Abstract
Purpose
We performed a multistage genome-wide association study to identify inherited genetic variants that predict outcome in diffuse large B-cell lymphoma patients treated with immunochemotherapy.
Methods
We conducted a meta-analysis of two genome-wide association study data sets, one from the LNH2003B trial (N = 540), a prospective clinical trial from the Lymphoma Study Association, and the other from the Molecular Epidemiology Resource study (N = 312), a prospective observational study from the University of Iowa–Mayo Clinic Lymphoma Specialized Program of Research Excellence. Top single nucleotide polymorphisms were then genotyped in independent cohorts of patients from the Specialized Program of Research Excellence (N = 391) and the Groupe Ouest-Est des Leucémies Aiguës et Maladies du Sang (GOELAMS) -075 randomized trial (N = 294). We calculated the hazard ratios (HRs) and 95% CIs for event-free survival (EFS) and overall survival (OS) using a log-additive genetic model with adjustment for age, sex, and age-adjusted International Prognostic Index.
Results
In a meta-analysis of the four studies, the top loci for EFS were marked by rs7712513 at 5q23.2 (near SNX2 and SNCAIP; HR, 1.39; 95% CI, 1.23 to 1.57; P = 2.08 × 10−7), and rs7765004 at 6q21 (near MARCKS and HDAC2; HR, 1.38; 95% CI, 1.22 to 1.57; P = 7.09 × 10−7), although they did not reach conventional genome-wide significance (P = 5 × 10−8). Both rs7712513 (HR, 1.49; 95% CI, 1.29 to 1.72; P = 3.53 × 10−8) and rs7765004 (HR, 1.47; 95% CI, 1.27 to 1.71; P = 5.36 × 10−7) were also associated with OS. In exploratory analyses, a two–single nucleotide polymorphism risk score was highly predictive of EFS (P = 1.78 × 10−12) and was independent of treatment, IPI, and cell-of-origin classification.
Conclusion
Our study provides encouraging evidence for associations between loci at 5q23.2 and 6q21 with EFS and OS in patients with diffuse large B-cell lymphoma treated with immunochemotherapy, suggesting novel biology and the potential contribution of host genetics to the prognosis of this aggressive malignancy.
INTRODUCTION
Diffuse large B-cell lymphoma (DLBCL) is the most common non-Hodgkin lymphoma subtype, and approximately 60% of patients with DLBCL are cured with rituximab, doxorubicin, cyclophosphamide, vincristine, and prednisone (R-CHOP) treatment.1 However, the clinical course is heterogeneous, and new biomarkers are needed to better delineate patient outcome, adapt treatment strategy, and identify novel treatment targets. The most commonly used tool for prognostication of patients with DLBCL is the International Prognostic Index (IPI), which is based on conventional clinical and pathology parameters.2 Although it has clinical utility, the IPI does not reflect the biologic heterogeneity of DLBCL. Gene expression profiling of DLBCL tumors from patients treated with R-CHOP has led to advances in the understanding of the pathogenesis, delineating the importance of cell of origin (germinal center v activated B-cell signature) and the potential role for non-neoplastic cells in the tumor microenvironment.3
The role of host genetic background (macroenvironment) in relation to patient outcome is less studied. Although there are promising leads for genetic variation in candidate genes and pathways related to metabolism, immune function, and DNA repair impacting outcomes,4–10 most studies to date have been limited by small sample sizes, have lacked robust replication, have had minimal clinical details, or were conducted in cohorts with unknown, old (prerituximab era), or highly heterogeneous treatments. Compared with the candidate gene approach, the agnostic genome-wide approach has been much more successful in identifying genetic variants linked to cancer risk, but to our knowledge, no comprehensive genome-wide association study (GWAS) has been conducted to identify genetic markers for DLBCL prognosis. In this context, we conducted a multistage GWAS to identify novel loci associated with DLBCL prognosis in patients treated with immunochemotherapy.
METHODS
Study Design and Populations
We performed a multistage analysis to discover genetic loci associated with DLBCL event-free survival (EFS; Data Supplement); a priori power calculations were not conducted. In the first stage, we conducted a meta-analysis of GWAS data from the Lymphoma Study Association (LYSA) prospective LNH03B clinical trial program (France) and the Molecular Epidemiology Resource from the University of Iowa/Mayo Clinic (United States) Lymphoma Specialized Program of Research Excellence (SPORE). The French cohort consisted of a subset of patients with DLBCL (N = 540) with GWAS data from the LNH03B program.11–15 The US cohort consisted of 312 patients with newly diagnosed DLBCL and treated with immunochemotherapy who were prospectively enrolled onto an observational cohort as part of the SPORE (SPORE-I).16 In the second stage, significant single nucleotide polymorphisms (SNPs) from the meta-analysis were evaluated in 391 additional patients with DLBCL from the SPORE (SPORE-II) and 294 patients with DLBCL included in the prospective Groupe Ouest-Est des Leucémies Aiguës et Maladies du Sang (GOELAMS) -075 trial.17 For all studies, diagnoses were reviewed and confirmed by study hematopathologists, and cell of origin was defined as germinal center B-cell (GCB) versus non-GCB DLBCL using a published algorithm.18 Further details are provided in the Data Supplement.
This study was conducted in accordance with the Declaration of Helsinki. The LYSA-03B and GOELAMS-075 studies were approved by the Ethics Committee Haute-Normandie and Nantes University Hospital, respectively. The SPORE studies were approved by the Human Subjects Review Boards at Mayo Clinic and the University of Iowa. All patients provided written consent for participation, including genetic analyses.
Genotyping and Imputation
The LYSA-03B samples were genotyped at Centre National de Genetique (Evry, France) using the Illumina HapMap 610K BeadChip (Illumina, San Diego, CA). The SPORE-I samples were genotyped at the Genotyping Shared Resource at the Mayo Clinic (Rochester, MN) using the Illumina 660Quad BeadChip. Standard quality control measures for genotyping were implemented (Data Supplement). Patients with less than 80% European ancestry were excluded after principal components analysis. To standardize genotyping across the GWAS and facilitate meta-analysis, we imputed participant-level genotyping data using MACH 1.0 software (http://csg.sph.umich.edu//abecasis/MACH/) with HapMap2 as a reference. We used the Illumina VeraCode custom platform to genotype the top SNPs for technical validation in SPORE-I and for stage 2 analyses in the SPORE-II and the GOELAMS-075 trial (Data Supplement).
Statistical Analyses
EFS was the primary end point and was defined as the time from date of random assignment (LYSA-03B and GOELAMS-075) or diagnosis (SPORE cohorts) to the date of disease progression, relapse, re-treatment, or death from any cause. For those SNPs of interest from the EFS analyses, we evaluated overall survival (OS) as a secondary end point. Hazard ratios (HRs) and 95% CIs for the association of SNPs with EFS and OS were estimated using Cox regression analysis and a log-additive (per allele) genetic model. All models were adjusted for age, sex, and age-adjusted IPI (aaIPI).
For the first stage, the LYSA-03B and SPORE-I studies were combined using the fixed-effects inverse variance method based on the β estimates and SEs from each study. From the top SNPs for EFS (genotyped or imputed), we selected 76 SNPs for evaluation along with 12 additional potentially functional SNPs based on a bioinformatics analysis of top regions (Data Supplement). Of the 76 SNPs genotyped in the second stage from the SPORE-II and GOELAMS-075 cohorts, two failed genotyping and one was monomorphic, leaving 73 SNPs for analysis. All 12 potentially functional SNPs were successfully genotyped. The final decision of statistical significance was based on the pooled analysis19 of all four cohorts and the current GWAS statistical significance threshold of P < 5 × 10−8. Meta-analyses were performed on Cox model summary statistics20 analogous to the approach used in the software package METAL (http://csg.sph.umich.edu//abecasis/Metal/).21
In secondary analyses of the replicated SNPs, we pooled the four data sets. We first assessed the SNP associations by cell of origin. Next, we created an SNP score by summing the number of deleterious alleles from the top SNPs that showed robust replication. Kaplan-Meier plots were generated to depict time to EFS by multi-SNP score categories. The multi-SNP score was then fit using a Cox model to adjust for potential confounding factors. We used c-statistics to compare the prognostic ability of the survival models.22 Wald tests using a multiplicative interaction term between the multi-SNP score and aaIPI or cell of origin were conducted. We correlated newly identified loci with gene expression and used multiple bioinformatics tools to assess potential functional impacts with public data sets (Data Supplement).
RESULTS
Study Patients
The four study cohorts had broadly similar clinical characteristics (Table 1), although the GOELAMS-075 cohort had a younger median age of 49 years (range, 18 to 60 years) as a result of the trial upper age eligibility. The majority of patients were treated with R-CHOP; in LYSA-03B, 37% of the patients were treated with rituximab, doxorubicin, vindesine, bleomycin, and prednisone, and in GOELAMS-075, 48% of the patients were treated with rituximab, cyclophosphamide, epirubicin, vindesine, and prednisone followed by transplantation (Data Supplement). The 2-year EFS rates were 72%, 71%, and 70% for SPORE-I, LYSA-03B, and SPORE-II, respectively, whereas in GOELAMS-075, the 2-year EFS rate was 80%.
Table 1.
Clinical Characteristics and Outcome of Patients in the First- and Second-Stage Cohorts
| Characteristic | No. of Patients (%) |
||||
|---|---|---|---|---|---|
| First-Stage Cohorts |
Second-Stage Cohorts |
Pooled (N = 1,537) | |||
| LYSA-03B (n = 540) | SPORE-I (n = 312) | SPORE-II (n = 391) | GOELAMS-075 (n = 294) | ||
| Age, years | |||||
| Median | 61 | 63 | 61 | 49 | 58 |
| Range | 18-93 | 20-92 | 18-88 | 18-60 | 18-93 |
| > 60 | 270 (50) | 185 (59) | 200 (51) | 0 (0) | 655 (43) |
| Male | 307 (57) | 156 (50) | 229 (59) | 174 (59) | 865 (56) |
| ECOG PS ≥ 2 | 101 (19) | 34 (11) | 79 (20) | 45 (15) | 259 (17) |
| LDH > ULN | 217 (40) | 164 (53) | 189 (548) | 213 (72) | 783 (51) |
| Stage III/IV | 394 (73) | 182 (58) | 244 (62) | 196 (67) | 830 (66) |
| ≥ 2 extranodal sites | 212 (39) | 72 (23) | 71 (18) | 120 (41) | 475 (31) |
| “B” symptoms | 193 (36) | 70 (22) | 96 (25) | 126 (43) | 485 (32) |
| BM involvement | 74 (14) | 49 (16) | 69 (18) | 45 (15) | 237 (15) |
| Bulky disease | 114 (21) | 38 (12) | 50 (13) | 180 (61) | 382 (25) |
| aaIPI | |||||
| 0-1 | 271 (50) | 186 (60) | 223 (56) | 121 (41) | 801 (52) |
| 2 | 199 (37) | 99 (32) | 123 (31) | 141 (48) | 562 (37) |
| 3 | 70 (13) | 27 (9) | 45 (12) | 32 (11) | 174 (11) |
| Cell of origin | |||||
| GCB | 132 (46) | 106 (62) | 128 (65) | 49 (50) | 415 (55) |
| Non-GCB | 152 (54) | 66 (38) | 69 (35) | 49 (50) | 336 (45) |
| Unknown | 256 | 140 | 194 | 196 | 786 |
| Treatment | |||||
| R-CHOP | 341 (63) | 274 (88) | 327 (84) | 154 (52) | |
| Others | 0 | 38 (12) | 64 (16) | 0 | |
| R-ACVBP | 199 (37) | 0 | 0 | 0 | |
| R-CEEP | 0 | 0 | 0 | 140 (48) | |
| Overall follow-up, months | |||||
| Median | 32 | 60 | 37 | 46 | 41 |
| Range | 1-85 | 1-127 | 1-129 | 1-92 | 1-129 |
| Follow-up of alive patients, months | |||||
| Median | 39 | 72 | 47 | 51 | 48 |
| Range | 1-85 | 8-126 | 6-129 | 5-92 | 1-129 |
| Events | 176 (33) | 133 (43) | 145 (37) | 75 (26) | 529 (34) |
| Deaths | 134 (25) | 104 (33) | 96 (25) | 50 (17) | 384 (25) |
| 2-Year EFS | 71 | 72 | 70 | 80 | |
| 95% CI | 66-76 | 67-77 | 66-75 | 75-84 | |
| 2-Year OS | 83 | 81 | 82 | 88 | |
| 95% CI | 79-87 | 77-85 | 79-86 | 84-91 | |
Abbreviations: aaIPI, age-adjusted International Prognostic Index; BM, bone marrow; ECOG PS, Eastern Cooperative Oncology Group performance status; EFS, event-free survival; GCB, germinal center B cell; GOELAMS, Groupe Ouest-Est des Leucémies Aiguës et Maladies du Sang; LDH, lactate dehydrogenase; LYSA, Lymphoma Study Association; OS, overall survival; R-ACVBP, rituximab, doxorubicin, vindesine, bleomycin, and prednisone; R-CEEP, rituximab, cyclophosphamide, epirubicin, vindesine, and prednisone; R-CHOP, rituximab, doxorubicin, cyclophosphamide, vincristine, and prednisone; SPORE, Specialized Program of Research Excellence; ULN, upper limit of normal.
SNPs Associated With EFS
In the first stage, the distribution of P values (observed and imputed) for the association of SNPs with EFS matched the expected distribution with a modest excess of significant associations for the LYSA-03B and SPORE-I studies (see quantile-quantile plots in Data Supplement). In the initial meta-analysis of these two studies only, one SNP (rs9298183) achieved genome-wide significance (P < 5 × 10−8).
The 73 top SNPs selected for second-stage analyses, along with 12 additional SNPs from the same regions selected for potential functional relevance, were genotyped in the SPORE-I samples (for technical validation) and in independent samples in SPORE-II and GOELAMS-075 (Data Supplement). The results for EFS, adjusted for age, sex, and aaIPI, in the second-stage cohorts and a meta-analysis of all four studies are provided in the Data Supplement. In the final meta-analysis of the four cohorts, no SNP exceeded the stringent threshold for genome-wide significance (P = 5 × 10−8), but rs7712513 (HR, 1.39; P = 2.08 × 10−7) showed a strongly suggestive P value and consistent HRs across all four studies, and rs7765004 (HR, 1.38; P = 7.09 × 10−7) was consistent in three of the four studies, with a null association in the GOELAMS-075 replication study (Table 2). Both SNPs were also strongly associated with OS, with rs7712513 (HR, 1.49; P = 3.53 × 10−8) reaching the genome-wide significance level. After pooling all four cohorts, these associations held in models adjusted for age, sex, aaIPI, study, and treatment, as well as in a subgroup analysis of patients only treated with R-CHOP (Data Supplement).
Table 2.
Associations of SNPs With Event-Free and Overall Survival in DLBCL in the First and Second Stages and Final Meta-Analysis
| SNP and Study | Chromosome | Position | Nearest Gene | Allele |
RAF | No. of Patients | Event-Free Survival |
Overall Survival |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk | Other | HR* | 95% CI | P | HR* | 95% CI | P | ||||||
| rs7712513 | 5 | 121918208 | ARGFXP1 | C | A | ||||||||
| First stage | |||||||||||||
| LYSA-03B | 0.35 | 540 | 1.40 | 1.14 to 1.73 | .0016 | 1.48 | 1.17 to 1.89 | .0013 | |||||
| SPORE-I | 0.34 | 312 | 1.46 | 1.14 to 1.88 | .0028 | 1.63 | 1.23 to 2.17 | .00077 | |||||
| Meta-analysis (Discovery) | 1.43 | 1.21 to 1.67 | 1.48 × 10−5 | 1.54 | 1.28 to 1.86 | 3.95 × 10−6 | |||||||
| Second stage | |||||||||||||
| SPORE-II | 0.31 | 391 | 1.33 | 1.05 to 1.68 | .020 | 1.50 | 1.13 to 1.98 | .0045 | |||||
| GOELAMS-075 | 0.35 | 294 | 1.33 | 0.97 to 1.84 | .079 | 1.28 | 0.87 to 1.88 | .20 | |||||
| Meta-analysis (all cohorts) | 1.39 | 1.23 to 1.57 | 2.08 × 10−7 | 1.49 | 1.29 to 1.72 | 3.53 × 10−8 | |||||||
| rs7765004 | 6 | 114071720 | LOC100652953 | C | A | ||||||||
| First stage | |||||||||||||
| LYSA-03B | 0.33 | 540 | 1.34 | 1.08 to 1.68 | .0085 | 1.43 | 1.11 to 1.84 | .0054 | |||||
| SPORE-I | 0.30 | 312 | 1.52 | 1.18 to 1.97 | .0013 | 1.43 | 1.06 to 1.92 | .018 | |||||
| Meta-analysis (Discovery) | 1.42 | 1.20 to 1.68 | 4.35 × 10−5 | 1.43 | 1.18 to 1.73 | 2.65 × 10−4 | |||||||
| Second stage | |||||||||||||
| SPORE-II | 0.30 | 391 | 1.44 | 1.13 to 1.84 | .0030 | 1.87 | 1.39 to 2.52 | 4.00 × 10−5 | |||||
| GOELAMS-075 | 0.32 | 294 | 1.14 | 0.80 to 1.62 | .48 | 1.04 | 0.68 to 1.60 | .86 | |||||
| Meta-analysis (all cohorts) | 1.38 | 1.22 to 1.57 | 7.09 × 10−7 | 1.47 | 1.27 to 1.71 | 5.36 × 10−7 | |||||||
Abbreviations: DLBCL, diffuse large B-cell lymphoma; GOELAMS, Groupe Ouest-Est des Leucémies Aiguës et Maladies du Sang; HR, hazard ratio; LYSA, Lymphoma Study Association; RAF, risk allele frequency; SNP, single nucleotide polymorphism; SPORE, Specialized Program of Research Excellence.
HR and 95% CIs adjusted for age, sex, and age-adjusted International Prognostic Index.
The SNP rs7712513 localizes to 5q23.2, which is near the pseudogene ARGFXP1 (arginine-fifty homeobox pseudogene 1) as well as SNCAIP (synuclein, α interacting protein) and SNX2 (sorting nexin 2; Fig 1A). rs7765004 localizes to 6q21, where MARCKS (gene encoding myristoylated alanine-rich protein kinase C substrate) and HDAC2 (gene encoding histone deacetylase 2) are located (Fig 1B).
Fig 1.
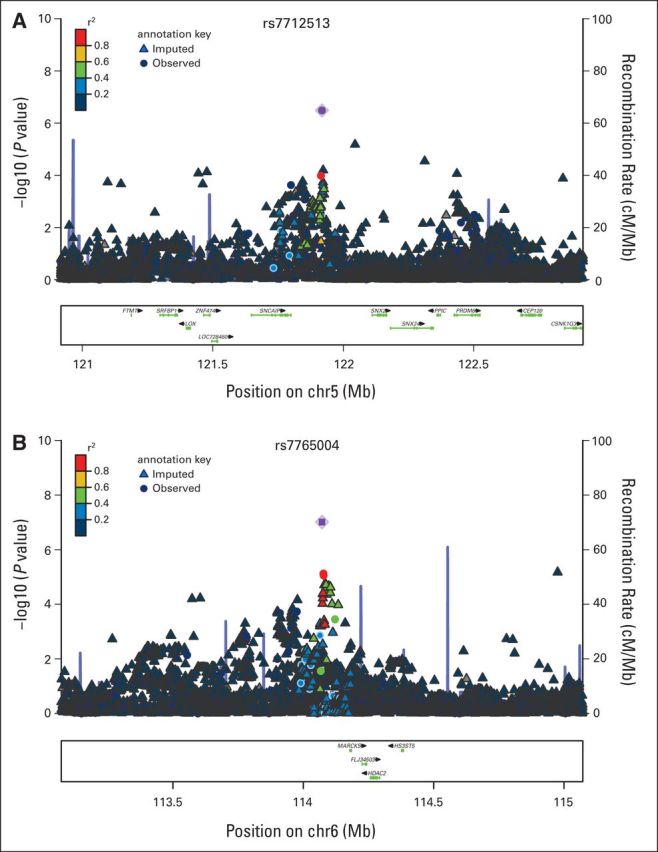
Association results, recombination hotspots, and linkage disequilibrium plots for the regions associated with event-free survival in diffuse large B-cell lymphoma in the discovery cohort. (A) 5q21. (B) 6q21.
Cell of Origin
In 695 patients with data on cell of origin, the associations for rs7712513 were similar for GCB DLBCL (HR, 1.38; 95% CI, 1.09 to 1.77; P = .0089) compared with non-GCB DLBCL (HR, 1.49; 95% CI, 1.15 to 1.93; P = .0029); the associations for rs7765004 were also similar for GCB DLBCL (HR, 1.30; 95% CI, 1.00 to 1.70; P = .053) and non-GCB DLBCL (HR, 1.19; 95% CI, 0.90 to 1.58; P = .21).
Multi-SNP Risk Score for EFS
In the pooled data set, we constructed a risk score based on the number of deleterious alleles from rs7712513 and rs7765004, which was strongly associated with EFS (P = 1.78 × 10−12; Fig 2A). Compared with no deleterious alleles, patients with one (HR, 1.23; 95% CI, 0.95 to 1.59), two (HR, 1.72; 95% CI, 1.32 to 2.23), or three or more (HR, 2.76; 95% CI, 2.03 to 3.73) deleterious alleles had inferior EFS, adjusted for age, sex, aaIPI, study, and treatment (Data Supplement). The c-statistic was 0.666 for the model with age, sex, aaIPI, study, and treatment, and the c-statistic was 0.689 after addition of the two-SNP risk score. In analyses stratified on aaIPI, the SNP score was associated with EFS for low-risk patients with DLBCL (score, 0 to 1; P = .02) and high-risk patients with DLBCL (score, 2 to 3; P = 1.09 × 10−11; Figs 2B and 2C). The two-SNP risk score also predicted EFS in both GCB (P = .001) and non-GCB (P = .0009) DLBCL (Data Supplement). No interactions were observed between multi-SNP risk score and aaIPI (P = .08) or cell of origin (P = .68) on EFS.
Fig 2.
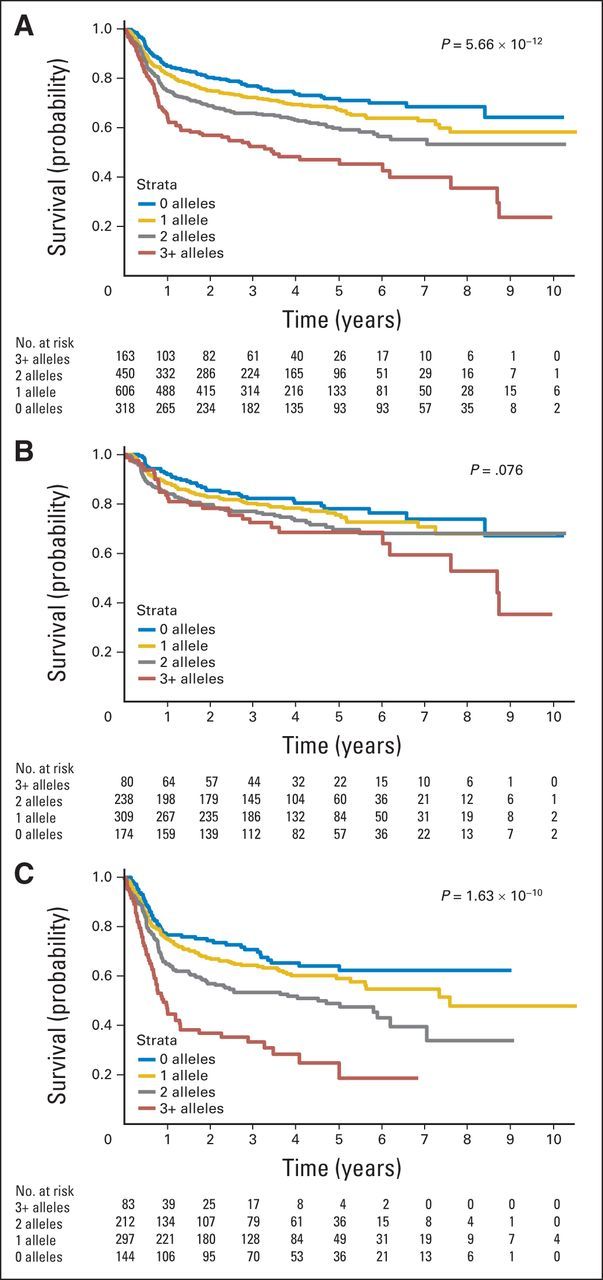
Multi–single nucleotide polymorphism (SNP) risk score for event-free survival; pooled analysis of all four cohorts. (A) All patients (N = 1,537). (B) Patients with low-risk age-adjusted International Prognostic Index (aaIPI; score 0 to 1; n = 801). (C) Patients with high-risk aaIPI (score 2 to 3; n = 736). P values shown are from the Cox proportional hazard models adjusted for age, sex, aaIPI (when not stratified on this value), and treatment regimen.
Correlation Between SNP Genotyping and Tumor Gene Expression
The SNPs at 5q23.2 and 6q21 are noncoding variants. To investigate possible functional consequences of these SNPs on nearby genes, we correlated the two sentinel SNPs with all probe sets annotated to a gene located within 500 kb of the SNP using tumor gene expression data from an Affymetrix U133 plus 2.0 experiment (Affymetrix, Santa Clara, CA) conducted using 73 patients from the LYSA-03B study (Data Supplement). A differential expression between the three genotypes of rs7712513 was observed for the following three genes of unknown function in the National Center for Biotechnology Information database: MGC32805 or LOC153163 (P = .013), PPIC or LOC101927357 (P = .014), and LOC100505841 (P = .034). We also found differential expression between the three genotypes of rs7765004 for the LOC101927794 (P = .037) and HDAC2 genes (P = .045); for the latter gene, there was significantly higher expression in the tumors from patients with the AA + AC genotype compared with the CC genotype (P = .020; Data Supplement).
Additional Bioinformatics Analyses
All variant positions in linkage disequilibrium (LD) (r2 ≥ 0.7) with rs7765004 on chromosome 6 (n = 20) and rs7712513 on chromosome 5 (n = 1) were identified (Data Supplement). One SNP in LD with rs7765004 is located in an evolutionarily conserved region, suggesting regulatory potential. More interestingly, both rs7765004 and another SNP in LD with rs7765004 (rs6923574) are cis-expression quantitative trait loci (eQTL) eSNPs regulating expression of MARCKS according to the lymphoblastoid cell line eQTL data set.23 In addition, all SNPs in LD with rs7765004 are located within the peak regions of the histone marker ChIP-Seq data in the B-lymphocyte cell line GM12878 according to ENCODE (Encyclopedia of DNA Elements; https://www.encodeproject.org). No functional roles for rs7712513 were identified using these bioinformatics approaches.
DISCUSSION
In this GWAS of prognosis in patients with DLBCL initially treated with immunochemotherapy regimens (the first of its kind, to our knowledge), we identified two loci that were suggestive of association with EFS and OS independent of IPI. A risk score constructed with these two loci showed a modest improvement in predictive ability over clinical characteristics (age, sex, aaIPI, and treatment) alone (c-statistic, 0.689 v 0.666, respectively). Potentially of more clinical significance, our two-SNP risk score identified patients with diverse outcomes within traditional low-risk (aaIPI of 0 to 1) and high-risk (aaIPI of 2 to 3) DLBCL patient groups and, in particular, identified a group of high-risk patients (aaIPI of 2 to 3) with three or more deleterious alleles who had a particularly poor prognosis (Fig 2C).
More fundamentally, our data are encouraging for the role of host genetic background on DLBCL prognosis, which can provide novel leads into lymphoma biology and perhaps new therapeutic targets. This concept was suggested by retrospective studies mainly conducted before the rituximab era and that used a candidate gene approach, with SNPs in immune genes,4–7,10 DNA repair and metabolism genes,8,9,24 and angiogenesis genes25 associated with outcome in patients with DLBCL. We explored the significant SNPs for DLBCL outcome from these studies in the first-stage meta-analysis and did not find any that approached conventional significance (Data Supplement).
The SNP at 5q23.2 (rs7712513) maps near two genes, SNCAIP and SNX2 (Fig 1A), implicated in pathogenic protein inclusions26 and cellular trafficking proteins,27 respectively. Tandem duplication of SNCAIP has been found in medulloblastoma,28 and one synphilin-1 partner, synuclein, was recently found to be implicated in B-cell development.29 SNX2 has been identified as a fusion partner of ABL1 in B-cell acute lymphoblastic leukemia,30 and as a member of membrane trafficking, SNX2 was found to interact with the receptors of platelet-derived growth factor or epidermal growth factor and could modulate target-drug sensitivity.27,31 However, we found no correlation between rs7712513 genotypes and either SNCAIP or SNX2 expression in DLBCL tumors (Data Supplement), and bioinformatics approaches did not identify any functional links to this SNP.
The SNP rs7765004 at 6q21 maps near the MARCKS and HDAC2 encoding genes (Fig 1B). MARCKS is a substrate for protein kinase C and has been implicated in the modulation of the metastatic phenotype in a colon carcinoma model,32 but a role of MARCKS in lymphoma pathogenesis has not been reported. Both rs7765004 and another SNP on chromosome 6, rs6923574, are SNPs regulating expression of MARCKS according to the eQTL data from HapMap human lymphoblastoid cell lines. MARCKS plays important roles in cell shape, cell motility, secretion, transmembrane transport, and regulation of the cell cycle. Recently, MARCKS has been implicated in the exocytosis of a number of vesicles and granules such as mucin and chromaffin.33–35 The search of the seeQTL database did not show an association of rs7765004 genotypes with HDAC2 expression, although HDAC2 has previously been found to be overexpressed in DLBCL.36,37 Perhaps unexpectedly, in our own gene expression data, we observed lower expression of HDAC2 in DLBCL tumors with the germline CC genotype compared with AA + AC genotype (P = .020; Data Supplement), suggesting that the risk allele (C) associated with poorer EFS and OS is also associated with lower HDAC expression by DLBCL tumors. Further functional work is needed to better interrogate this region.
Strengths of this study include the use of two independent first-stage cohorts combined with additional independent cohorts in the second stage, all with detailed and prospectively acquired clinical and outcome data. Further advantages include use of both observational and randomized clinical trial study designs and the fact that all treatments were based on immunochemotherapy regimens, enhancing generalizability. Sensitivity analysis suggested minimal impact of the type of immunochemotherapy used. A potential limitation was that one of the second-stage cohorts (GOELAMS-075) had more unique clinical characteristics, restricting patients to age ≤ 60 years. Our primary EFS end point results did not reach the stringent GWAS threshold for statistical significance of P < 5 × 10−8. We were only powered to detect effect sizes as small as 1.75 assuming minor allele frequency of 30% or higher with 80% power. However, given the consistency of effect sizes across the cohorts, a statistical significance that was just less than the stringent GWAS statistical threshold, and biologic plausibility, our results suggest true findings rather than false-negative results. Although our approach seems to be promising, in the future, even larger studies will be required to identify additional loci.
From a therapeutic perspective, we recognize that our results cannot immediately lead to proposing targeted agents, but the identification of an SNP modulating HDAC expression emphasizes again the role of epigenetics in the biology of DLBCL, but now at the host germline level as well, and perhaps, in the future, this may help to choose patients in which epigenetic modifiers might be more efficient. The other locus draws attention to cellular trafficking pathways that might be targetable if additional work confirms the importance of these pathways in DLBCL, considering the recent example of selective inhibition of nuclear export.38
In summary, our GWAS provides encouraging evidence for germline susceptibility loci of prognostic impact in patients with DLBCL treated with immunochemotherapy. These data further suggest the contribution of inherited genetic background to the clinical behavior of lymphoma. The assessment of the allelic status of these novel germline SNPs offers promise as a prognostic marker and a new approach to uncovering novel biology.
Supplementary Material
Acknowledgment
We thank Anne-Laure Borrel and Aurelie Verney for their technical support; Catherine Maingonnat, Philippe Bertrand, and Elodie Bohers for the genotyping experiments in the Groupe Ouest-Est des Leucémies Aiguës et Maladies du Sang (GOELAMS) cohort; and all the Lymphoma Study Association (LYSA) and Molecular Epidemiology Resource (MER) staff for data collection. We thank Sondra Buehler for editorial assistance. We thank Diana Zelenika, PhD, for genotyping of the Groupe d'Etude des Lymphomes de l'Adulte samples.
Footnotes
Supported by National Cancer Institute Specialized Programs of Research Excellence (SPORE) in Human Cancer (Grant No. P50 CA97274), Molecular Epidemiology of Non-Hodgkin Lymphoma Survival (Grant No. R01 CA129539), National Center for Advancing Translational Science (Grant No. UL1 TR000135), Henry J. Predolin Foundation, Institut National du Cancer (INCa, PAIR Lymphome, 2008-020), Lymphoma Study Association, Fondation de France, and Philippe Foundation.
Authors' disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Disclosures provided by the authors are available with this article at www.jco.org.
AUTHOR CONTRIBUTIONS
Conception and design: Hervé Ghesquieres, Matthew J. Maurer, Gilles A. Salles, James R. Cerhan
Financial support: Gilles A. Salles, James R. Cerhan
Administrative support: Gilles A. Salles, James R. Cerhan
Provision of study materials or patients: Amelie S. Veron, Thierry Fest, Thomas M. Habermann, Marie C. Bene, Sylvain Mareschal, Corinne Haioun, Thierry Lamy, Stephen M. Ansell, Herve Tilly, Thomas E. Witzig, George J. Weiner, Andrew L. Feldman, Ahmet Dogan, Thierry Jo Molina, Brian K. Link, Noel Milpied, Gilles A. Salles
Collection and assembly of data: Hervé Ghesquieres, Susan L. Slager, Fabrice Jardin, Amelie S. Veron, Matthew J. Maurer, Thierry Fest, Thomas M. Habermann, Marie C. Bene, Anne J. Novak, Sylvain Mareschal, Corinne Haioun, Thierry Lamy, Stephen M. Ansell, Herve Tilly, Thomas E. Witzig, George J. Weiner, Andrew L. Feldman, Ahmet Dogan, Julie M. Cunningham, Curtis L. Olswold, Thierry Jo Molina, Brian K. Link, Noel Milpied, David G. Cox, Gilles A. Salles, James R. Cerhan
Data analysis and interpretation: Hervé Ghesquieres, Susan L. Slager, Yan W. Asmann, Matthew J. Maurer, Curtis L. Olswold, David G. Cox, Gilles A. Salles, James R. Cerhan
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Genome-Wide Association Study of Event-Free Survival in Diffuse Large B-Cell Lymphoma Treated With Immunochemotherapy
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Hervé Ghesquieres
No relationship to disclose
Susan L. Slager
No relationship to disclose
Fabrice Jardin
No relationship to disclose
Amelie S. Veron
No relationship to disclose
Yan W. Asmann
No relationship to disclose
Matthew J. Maurer
No relationship to disclose
Thierry Fest
No relationship to disclose
Thomas M. Habermann
No relationship to disclose
Marie C. Bene
Research Funding: Beckman Coulter, sponsoring of Harmonemia Project
Travel, Accommodations, Expenses: Celgene, American Society of Hematology Meeting
Anne J. Novak
No relationship to disclose
Sylvain Mareschal
No relationship to disclose
Corinne Haioun
No relationship to disclose
Thierry Lamy
No relationship to disclose
Stephen M. Ansell
Consulting or Advisory Role: Research to Practice
Research Funding: Bristol-Myers Squibb, Celldex, Seattle Genetics
Herve Tilly
Honoraria: Celgene, Roche/Genentech, Janssen Pharmaceuticals
Consulting or Advisory Role: Takeda, Immunogen
Research Funding: Celgene (Inst)
Travel, Accommodations, Expenses: Roche
Thomas E. Witzig
Stock or Other Ownership: Valeant Pharmaceuticals International
Consulting or Advisory Role: Amgen
Research Funding: Celgene (Inst), Novartis (Inst), Spectrum Pharmaceuticals (Inst), Acerta Pharma (Inst)
George J. Weiner
No relationship to disclose
Andrew L. Feldman
No relationship to disclose
Ahmet Dogan
No relationship to disclose
Julie M. Cunningham
No relationship to disclose
Curtis L. Olswold
No relationship to disclose
Thierry Jo Molina
Honoraria: Merck Serono
Travel, Accommodations, Expenses: Mundipharma, Gilead Sciences
Brian K. Link
Consulting or Advisory Role: Genentech/Roche, Abbvie, Gilead Sciences, Pharmacyclics
Research Funding: Genentech/Roche (Inst), Millennium Takeda (Inst), Bristol-Myers Squibb (Inst), Seattle Cancer Care Alliance (Inst), Pharmacyclics (Inst)
Travel, Accommodations, Expenses: Genentech/Roche, Gilead Sciences
Noel Milpied
No relationship to disclose
David G. Cox
Honoraria: Genomic Health
Patents, Royalties, Other Intellectual Property: Patent EP2764365A1
Gilles A. Salles
Honoraria: Roche/Genentech, Mundipharma, Janssen, Celgene
Consulting or Advisory Role: Roche/Genentech, Gilead Sciences, Janssen Pharmaceuticals, Celgene, Amgen
Research Funding: Roche/Genentech (Inst)
Travel, Accommodations, Expenses: Roche/Genentech
James R. Cerhan
No relationship to disclose
REFERENCES
- 1.Roschewski M, Staudt LM, Wilson WH. Diffuse large B-cell lymphoma-treatment approaches in the molecular era. Nat Rev Clin Oncol. 2014;11:12–23. doi: 10.1038/nrclinonc.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shipp MA, Harrington DP, Anderson JR, et al. A predictive model for aggressive non-Hodgkin's lymphoma: The International Non-Hodgkin's Lymphoma Prognostic Factors Project. N Engl J Med. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 3.Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362:1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aschebrook-Kilfoy B, Zheng T, Foss F, et al. Polymorphisms in immune function genes and non-Hodgkin lymphoma survival. J Cancer Surviv. 2012;6:102–114. doi: 10.1007/s11764-010-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charbonneau B, Maurer MJ, Fredericksen ZS, et al. Germline variation in complement genes and event-free survival in follicular and diffuse large B-cell lymphoma. Am J Hematol. 2012;87:880–885. doi: 10.1002/ajh.23273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Habermann TM, Wang SS, Maurer MJ, et al. Host immune gene polymorphisms in combination with clinical and demographic factors predict late survival in diffuse large B-cell lymphoma patients in the pre-rituximab era. Blood. 2008;112:2694–2702. doi: 10.1182/blood-2007-09-111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lech-Maranda E, Baseggio L, Bienvenu J, et al. Interleukin-10 gene promoter polymorphisms influence the clinical outcome of diffuse large B-cell lymphoma. Blood. 2004;103:3529–3534. doi: 10.1182/blood-2003-06-1850. [DOI] [PubMed] [Google Scholar]
- 8.Rossi D, Rasi S, Di Rocco A, et al. The host genetic background of DNA repair mechanisms is an independent predictor of survival in diffuse large B-cell lymphoma. Blood. 2011;117:2405–2413. doi: 10.1182/blood-2010-07-296244. [DOI] [PubMed] [Google Scholar]
- 9.Wang SS, Maurer MJ, Morton LM, et al. Polymorphisms in DNA repair and one-carbon metabolism genes and overall survival in diffuse large B-cell lymphoma and follicular lymphoma. Leukemia. 2009;23:596–602. doi: 10.1038/leu.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warzocha K, Ribeiro P, Bienvenu J, et al. Genetic polymorphisms in the tumor necrosis factor locus influence non-Hodgkin's lymphoma outcome. Blood. 1998;91:3574–3581. [PubMed] [Google Scholar]
- 11.Delarue R, Tilly H, Mounier N, et al. Dose-dense rituximab-CHOP compared with standard rituximab-CHOP in elderly patients with diffuse large B-cell lymphoma (the LNH03-6B study): A randomised phase 3 trial. Lancet Oncol. 2013;14:525–533. doi: 10.1016/S1470-2045(13)70122-0. [DOI] [PubMed] [Google Scholar]
- 12.Fitoussi O, Belhadj K, Mounier N, et al. Survival impact of rituximab combined with ACVBP and upfront consolidation autotransplantation in high-risk diffuse large B-cell lymphoma for GELA. Haematologica. 2011;96:1136–1143. doi: 10.3324/haematol.2010.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ketterer N, Coiffier B, Thieblemont C, et al. Phase III study of ACVBP versus ACVBP plus rituximab for patients with localized low-risk diffuse large B-cell lymphoma (LNH03-1B) Ann Oncol. 2013;24:1032–1037. doi: 10.1093/annonc/mds600. [DOI] [PubMed] [Google Scholar]
- 14.Peyrade F, Jardin F, Thieblemont C, et al. Attenuated immunochemotherapy regimen (R-miniCHOP) in elderly patients older than 80 years with diffuse large B-cell lymphoma: A multicentre, single-arm, phase 2 trial. Lancet Oncol. 2011;12:460–468. doi: 10.1016/S1470-2045(11)70069-9. [DOI] [PubMed] [Google Scholar]
- 15.Récher C, Coiffier B, Haioun C, et al. Intensified chemotherapy with ACVBP plus rituximab versus standard CHOP plus rituximab for the treatment of diffuse large B-cell lymphoma (LNH03-2B): An open-label randomised phase 3 trial. Lancet. 2011;378:1858–1867. doi: 10.1016/S0140-6736(11)61040-4. [DOI] [PubMed] [Google Scholar]
- 16.Drake MT, Maurer MJ, Link BK, et al. Vitamin D insufficiency and prognosis in non-Hodgkin's lymphoma. J Clin Oncol. 2010;28:4191–4198. doi: 10.1200/JCO.2010.28.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milpied NJ, Legouill S, Lamy T, et al. No benefit of first-line rituximab (R) high-dose therapy (R-HDT) over R-CHOP14 for young adults with diffuse large B-cell lymphoma: Preliminary results of the GOELAMS 075 prospective multicentre randomized trial. Blood. 2010;116:685. (abstr) [Google Scholar]
- 18.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 19.Skol AD, Scott LJ, Abecasis GR, et al. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 20.Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 23.Xia K, Shabalin AA, Huang S, et al. SeeQTL: A searchable database for human eQTLs. Bioinformatics. 2012;28:451–452. doi: 10.1093/bioinformatics/btr678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustafson HL, Yao S, Goldman BH, et al. Genetic polymorphisms in oxidative stress-related genes are associated with outcomes following treatment for aggressive B-cell non-Hodgkin lymphoma. Am J Hematol. 2014;89:639–645. doi: 10.1002/ajh.23709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim MK, Suh C, Chi HS, et al. VEGFA and VEGFR2 genetic polymorphisms and survival in patients with diffuse large B cell lymphoma. Cancer Science. 2012;103:497–503. doi: 10.1111/j.1349-7006.2011.02168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong E, Bejarano E, Rakshit M, et al. Molecular determinants of selective clearance of protein inclusions by autophagy. Nat Commun. 2012;3:1240. doi: 10.1038/ncomms2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol. 2002;3:919–931. doi: 10.1038/nrm974. [DOI] [PubMed] [Google Scholar]
- 28.Northcott PA, Shih DJ, Peacock J, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao W, Shameli A, Harding C, et al. B cell development is regulated by a-synuclein, a key player in Parkinson's disease. Blood. 2013;122:785. (abstr) [Google Scholar]
- 30.Ernst T, Score J, Deininger M, et al. Identification of FOXP1 and SNX2 as novel ABL1 fusion partners in acute lymphoblastic leukaemia. Br J Haematol. 2011;153:43–46. doi: 10.1111/j.1365-2141.2010.08457.x. [DOI] [PubMed] [Google Scholar]
- 31.Ogi S, Fujita H, Kashihara M, et al. Sorting nexin 2-mediated membrane trafficking of c-Met contributes to sensitivity of molecular-targeted drugs. Cancer Sci. 2013;104:573–583. doi: 10.1111/cas.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rombouts K, Carloni V, Mello T, et al. Myristoylated alanine-rich protein kinase C substrate (MARCKS) expression modulates the metastatic phenotype in human and murine colon carcinoma in vitro and in vivo. Cancer Lett. 2013;333:244–252. doi: 10.1016/j.canlet.2013.01.040. [DOI] [PubMed] [Google Scholar]
- 33.Hartwig JH, Thelen M, Rosen A, et al. MARCKS is an actin filament crosslinking protein regulated by protein kinase C and calcium-calmodulin. Nature. 1992;356:618–622. doi: 10.1038/356618a0. [DOI] [PubMed] [Google Scholar]
- 34.Blackshear PJ. The MARCKS family of cellular protein kinase C substrates. J Biol Chem. 1993;268:1501–1504. [PubMed] [Google Scholar]
- 35.Jin Cho S, La M, Ahn JK, et al. Tob-mediated cross-talk between MARCKS phosphorylation and ErbB-2 activation. Biochem Biophys Res Commun. 2001;283:273–277. doi: 10.1006/bbrc.2001.4773. [DOI] [PubMed] [Google Scholar]
- 36.Min SK, Koh YH, Park Y, et al. Expression of HAT1 and HDAC1, 2, 3 in diffuse large B-cell lymphomas, peripheral T-cell lymphomas, and NK/T-cell lymphomas. Korean J Pathol. 2012;46:142–150. doi: 10.4132/KoreanJPathol.2012.46.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marquard L, Poulsen CB, Gjerdrum LM, et al. Histone deacetylase 1, 2, 6 and acetylated histone H4 in B- and T-cell lymphomas. Histopathology. 2009;54:688–698. doi: 10.1111/j.1365-2559.2009.03290.x. [DOI] [PubMed] [Google Scholar]
- 38.Parikh K, Cang S, Sekhri A, et al. Selective inhibitors of nuclear export (SINE): A novel class of anti-cancer agents. J Hematol Oncol. 2014;7:78. doi: 10.1186/s13045-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


