation across karyotype groups, both imply a spatial patterning of chromosomal and gonadal influences on murine brain volume. Specifically, Orange, Blue and Red clusters are arranged such that Orange and Red regions are always separated from each other by interposing Blue. We speculate that this consistent motif reflects the action of spatially graded gene dosage and hormonal effects across Orange, Blue and Red clusters. The pattern of karyotype group differences in volume across Orange, Blue and Red clusters identifies suggests two such graded effects (Table 1; Fig. 3).
First, the penetrance of factors differentiating gonadally male mice from gonadally female mice decreases from Orange to Blue to Red. Given convergent experimental evidence that male–female differences on the volume of Orange cluster components is androgen dependent (Forger 2009), we hypothesize that this gradient is most likely to reflect regional differences in sensitivity to circulating androgens during perinatal sexual differentiation. Moreover, under this hypothesis, documented androgenic deficits in XXY mice (Lue et al. 2005) could account for the fact that the Orange cluster volume is reduced in XXY as compared to XY males.
Second, the penetrance of factors differentiating XO and XX mice increases from Orange to Blue to Red. This gradient would be most parsimoniously accounted for the effects of PAR genes, or non-PAR X-linked genes that escape XCI. Our proposed model for the karyotype group trends observed across Orange, Blue and Red clusters allows for (1) interaction between gradients of androgen-dependent masculinization and X-dosage effects within females (e.g., a strongly deterministic effect of androgens on cell-fate could preventing penetrance of X chromosome gene-dosage effects), and (2) “non-reciprocal” phenotypic differences in XO-XX vs. XXY-XY contrasts (e.g., X chromosome dosage effects may be non-linear or modified by endocrine and/or Y chromosome context) (Ottesen et al. 2010; Arnold 2014).
The two gradients hypothesized above also provide a parsimonious basis for the contrasting pattern of karyotype group differences in brain volume observed between the Yellow and Turquoise clusters. These frequently confluent clusters both show volume increases in XX females relative to XY males, but differ in the penetrance of differences between gonadally male mice from gonadally female mice (Turquoise > Yellow) and differences between XO and XX mice (Yellow > Turquoise).
Limitations
The findings of our study should be considered in light of the following limitations. First, XO mice modeled haploinsufficiency of a maternally inherited X chromosome whereas XXY mice modeled supernumeracy of a paternally inherited X chromosome. Thus, these two murine SCAs do not provide models for reciprocal dosage alterations of X chromosome genes that are subject to paternal imprinting, which differ as a function of brain regions and gonadal sex (Gregg et al. 2010a, b; Raefski and O’Neill 2005; Davies et al. 2006). Second, X chromosome parent of origin in these murine SCA models differs from that most commonly seen in their human counterparts of XO (haploinsufficiency for paternal X) and XXY (supernumeracy of a maternally inherited X). Mice also have different sex chromosome gene content to humans, and the proportion of X chromosome genes that are known to undergo X-inactivation in mice is much smaller than that in humans (Berletch et al. 2011). These factors complicate analysis of sex chromosome dosage effects across the two species. Third, the two cohorts of mice examined in this study were raised in different labs and sacrificed at different points in adulthood. However, the only component of our study that was capable of being confounded by cohort effects (clustering by anatomical trend across all four karyotype groups) used data that had been stripped of any anatomical differences between cohorts in a voxel-wise manner. Fourth, the cross-sectional nature of our study prevents us from specifying when in development the observed anatomical differences emerge. Fifth, our observational neuroimaging study does not provide information about the cellular or molecular underpinnings of reported anatomical differences. However, our approach yields a spatially comprehensive and methodologically homogenous map of chromosomal and gonadal influences on brain development, which would be unfeasible using cellular-level assays within an experimental paradigm. Rather, our study generates a series of novel, highly falsifiable, spatially specific and empirically based hypotheses for examination in future experimental work.
Supplementary Material
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00429-014-0875-9) contains supplementary material, which is available to authorized users.
Contributor Information
Armin Raznahan, Child Psychiatry Branch, National Institute of Mental Health, National Institutes of Health, Rm 4C108, Building 20, 10 Center Drive, Bethesda, MD 20815, USA, raznahana@mail.nih.gov.
YanHe Lue, Division of Endocrinology, Department of Medicine, Los Angele Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA.
Frank Probst, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA.
Deanna Greenstein, Child Psychiatry Branch, National Institute of Mental Health, National Institutes of Health, Rm 4C108, Building 20, 10 Center Drive, Bethesda, MD 20815, USA.
Jay Giedd, Child Psychiatry Branch, National Institute of Mental Health, National Institutes of Health, Rm 4C108, Building 20, 10 Center Drive, Bethesda, MD 20815, USA.
Christina Wang, Division of Endocrinology, Department of Medicine, Los Angele Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA.
Jason Lerch, Mouse Imaging Centre and Program in Neuroscience and Mental, The Hospital for Sick Children Hospital, Toronto, ON, Canada.
Ronald Swerdloff, Division of Endocrinology, Department of Medicine, Los Angele Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA.
References
- Arnold AP. Conceptual frameworks and mouse models for studying sex differences in physiology and disease: why compensation changes the game. Exp Neurol. 2014 doi: 10.1016/j.expneurol.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Genes that escape from X inactivation. Hum Genet. 2011;130(2):237–245. doi: 10.1007/s00439-011-1011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DM, Hoeft F, Lai S, Lackey J, Roeltgen D, Ross J, Reiss AL. Neuroanatomical phenotype of Klinefelter syndrome in childhood: a voxel-based morphometry study. J Neurosci. 2011;31(18):6654–6660. doi: 10.1523/JNEUROSCI.5899-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne PS, Baker TG. Perinatal oocyte loss in XO mice and its implications for the aetiology of gonadal dysgenesis in XO women. J Reprod Fertil. 1985;75(2):633–645. doi: 10.1530/jrf.0.0750633. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Ojarikre OA, Turner JM. Evidence that postnatal growth retardation in XO mice is due to haploinsufficiency for a non-PAR X gene. Cytogenet Genome Res. 2002;99(1–4):252–256. doi: 10.1159/000071601. [DOI] [PubMed] [Google Scholar]
- Cahill LS, Laliberte CL, Ellegood J, Spring S, Gleave JA, Eede MC, Lerch JP, Henkelman RM. Preparation of fixed mouse brains for MRI. Neuroimage. 2012;60(2):933–939. doi: 10.1016/j.neuroimage.2012.01.100. [DOI] [PubMed] [Google Scholar]
- Calandreau L, Jaffard R, Desmedt A. Dissociated roles for the lateral and medial septum in elemental and contextual fear conditioning. Learn Mem. 2007;14(6):422–429. doi: 10.1101/lm.531407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, Ojarikre O, Biggin C, Skuse D, Burgoyne P, Wilkinson L. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat Genet. 2005;37(6):625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- Davies W, Isles AR, Burgoyne PS, Wilkinson LS. X-linked imprinting: effects on brain and behaviour. [Review] [82 refs] BioEssays. 2006;28(1):35–44. doi: 10.1002/bies.20341. [DOI] [PubMed] [Google Scholar]
- De Vries GJ, Rissman EF, Simerly RB, Yang LY, Scordalakes EM, Auger CJ, Swain A, Lovell-Badge R, Burgoyne PS, Arnold AP. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J Neurosci. 2002;22(20):9005–9014. doi: 10.1523/JNEUROSCI.22-20-09005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HW, Petrovich GD, Swanson LW. Topography of projections from amygdala to bed nuclei of the stria terminalis. Brain Res Brain Res Rev. 2001;38(1–2):192–246. doi: 10.1016/s0165-0173(01)00079-0. [DOI] [PubMed] [Google Scholar]
- Dumais KM, Bredewold R, Mayer TE, Veenema AH. Sex differences in oxytocin receptor binding in forebrain regions: correlations with social interest in brain region- and sex- specific ways. Horm Behav. 2013;64(4):693–701. doi: 10.1016/j.yhbeh.2013.08.012. [DOI] [PubMed] [Google Scholar]
- Felix-Ortiz AC, Tye KM. Amygdala inputs to the ventral hippocampus bidirectionally modulate social behavior. J Neurosci. 2014;34(2):586–595. doi: 10.1523/JNEUROSCI.4257-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando AB, Murray JE, Milton AL. The amygdala: securing pleasure and avoiding pain. Front Behav Neurosci. 2013;7:190. doi: 10.3389/fnbeh.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger NG. Control of cell number in the sexually dimorphic brain and spinal cord. J Neuroendocrinol. 2009;21(4):393–399. doi: 10.1111/j.1365-2826.2009.01825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Quevedo L, Blanco J, Sarrate Z, Catala V, Bassas L, Vidal F. Hidden mosaicism in patients with Klinefelter’s syndrome: implications for genetic reproductive counselling. Hum Reprod. 2011;26(12):3486–3493. doi: 10.1093/humrep/der351. [DOI] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15(4):870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Raznahan A, Mills K, Lenroot RK. Review: magnetic resonance imaging of male/female differences in human adolescent brain anatomy. Biol Sex Differ. 2012;3(1):19. doi: 10.1186/2042-6410-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010a;329(5992):682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010b;329(5992):643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimer L, Harlan RE, Alheid GF, Garcia MM, de Olmos J. Substantia innominata: a notion which impedes clinical-anatomical correlations in neuropsychiatric disorders. Neuroscience. 1997;76(4):957–1006. doi: 10.1016/s0306-4522(96)00405-8. [DOI] [PubMed] [Google Scholar]
- Hines M, Allen LS, Gorski RA. Sex differences in subregions of the medial nucleus of the amygdala and the bed nucleus of the stria terminalis of the rat. Brain Res. 1992;579(2):321–326. doi: 10.1016/0006-8993(92)90068-k. [DOI] [PubMed] [Google Scholar]
- Itti E, Gaw Gonzalo IT, Pawlikowska-Haddal A, Boone KB, Mlikotic A, Itti L, Mishkin FS, Swerdloff RS. The structural brain correlates of cognitive deficits in adults with Klinefelter’s syndrome. J Clin Endocrinol Metab. 2006;91(4):1423–1427. doi: 10.1210/jc.2005-1596. [DOI] [PubMed] [Google Scholar]
- Koolhaas JM, Everts H, de Ruiter AJ, de Boer SF, Bohus B. Coping with stress in rats and mice: differential peptidergic modulation of the amygdala-lateral septum complex. Prog Brain Res. 1998;119:437–448. doi: 10.1016/s0079-6123(08)61586-1. [DOI] [PubMed] [Google Scholar]
- Krzanowska EK, Ogawa S, Pfaff DW, Bodnar RJ. Reversal of sex differences in morphine analgesia elicited from the ventrolateral periaqueductal gray in rats by neonatal hormone manipulations. Brain Res. 2002;929(1):1–9. doi: 10.1016/s0006-8993(01)03350-9. [DOI] [PubMed] [Google Scholar]
- Lee NR, Wallace GL, Adeyemi EI, Lopez KC, Blumenthal JD, Clasen LS, Giedd JN. Dosage effects of X and Y chromosomes on language and social functioning in children with supernumerary sex chromosome aneuploidies: implications for idiopathic language impairment and autism spectrum disorders. J Child Psychol Psychiatry. 2012;53(10):1072–1081. doi: 10.1111/j.1469-7610.2012.02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Lee NR, Giedd JN. Effects of sex chromosome aneuploidies on brain development: evidence from neuroimaging studies. Dev Disabil Res Rev. 2009;15(4):318–327. doi: 10.1002/ddrr.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage JF, Mazaika PK, Hong DS, Raman M, Reiss AL. Cortical brain morphology in young, estrogen-naive, and adolescent, estrogen-treated girls with turner syndrome. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage JF, Hong DS, Mazaika PK, Raman M, Sheau K, Marzelli MJ, Hallmayer J, Reiss AL. Genomic imprinting effects of the X chromosome on brain morphology. J Neurosci. 2013;33(19):8567–8574. doi: 10.1523/JNEUROSCI.5810-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Sled JG, Henkelman RM. MRI phenotyping of genetically altered mice. Methods Mol Biol. 2011;711:349–361. doi: 10.1007/978-1-61737-992-5_17. [DOI] [PubMed] [Google Scholar]
- Liu PY, Erkkila K, Lue Y, Jentsch JD, Schwarcz MD, Abuyounes D, Hikim AS, Wang C, Lee PW, Swerdloff RS. Genetic, hormonal, and metabolomic influences on social behavior and sex preference of XXY mice. Am J Physiol Endocrinol Metab. 2010;299(3):E446–E455. doi: 10.1152/ajpendo.00085.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes AM, Burgoyne PS, Ojarikre A, Bauer J, Sargent CA, Amorim A, Affara NA. Transcriptional changes in response to X chromosome dosage in the mouse: implications for X inactivation and the molecular basis of Turner syndrome. BMC Genom. 2010;11:82. doi: 10.1186/1471-2164-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyd DR, Murphy AZ. Sex differences in the anatomical and functional organization of the periaqueductal gray-rostral ventromedial medullary pathway in the rat: a potential circuit mediating the sexually dimorphic actions of morphine. J Comp Neurol. 2006;496(5):723–738. doi: 10.1002/cne.20962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue Y, Jentsch JD, Wang C, Rao PN, Hikim AP, Salameh W, Swerdloff RS. XXY mice exhibit gonadal and behavioral phenotypes similar to Klinefelter syndrome. Endocrinology. 2005;146(9):4148–4154. doi: 10.1210/en.2005-0278. [DOI] [PubMed] [Google Scholar]
- Lynn PM, Davies W. The 39, XO mouse as a model for the neurobiology of Turner syndrome and sex-biased neuropsychiatric disorders. Behav Brain Res. 2007;179(2):173–182. doi: 10.1016/j.bbr.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Mallar Chakravarty M, Steadman P, van Eede MC, Calcott RD, Gu V, Shaw P, Raznahan A, Louis Collins D, Lerch JP. Performing label-fusion-based segmentation using multiple automatically generated templates. Hum Brain Mapp. 2012 doi: 10.1002/hbm.22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesulam MM, Mufson EJ, Wainer BH, Levey AI. Central cholinergic pathways in the rat: an overview based on an alternative nomenclature (Ch1-Ch6) Neuroscience. 1983;10(4):1185–1201. doi: 10.1016/0306-4522(83)90108-2. [DOI] [PubMed] [Google Scholar]
- Morris JA, Jordan CL, Breedlove SM. Sexual differentiation of the vertebrate nervous system. Nat Neurosci. 2004;7(10):1034–1039. doi: 10.1038/nn1325. [DOI] [PubMed] [Google Scholar]
- Ono T, Luiten PG, Nishijo H, Fukuda M, Nishino H. Topographic organization of projections from the amygdala to the hypothalamus of the rat. Neurosci Res. 1985;2(4):221–238. doi: 10.1016/0168-0102(85)90002-1. [DOI] [PubMed] [Google Scholar]
- Ottesen AM, Aksglaede L, Garn I, Tartaglia N, Tassone F, Gravholt CH, Bojesen A, Sorensen K, Jorgensen N, Rajpert-De Meyts E, Gerdes T, Lind AM, Kjaergaard S, Juul A. Increased number of sex chromosomes affects height in a nonlinear fashion: a study of 305 patients with sex chromosome aneuploidy. Am J Med Genet A. 2010;152A(5):1206–1212. doi: 10.1002/ajmg.a.33334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Pannell JR, Peichel CL, Ashman TL, Charlesworth D, Chippindale AK, Delph LF, Guerrero RF, Scarpino SV, McAllister BF. About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 2011;27(9):358–367. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Palmer S, Perry J, Kipling D, Ashworth A. A gene spans the pseudoautosomal boundary in mice. Proc Natl Acad Sci USA. 1997;94(22):12030–12035. doi: 10.1073/pnas.94.22.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Bellver C, Cadiz-Moretti B, Novejarque A, Martinez-Garcia F, Lanuza E. Differential efferent projections of the anterior, posteroventral, and posterodorsal subdivisions of the medial amygdala in mice. Front Neuroanat. 2012;6:33. doi: 10.3389/fnana.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst FJ, Cooper ML, Cheung SW, Justice MJ. Genotype, phenotype, and karyotype correlation in the XO mouse model of Turner Syndrome. J Hered. 2008;99(5):512–517. doi: 10.1093/jhered/esn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raefski AS, O’Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nat Genet. 2005;37(6):620–624. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- Raznahan A, Cutter W, Lalonde F, Robertson D, Daly E, Conway GS, Skuse DH, Ross J, Lerch JP, Giedd JN, Murphy DD. Cortical anatomy in human X monosomy. Neuroimage. 2010;49(4):2915–2923. doi: 10.1016/j.neuroimage.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Probst F, Palmert MR, Giedd JN, Lerch JP. High resolution whole brain imaging of anatomical variation in XO, XX, and XY mice. Neuroimage. 2013;83:962–968. doi: 10.1016/j.neuroimage.2013.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risold PY, Swanson LW. Connections of the rat lateral septal complex. Brain Res Brain Res Rev. 1997;24(2–3):115–195. doi: 10.1016/s0165-0173(97)00009-x. [DOI] [PubMed] [Google Scholar]
- Rizvi TA, Ennis M, Behbehani MM, Shipley MT. Connections between the central nucleus of the amygdala and the midbrain periaqueductal gray: topography and reciprocity. J Comp Neurol. 1991;303(1):121–131. doi: 10.1002/cne.903030111. [DOI] [PubMed] [Google Scholar]
- Rutter M, Caspi A, Moffitt TE. Using sex differences in psychopathology to study causal mechanisms: unifying issues and research strategies. [Review] [219 refs] J Child Psychol Psychiatry. 2003;44(8):1092–1115. doi: 10.1111/1469-7610.00194. [DOI] [PubMed] [Google Scholar]
- Segovia S, Guillamon A. Sexual dimorphism in the vomeronasal pathway and sex differences in reproductive behaviors. Brain Res Brain Res Rev. 1993;18(1):51–74. doi: 10.1016/0165-0173(93)90007-m. [DOI] [PubMed] [Google Scholar]
- Shah NM, Pisapia DJ, Maniatis S, Mendelsohn MM, Nemes A, Axel R. Visualizing sexual dimorphism in the brain. Neuron. 2004;43(3):313–319. doi: 10.1016/j.neuron.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Shinoda Y, Kakei S, Futami T, Wannier T. Thalamocortical organization in the cerebello-thalamo-cortical system. Cereb Cortex. 1993;3(5):421–429. doi: 10.1093/cercor/3.5.421. [DOI] [PubMed] [Google Scholar]
- Simpson JL, de la Cruz F, Swerdloff RS, Samango-Sprouse C, Skakkebaek NE, Graham JM, Jr, Hassold T, Aylstock M, Meyer-Bahlburg HF, Willard HF, Hall JG, Salameh W, Boone K, Staessen C, Geschwind D, Giedd J, Dobs AS, Rogol A, Brinton B, Paulsen CA. Klinefelter syndrome: expanding the phenotype and identifying new research directions. Genet Med. 2003;5(6):460–468. doi: 10.1097/01.gim.0000095626.54201.d0. [DOI] [PubMed] [Google Scholar]
- Skakkebaek A, Gravholt CH, Rasmussen PM, Bojesen A, Jensen JS, Fedder J, Laurberg P, Hertz JM, Ostergaard JR, Pedersen AD, Wallentin M. Neuroanatomical correlates of Klinefelter syndrome studied in relation to the neuropsychological profile. NeuroImage Clin. 2013;4:1–9. doi: 10.1016/j.nicl.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring S, Lerch JP, Henkelman RM. Sexual dimorphism revealed in the structure of the mouse brain using three-dimensional magnetic resonance imaging. Neuroimage. 2007;35(4):1424–1433. doi: 10.1016/j.neuroimage.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Steinman K, Ross J, Lai S, Reiss A, Hoeft F. Structural and functional neuroimaging in Klinefelter (47, XXY) syndrome: a review of the literature and preliminary results from a functional magnetic resonance imaging study of language. Dev Disabil Res Rev. 2009;15(4):295–308. doi: 10.1002/ddrr.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdloff RS, Lue Y, Liu PY, Erkkila K, Wang C. Mouse model for men with klinefelter syndrome: a multifaceted fit for a complex disorder. Acta Paediatr. 2011;100(6):892–899. doi: 10.1111/j.1651-2227.2011.02149.x. [DOI] [PubMed] [Google Scholar]
- Visootsak J, Rosner B, Dykens E, Tartaglia N, Graham JM., Jr Behavioral phenotype of sex chromosome aneuploidies: 48, XXYY, 48, XXXY, and 49 XXXXY. Am J Med Genet A. 2007;143A(11):1198–1203. doi: 10.1002/ajmg.a.31746. [DOI] [PubMed] [Google Scholar]
- Werler S, Poplinski A, Gromoll J, Wistuba J. Expression of selected genes escaping from X inactivation in the 41, XX(Y)* mouse model for Klinefelter’s syndrome. Acta Paediatr. 2011;100(6):885–891. doi: 10.1111/j.1651-2227.2010.02112.x. [DOI] [PubMed] [Google Scholar]
- Wolff DJ, Van Dyke DL, Powell CM Working Group of the ALQAC. Laboratory guideline for Turner syndrome. Genet Med. 2010;12(1):52–55. doi: 10.1097/GIM.0b013e3181c684b2. [DOI] [PubMed] [Google Scholar]
- Wu MV, Manoli DS, Fraser EJ, Coats JK, Tollkuhn J, Honda S, Harada N, Shah NM. Estrogen masculinizes neural pathways and sex-specific behaviors. Cell. 2009;139(1):61–72. doi: 10.1016/j.cell.2009.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Luo J, Yu H, Rattner A, Mo A, Wang Y, Smallwood PM, Erlanger B, Wheelan SJ, Nathans J. Cellular resolution maps of x chromosome inactivation: implications for neural development, function, and disease. Neuron. 2014;81(1):103–119. doi: 10.1016/j.neuron.2013.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuloaga DG, Puts DA, Jordan CL, Breedlove SM. The role of androgen receptors in the masculinization of brain and behavior: what we’ve learned from the testicular feminization mutation. Horm Behav. 2008;53(5):613–626. doi: 10.1016/j.yhbeh.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.