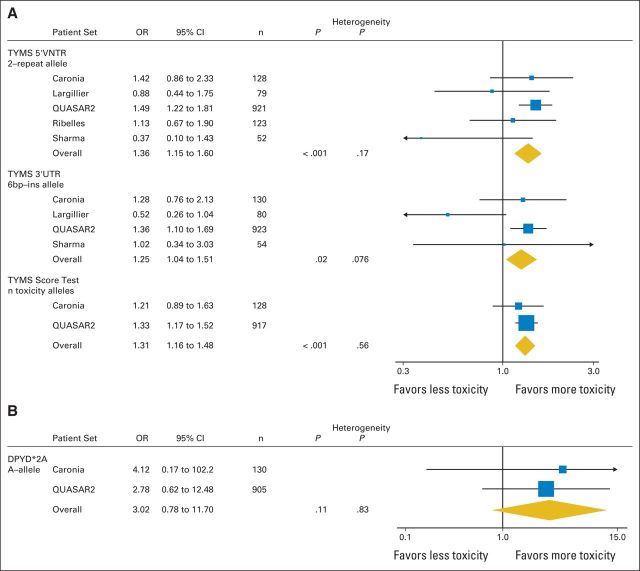
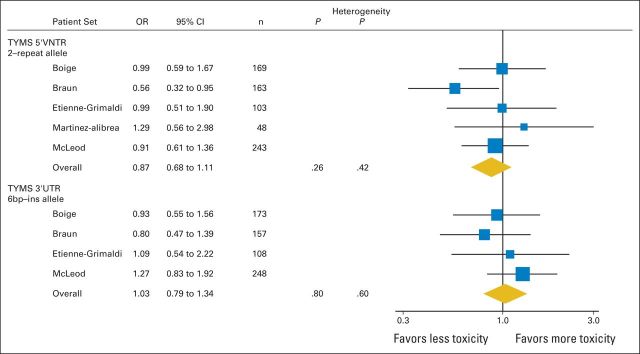
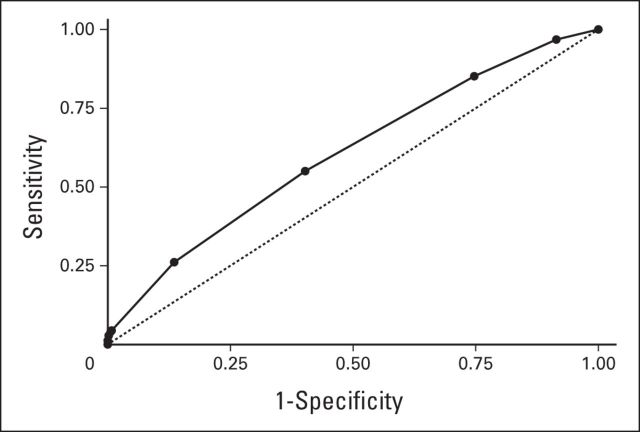
Dan Rosmarin
Dan Rosmarin
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Claire Palles
Claire Palles
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
David Church
David Church
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Enric Domingo
Enric Domingo
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Angela Jones
Angela Jones
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Elaine Johnstone
Elaine Johnstone
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Haitao Wang
Haitao Wang
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Sharon Love
Sharon Love
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Patrick Julier
Patrick Julier
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Claire Scudder
Claire Scudder
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
George Nicholson
George Nicholson
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Anna Gonzalez-Neira
Anna Gonzalez-Neira
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Miguel Martin
Miguel Martin
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Daniel Sargent
Daniel Sargent
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Erin Green
Erin Green
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Howard McLeod
Howard McLeod
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Ulrich M Zanger
Ulrich M Zanger
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Matthias Schwab
Matthias Schwab
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Michael Braun
Michael Braun
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Matthew Seymour
Matthew Seymour
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Lindsay Thompson
Lindsay Thompson
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Benjamin Lacas
Benjamin Lacas
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Valérie Boige
Valérie Boige
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Nuria Ribelles
Nuria Ribelles
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Shoaib Afzal
Shoaib Afzal
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Henrik Enghusen
Henrik Enghusen
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Søren Astrup Jensen
Søren Astrup Jensen
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Marie-Christine Etienne-Grimaldi
Marie-Christine Etienne-Grimaldi
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Gérard Milano
Gérard Milano
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Mia Wadelius
Mia Wadelius
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Bengt Glimelius
Bengt Glimelius
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Hans Garmo
Hans Garmo
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Milena Gusella
Milena Gusella
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Thierry Lecomte
Thierry Lecomte
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Pierre Laurent-Puig
Pierre Laurent-Puig
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Eva Martinez-Balibrea
Eva Martinez-Balibrea
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Rohini Sharma
Rohini Sharma
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Jesus Garcia-Foncillas
Jesus Garcia-Foncillas
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Zdenek Kleibl
Zdenek Kleibl
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Alain Morel
Alain Morel
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Jean-Pierre Pignon
Jean-Pierre Pignon
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Rachel Midgley
Rachel Midgley
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
David Kerr
David Kerr
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,
Ian Tomlinson
Ian Tomlinson
1Dan Rosmarin, Claire Palles, David Church, Enric Domingo, Angela Jones, Ian Tomlinson, Wellcome Trust Centre for Human Genetics, NIHR Comprehensive Biomedical Research Centre, University of Oxford; Dan Rosmarin, David Church, Elaine Johnstone, Haitao Wang, Sharon Love, Patrick Julier, Claire Scudder, George Nicholson, Rachel Midgley, David Kerr, and George Nicholson, University of Oxford, Oxford; Michael Braun, The Christie NHS Foundation Trust, Manchester; Matthew Seymour, St James's University Hospital, Leeds; Lindsay Thompson, Medical Research Council Clinical Trials Unit; Rohini Sharma, Imperial College Healthcare NHS Trust, Hammersmith and Charing Cross Hospitals, London, United Kingdom; Anna Gonzalez-Neira, Spanish National Cancer Research Centre; Miguel Martin, Instituto de Investigacion Sanitaria Hospital General Universitario Gregorio Marañón, Universidad Complutense, Madrid; Nuria Ribelles, Hospital Universitario Virgen de la Victoria, Málaga; Eva Martinez-Balibrea, Institut Català d'Oncologia, Institut de Recerca Sanitària Germans Trias i Pujol, Barcelona; Jesus Garcia-Foncillas, University Clinic of Navarra, University of Navarra, Pamplona, Spain; Daniel Sargent, Erin Green, Mayo Clinic, Rochester, MN; Howard McLeod, University of North Carolina, Chapel Hill, NC; Ulrich M. Zanger, Matthias Schwab, Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; Ulrich M. Zanger, Matthias Schwab, University Hospital Tuebingenl, Tuebingen, Germany; Benjamin Lacas, Valérie Boige, Jean-Pierre Pignon, Institut Gustave Roussy, Villejuif; Marie-Christine Etienne-Grimaldi, Gérard Milano, Centre Antoine Lacassagne, Nice; Thierry Lecomte, Pierre Laurent-Puig, Assistance Publique Hopitaux de Paris, Hopital Européen Georges Pompidou, Paris; Alain Morel, Institut National de la Sante et de la Recherche Medicale U564, Centre Paul Papin, Angers, France; Shoaib Afzal, Henrik Enghusen, Laboratory of Clinical Pharmacology, Rigshospitalet, Copenhagen; Søren Astrup Jensen, Herlev Hospital, Herlev, Denmark; Mia Wadelius, Bengt Glimelius, Hans Garmo, Uppsala University, Uppsala, Sweden; Milena Gusella, Azienda ULSS 18 di Rovigo, Trecenta, Italy; Zdenek Kleibl, Charles University in Prague, Prague, Czech Republic.
1,✉