Abstract
Background:
Globally, methicillin resistant Staphylococcus aureus (MRSA) is one of the most common pathogens that causes hospital- and community-acquired infections. The use of molecular typing methods is essential for determining the origin of the isolates, their clonal relations, and also epidemiological investigations.
Objective:
The purpose of this study was to determine the prevalence of antibiotic-resistant MRSA investigate the accessory gene regulator (agr) and staphylococcal cassette chromosome mec (SCCmec) types and perform multilocus sequence typing (MLST). Furthermore, the minimum inhibitory concentration of MRSA isolates was determined for vancomycin and daptomycin.
Materials and Methods:
Two hundred and fifty-nine MRSA isolates were collected from Tertiary Care Hospitals in Coimbatore. Disk diffusion method was employed to assess the sensitivity of MRSA isolates to selected antibiotics and genetic analysis was performed using SCCmec, agr, and MLST typing by multiplex-polymerase chain reaction strategy. Minimal inhibitory concentration (MIC) was determined using Ezy MIC (vancomycin) and Biomerieux (daptomycin) E-test strip.
Results:
Of 259 MRSA isolates, 209 (80.7%) were confirmed as methicillin resistant. Antibiotic susceptibility pattern revealed that all the MRSA isolates were 100% sensitive to linezolid, rifampicin, teicoplanin, and vancomycin. MIC results showed that of 209 MRSA isolates, 10 were found to be vancomycin intermediate S. aureus and 100% of the MRSA isolates were daptomycin-susceptible. The agr group I and SCCmec Type III were the major type among MRSA isolates. In addition to these MLST typing revealed the prevalence of sequence type (ST) 239 (SLV of ST8) among the MRSA isolates.
Conclusion:
This study confirms that ST239 (Brazilian clone) of MRSA is predominant in this region which is responsible for the hospital-acquired MRSA infections. Thus, the study also suggests that vancomycin and daptomycin can still be used as an alternative drug for treating severe MRSA infections.
Keywords: Antibiotic resistance, daptomycin, methicillin resistant Staphylococcus aureus, sequence type 239, vancomycin intermediate Staphylococcus aureus
INTRODUCTION
Staphylococcus aureus is one of the most prevalent pathogens that cause both community and hospital acquired infections ranging from skin surface infections, such as folliculitis and furunculosis, to life-threatening conditions, such as endocarditis, pneumonia, and septicemia.[1] Immediately after the introduction of methicillin into clinical practice, the staphylococci isolates developed resistance to it and MRSA was initially identified for the 1st time in 1961 by Jevons.[2] Since then, isolates of MRSA have spread worldwide and have become established inside and outside the hospital environments.[3] It is known that the infections caused by this pathogen are acquired by the expression of many virulence factors which include an agr system and so far, four major agr types in S. aureus have been recognized.[4] Methicillin resistance is due to the acquisition of mecA gene which is a part of the staphylococcal cassette chromosome (SCC). This gene encodes the protein PBP2A (protein binding to penicillin) that inhibits the action of β-lactam antibiotics. SCCmec elements have been classified into eight different types (I-VIII) and some of them are divided further into subtypes.[5]
MRSA is one of the major causes of hospital-acquired infections in many Indian hospitals and around ≥40% of MRSA cases have been reported from various regions based on their antimicrobial susceptibility patterns.[6,7] This increase in methicillin resistance leaves only vancomycin as the drug of choice. Most commonly used antimicrobial agents for the treatment of MRSA infections across the globe include glycopeptides, linezolid, vancomycin and teicoplanin. Moreover, MRSA isolates are resistant to a wide range of antimicrobial agents. As a consequence, selective pressure was exerted that eventually led to the emergence of isolates of S. aureus and other species of staphylococci with decreased susceptibility to vancomycin and other glycopeptides.[8] The first clinical isolate of vancomycin-resistant S. aureus (VRSA) was reported globally[9,10] in subsequent years.
In India, MRSA with reduced susceptibility to vancomycin was reported as early as 2003.[11] The emergence of vancomycin-intermediate S. aureus (VISA) and VRSA have also been reported from India; particularly from the Northern[12,13,14] and Southern[15,16] parts. Concerns over the increase in the rates of heteroresistance and tolerance to this antibiotic, combined with it's pharmacodynamic as well as clinical shortcomings with the progressive role of Gram-positive bacterial infections in clinical medicine, have thus inspired the development of novel antibiotics. Daptomycin is a cyclic lipopeptide antibiotic that is rapidly bactericidal in vitro against a broad spectrum of Gram-positive bacteria. Its unique mechanism of action involves calcium-dependent binding to the bacterial plasma membrane and disruption of membrane function.[17]
Overall, in view of the above facts, this study was aimed to investigate the prevalence and persistence of MRSA clinical isolates obtained from various Tertiary Care Hospitals in Coimbatore, South India. The study also investigated the SCCmec, agr types and determined the clonal relationships among MRSA isolates by multilocus sequence typing (MLST). Further, the in vitro antimicrobial susceptibility to vancomycin and daptomycin was also characterized against MRSA isolates.
MATERIALS AND METHODS
Bacterial isolates
Two hundred and fifty-nine MRSA isolates were obtained from various Tertiary Care Hospitals in and around Coimbatore, South India during August 2010 to December 2014. The MRSA isolates were reconfirmed in the Microbiology Laboratory, Karunya University by standard laboratory methods.[18] Methicillin resistance was confirmed by using cephoxitin (CX) disc (30 mcg) and also by the presence of mecA gene.[19] After identification, the isolates were maintained in tryptone soya broth to which 15% glycerol was added and stored at −20°C. The species identification was conducted by 16srRNA sequencing using the primers 16srRNAF 5’-AGTTTGATCCTGGCTCAG-3’; 16srRNAR 5’-AGGCCCGGGAACGTATTCAC-3’ of 1500 bp and was submitted to NCBI and the accession ID's was obtained (KP121396 and KP121397).
Antimicrobial susceptibility tests
The in vitro antimicrobial susceptibility test for all the MRSA isolates were performed using ampicillin (A), ampicillin/sulfbactam (A/S), chloramphenicol (CL), ciprofloxacin (CP), clindamycin (CD), co-trimoxazole (COT), levofloxacin (LE), linezolid (LZ), fusidic acid (FC), minocycline (MIN), mupirocin (MU), ofloxacin (OF), oxacillin (OX), gentamicin (G), tetracycline (TET), rifampicin (RIF), teicoplanin (TEI), and vancomycin (V) (HiMedia Laboratories, India). Minimal inhibitory concentration (MIC) of daptomycin and vancomycin were determined by E-test method (Biomerieux, New Delhi; Ezy MIC strips, HiMedia Laboratories, India). The MRSA isolate that exhibited a vancomycin MIC of 4-8 mg/L was considered a VISA isolate. All assays were performed in accordance with Clinical and Laboratory Standards Institute guidelines.[18]
Staphylococcal cassette chromosome mec molecular typing
SCCmec typing of MRSA isolates was carried out as described elsewhere.[19] Briefly, the SCCmec multiplex-polymerase chain reaction (PCR) assay contained nine pairs of primers including the unique and specific primers for SCCmec types, Subtypes I, II, III, IVa, IVb, IVc, IVd, V and also the primers for mecA gene. An aliquot of 2 μl of DNA template (~10 ng), 1 μl of each primer (10 μM) and 9 μl of PCR grade water was added to 23 μl of PCR master mix (Ampliqon, Denmark). The thermal cycles performed are as follows: An initial denaturation at 95°C for 5 min, followed by 40 cycles at 94°C for 45 s, 55°C for 1 min, 72°C for 90 s and a final extension at 72°C for 5 min.
Detection of accessory gene regulator alleles
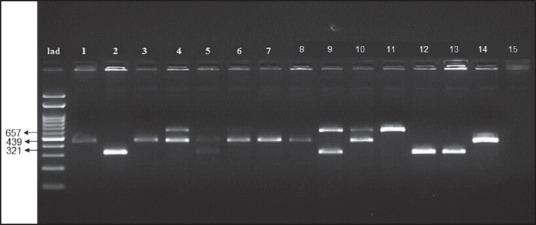
The MRSA isolates were subjected for amplification of agr alleles using the group-specific primers.[20] An aliquot of 2 μl of DNA template (~10 ng), 1 μl of each primer (10 μM) and 11 μl of PCR grade water was added to 23 μl of PCR mastermix (Ampliqon, Denmark). Amplifications were carried out through the following temperature program: 1 cycle of 5 min at 94°C; 26 cycles of 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C; and finally 1 cycle at 72°C for 10 min. Amplification products were electrophoresed in a 1.5% agarose gel containing ethidium bromide and visualized by transillumination under ultraviolet. These primers allowed amplification of 439-, 572-, 320-, and 657-bp DNA fragments of the agr group's 1-4, respectively.
Multilocus sequence typing
MLST was performed as previously described elsewhere.[21] Internal fragments of seven housekeeping genes for carbamate kinase (arcC), shikimate dehydrogenase (aroE), glycerol kinase (glpF), guanylate kinase (gmK), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi), and acetyl coenzyme A acetyltransferase (yqiL) were amplified by M-PCR with the specified primers and the amplified products were purified and sequenced. Consensus sequences were assembled from both orientations.
RESULTS
Bacterial isolates
Of 259 MRSA isolates, 209 (80.7%) were confirmed as MRSA which had been recovered from 197 (94.3%) pus to 12 (5.7%) urine specimens, respectively. Among the obtained MRSA isolates 122 (58.4%) and 87 (41.6%) were recovered from male and female patients respectively [Table 1]. All the 209 MRSA isolates exhibited resistance to methicillin phenotypically and were confirmed by the presence of mecA gene.
Table 1.
Prevalence of methicillin resistant Staphylococcus aureus strains among clinical specimens in relationship with specimen type

Antimicrobial susceptibility
The resistance rates to antibiotics tested were, 79.42% to ampicillin/sulbactam, 78.46% to chloramphenicol, 76.08% to clindamycin, 72.72% to ofloxacin, 59.33% to mupirocin, 56.46% to levofloxacin, 42.58% to fusidic acid, and 31.1% to minocycline. The MRSA isolates showed 100% resistance to oxacillin, ciprofloxacin, co-trimoxazole, ampicillin, gentamicin, and tetracycline [Figure 1]. The MIC assay revealed that 10 (4.8%) of the 209 MRSA isolates were found to be VISA (with the vancomycin MIC of 4 μg/mL) and all the subjected MRSA isolates were susceptible to daptomycin [Figures 2]. Figure 3 depicts the multi-drug resistant pattern among the MRSA isolates and it was seen that the isolates were resistant to ≥ 6 antibiotics.
Figure 1.
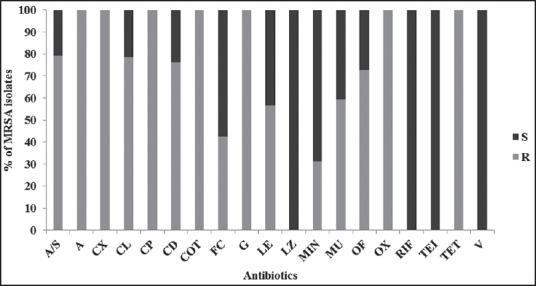
Antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus isolates (n = 209) (S-Sensitive; R-Resistant)
Figure 2.
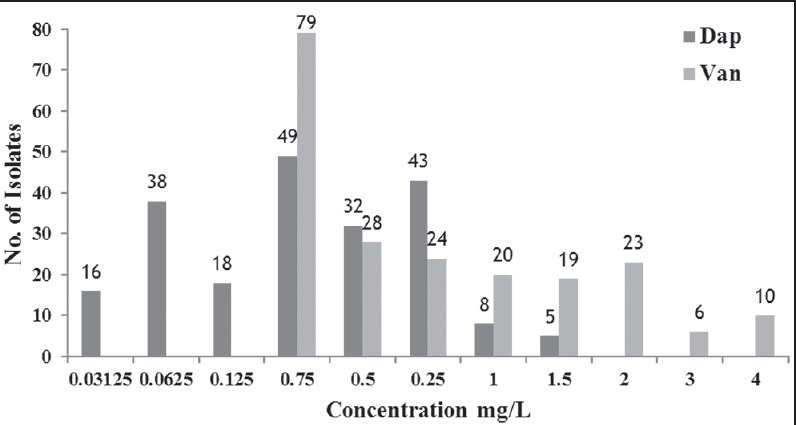
Minimal inhibitory concentration of daptomycin (dap) and vancomycin (van) among the methicillin resistant Staphylococcus aureus isolates (n = 209)
Figure 3.
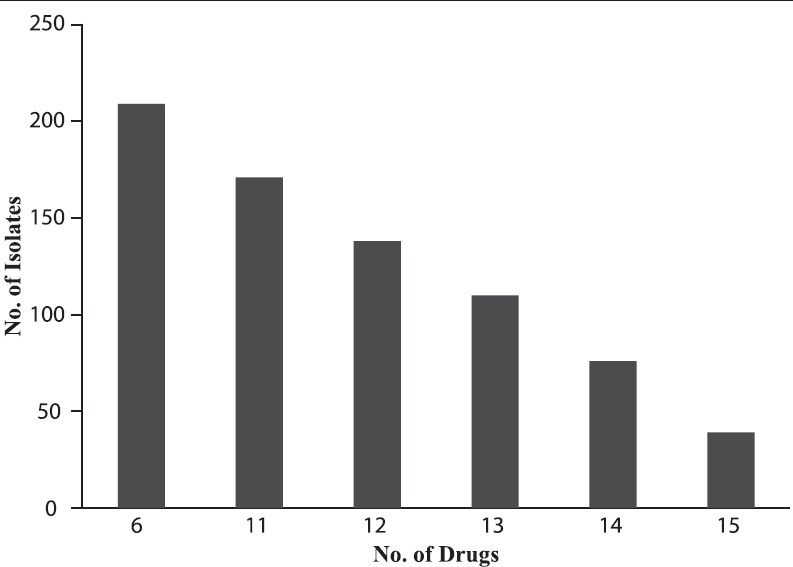
Multi-drug resistant pattern of methicillin resistant Staphylococcus aureus (n = 209)
Staphylococcal cassette chromosome mec typing
All the MRSA isolates were analyzed through SCCmec typing, in which the SCCmec Type III were found to be predominant with a frequency of 94.8% and SCCmec Type IVa was found to be 5.3%, respectively [Figure 4].
Figure 4.
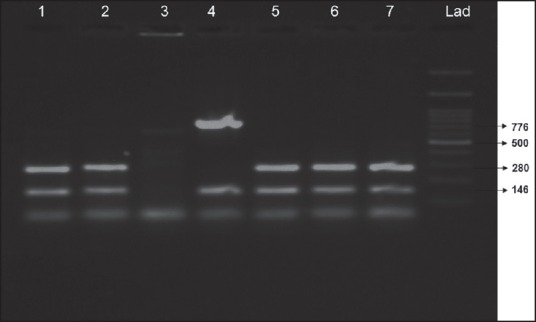
Staphylococcal cassette chromosome mec typing in methicillin resistant Staphylococcus aureus isolates (Lanes 1, 2, 5, 6, 7 shows staphylococcal cassette chromosome mec III-280 bp and mecA-146bp; Lane 4 shows staphylococcal cassette chromosome mec IVa-776 bp and mecA-146bp)
Accessory gene regulator typing
The predominant agr group among the MRSA isolates was group I (100%) followed by Group III (63.6 %) and Group IV (57.6%), respectively. Furthermore, agr Groups I and IV were observed in 42.4% of isolates, groups I and III in 33.3% isolates and groups III and IV in 27.3 % of isolates. None of the MRSA isolates harbored agr Group II. Varying patterns of the agr types among the 33 MRSA isolates with relation to specimen type is shown in Table 2 and Figure 5.
Table 2.
Distribution of methicillin resistant Staphylococcus aureus accessory gene regulator types with relation to the source of infection
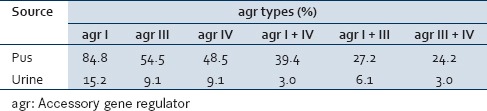
Figure 5.
Accessory gene regulator specific types among the methicillin resistant Staphylococcus aureus isolates
Multilocus sequence typing
MLST was performed on 33 representative isolates with differences in antimicrobial susceptibility pattern. All MRSA isolates were sequence type (ST) 239, with the allelic profile 2-3-1-1-4-4-3 and CC-8 confirming the epidemic clonal origin of the MRSA isolates to be Brazilian clone (SLV of ST8) [Table 3].
Table 3.
Molecular characteristics of ST239 methicillin resistant Staphylococcus aureus isolates

DISCUSSION
Due to the diverse pathogenicity and high antibiotic resistance of MRSA, drug therapy has been problematic causing a great burden for patients and healthcare providers. Molecular typing is thus essential to determine the origin of the isolates, its clonal relationship and also for epidemiological investigations that play an important role in the prevention and treatment of infections.[22] The present study confirmed the presence of 209 (80.7%) MRSA isolates which is found to be much higher than previous findings.[6,16] The antibiotic resistance rates of MRSA to various classes of antibiotics used in this study were higher than the global average.[23,24,25] Antimicrobial susceptibility results showed that all MRSA isolates were susceptible to rifampicin, teicoplanin, linezolid, and vancomycin, which corroborates with the previous findings.[6,26]
Several different phenotypic and genotypic methods are used to classify isolates in epidemiological investigations and also to monitor the nosocomial outbreaks. To attain these objectives, this study used SCCmec, agr, and MLST typing.[25] MRSA classification requires a thorough understanding of their genetic structure as well as detection of SCCmec types. SCCmec typing provides important information about the movable genetic components responsible for resistance to methicillin and it is a marker for differentiation between hospital-acquired MRSA (HA-MRSA) and community-acquired MRSA (CA-MRSA) isolates.[27,28] SCCmec typing was performed on MRSA isolates in which SCCmec Type III (94.8%) was predominant and few isolates were SCCmec Type IVa (5.3%) which are similar with the previously published reports.[16,29,30,31,32] SCCmec Type III has been reported to be the predominant MRSA in Asian continent[33,34] whereas D'souza et al.[30] and Bhutia et al.[32] have reported a lower prevalence of SCCmec Type III and more prevalence of SCCmec Type V in the hospital settings of the Indian subcontinent. However, Deurenberg and Stobberingh[27] have reported that the low frequency of SCCmec Type IV in the many studies may be due to their small size that facilitates their spread among S. aureus isolates.
Various pathogenic factors from the nosocomial isolates of S. aureus develop staphylococcal clinical symptoms.[35] Virulence factors of S. aureus are regulated by various mechanisms and one among them is the agr system. In this study, all of the MRSA isolates belonged to agr Group I (100%) which was comparatively higher than the previous reports.[36,37,38,39,40] Thus, it can be concluded that agr Group I may have a crucial role in the regulation of staphylococcal virulence. In the current study, we noticed the prevalence of agr Groups III and IV to be 21 (63.6%) and 19 (57.6%). The varying agr patterns observed in several studies in different areas are potentially due to some factors, such as geography, origin of specimens and the time of conduction of studies. The rate of agr Group III was 63.6% which was found to be higher than as reported by Kolawole et al.[40] There was no significant relationship between each specific agr group prevalence and antibiotic susceptibility pattern of MRSA isolates which coincides well with the reports of Indrawattana et al. (2013).[39] It was also noted that agr I and III type were documented in VISA isolates which is in line with the results by Ho et al.[37] In addition, investigations are required to find the role of agr on the expression of antibiotic resistance genes.
MRSA ST239 is an invasive clone of multi-drug resistant MRSA producing potential exotoxins thereby cause a wide range of life-threatening infections. It is reported that MRSA ST239 isolates from Asian countries are resistant to additional antibiotics compared to their counterparts in Western countries. MRSA ST239 isolates with varying resistances and susceptibilities to mupirocin, amikacin, and co-trimoxazole have been reported from different parts of the world.[41] Even in this study, erratic patterns of multidrug resistance were observed to different classes of antibiotics. Thus, it can be concluded that ST239 isolates have become endemic in this region. One possible reason for this presumed survival advantage was the significantly higher resistance rates to gentamicin, cefoxitin, ampicillin, clindamycin, and levofloxacin which is supported by Song et al.[42] This report is higher than the results obtained by Ito et al.[43] and Zafar et al.[44] where ST239-MRSA-III isolates were resistant to tetracycline and erythromycin. In India, Nagarajan et al.[45] reported the predominant clone among the MRSA to be ST239 in HA-MRSA and resistance to mupirocin and clindamycin which is also observed in this study. However, Gowrishankar et al.[16] have reported the prevalence of ST239 in South India among the CA-MRSA patients. The MLST data, along with the molecular characteristic studies, illustrate the predominant clonal expansion of ST239 lineages among the MRSA isolates in Tamil Nadu.
Globally, the prevalence of VISA and heterogenous VISA globally was observed in the range of 0.1-11%.[16] The emergence of VRSA from tertiary care hospitals had also been well-documented from North India.[14,15] In South India, recently the emergence of VISA (15.9%) was documented among CA-MRSA patients.[16] However, the present study showed the prevalence rate of VISA to be 4.8% and thereby suggesting the need to develop newer antibiotics. It was also noticed that 100% MRSA isolates exhibited susceptibility to daptomycin suggesting its role in combating MRSA infections. However, daptomycin resistance is also reported among the MRSA isolates worldwide[46,47] and also in India.[48,49,50]
CONCLUSION
The current study suggests that when treatment failure occurs by vancomycin, daptomycin can be prescribed as an alternative treatment option. Thus, the current study highlight the potent use of daptomycin and also suggest that the MIC of daptomycin should be regularly monitored to prevent treatment failure from the possible emergence of isolates with reduced susceptibility. Concomitantly, this study also indicates that the prevalent MRSA clones associated with the hospital settings in Coimbatore, Tamil Nadu are ST239-MRSA-III. However, it should be noted that selection of isolates was narrowed in this study, covering a tapered geographical area over an equitably short period of time. Nevertheless, the data obtained is imperative as it empowers us to form a foundation against what to monitor, the future spread or advent of isolates in this region.
Financial support and sponsorship
UGC Maulana Azad National Fellowship (F1-17.1/2011/MANF-CHR-GUJ-341/(SA-III/Website) 02 Jan 2012).
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The infrastructure and facilities provided by the Department of Biotechnology, Karunya University is gratefully acknowledged. The first author is the recipient of Maulana Azad National Senior Research Fellowship (MANSRF) from University Grants Commission, New Delhi.
REFERENCES
- 1.Kilic A, Guclu AU, Senses Z, Bedir O, Aydogan H, Basustaoglu AC. Staphylococcal cassette chromosome mec (SCCmec) characterization and panton-valentine leukocidin gene occurrence for methicillin-resistant Staphylococcus aureus in Turkey, from 2003 to 2006. Antonie Van Leeuwenhoek. 2008;94:607–14. doi: 10.1007/s10482-008-9278-3. [DOI] [PubMed] [Google Scholar]
- 2.Jevons MP. Celbenin-resistant Staphylococci. Br Med J. 1961;1:b124–5. [Google Scholar]
- 3.Waldvogel FA. New resistance in Staphylococcus aureus. N Engl J Med. 1999;340:556–7. doi: 10.1056/NEJM199902183400709. [DOI] [PubMed] [Google Scholar]
- 4.Azimian A, Najar-Pirayeh S, Mirab-Samiee S, Naderi M. Occurrence of methicillin resistant Staphylococcus aureus (MRSA) among clinical samples in tehran-iran and its correlation with polymorphism of specific accessory gene regulator (AGR) groups. Braz J Microbiol. 2012;43:779–85. doi: 10.1590/S1517-83822012000200043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong H, Louie L, Lo RY, Simor AE. Characterization of Staphylococcus aureus isolates with a partial or complete absence of staphylococcal cassette chromosome elements. J Clin Microbiol. 2010;48:3525–31. doi: 10.1128/JCM.00775-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Indian Network for Surveillance of Antimicrobial Resistance (INSAR) group, India. Methicillin resistant Staphylococcus aureus (MRSA) in India: Prevalence & susceptibility pattern. Indian J Med Res. 2013;137:363–9. [PMC free article] [PubMed] [Google Scholar]
- 7.Neetu PJ, Murugan S. Biofilm formation by methicillin resistant Staphylococcus aureus and their antibiotic susceptibility pattern: An in vitro study. Curr Res Bacteriol. 2014;7:1–11. [Google Scholar]
- 8.Tenover FC. Development and spread of bacterial resistance to antimicrobial agents: An overview. Clin Infect Dis. 2001;33(Suppl 3):S108–15. doi: 10.1086/321834. [DOI] [PubMed] [Google Scholar]
- 9.Palazzo IC, Araujo ML. Darini AL First report of vancomycin-resistant staphylococci isolated from healthy carriers in Brazil. J Clin Microbiol. 2005;43:179–85. doi: 10.1128/JCM.43.1.179-185.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bataineh HA. Resistance of Staphylococcus aureus to vancomycin in Zarqa, Jordan. Pak J Med Sci. 2006;22:144–8. [Google Scholar]
- 11.Assadullah S, Kakru DK, Thoker MA, Bhat FA, Hussain N, Shah A. Emergence of low level vancomycin resistance in MRSA. Indian J Med Microbiol. 2003;21:196–8. [PubMed] [Google Scholar]
- 12.Bhateja P, Mathur T, Pandya M, Fatma T, Rattan A. Detection of vancomycin resistant Staphylococcus aureus: A comparative study of three different phenotypic screening methods. Indian J Med Microbiol. 2005;23:52–5. doi: 10.4103/0255-0857.13875. [DOI] [PubMed] [Google Scholar]
- 13.Tiwari HK, Sen MR. Emergence of vancomycin resistant Staphylococcus aureus (VRSA) from a tertiary care hospital from northern part of India. BMC Infect Dis. 2006;6:156. doi: 10.1186/1471-2334-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saha B, Singh AK, Ghosh A, Bal M. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia) J Med Microbiol. 2008;57:72–9. doi: 10.1099/jmm.0.47144-0. [DOI] [PubMed] [Google Scholar]
- 15.Thati V, Shivannavar CT, Gaddad SM. Vancomycin resistance among methicillin resistant Staphylococcus aureus isolates from intensive care units of tertiary care hospitals in Hyderabad. Indian J Med Res. 2011;134:704–8. doi: 10.4103/0971-5916.91001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gowrishankar S, Thenmozhi R, Balaji K, Pandian SK. Emergence of methicillin-resistant, vancomycin-intermediate Staphylococcus aureus among patients associated with group A Streptococcal pharyngitis infection in southern India. Infect Genet Evol. 2013;14:383–9. doi: 10.1016/j.meegid.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Carpenter CF, Chambers HF. Daptomycin: Another novel agent for treating infections due to drug-resistant gram-positive pathogens. Clin Infect Dis. 2004;38:994–1000. doi: 10.1086/383472. [DOI] [PubMed] [Google Scholar]
- 18.Wayne, PA, USA: CLSI; 2013. CLSI (Clinical and Laboratory Standards Institute). Performance Standards for Antimicrobial Susceptibility Testing: Twenty- first Informational Supplement M100-S21. [Google Scholar]
- 19.Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–33. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilot P, Lina G, Cochard T, Poutrel B. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J Clin Microbiol. 2002;40:4060–7. doi: 10.1128/JCM.40.11.4060-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wildemauwe C, De Brouwer D, Godard C, Buyssens P, Dewit J, Joseph R, et al. The use of spa and phage typing for characterization of a MRSA population in a Belgian hospital: Comparison between 2002 and 2007. Pathol Biol (Paris) 2010;58:70–2. doi: 10.1016/j.patbio.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 23.Peerayeh SN, Azimian A, Nejad QB, Kashi M. Prevalence of agr specificity groups among Staphylococcus aureus isolates from university hospitals in Tehran. Lab Med. 2009;40:27–9. [Google Scholar]
- 24.Vindel A, Cuevas O, Cercenado E, Marcos C, Bautista V, Castellares C, et al. Methicillin-resistant Staphylococcus aureus in Spain: Molecular epidemiology and utility of different typing methods. J Clin Microbiol. 2009;47:1620–7. doi: 10.1128/JCM.01579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tokajian ST, Khalil PA, Jabbour D, Rizk M, Farah MJ, Hashwa FA, et al. Molecular characterization of Staphylococcus aureus in Lebanon. Epidemiol Infect. 2010;138:707–12. doi: 10.1017/S0950268810000440. [DOI] [PubMed] [Google Scholar]
- 26.Wang JL, Chen SY, Wang JT, Wu GH, Chiang WC, Hsueh PR, et al. Comparison of both clinical features and mortality risk associated with bacteremia due to community-acquired methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus. Clin Infect Dis. 2008;46:799–806. doi: 10.1086/527389. [DOI] [PubMed] [Google Scholar]
- 27.Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infect Genet Evol. 2008;8:747–63. doi: 10.1016/j.meegid.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Vainio A, Kardén-Lilja M, Ibrahem S, Kerttula AM, Salmenlinna S, Virolainen A, et al. Clonality of epidemic methicillin-resistant Staphylococcus aureus strains in Finland as defined by several molecular methods. Eur J Clin Microbiol Infect Dis. 2008;27:545–55. doi: 10.1007/s10096-008-0470-1. [DOI] [PubMed] [Google Scholar]
- 29.Fatholahzadeh B, Emaneini M, Gilbert G, Udo E, Aligholi M, Modarressi MH, et al. Staphylococcal cassette chromosome mec (SCC mec) analysis and antimicrobial susceptibility patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolates in Tehran, Iran. Microb Drug Resist. 2008;14:217–20. doi: 10.1089/mdr.2008.0822. [DOI] [PubMed] [Google Scholar]
- 30.D’souza N, Rodrigues C, Mehta A. Molecular characterization of methicillin-resistant Staphylococcus aureus with emergence of epidemic clones of sequence type (ST) 22 and ST 772 in Mumbai, India. J Clin Microbiol. 2010;48:1806–11. doi: 10.1128/JCM.01867-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Japoni A, Jamalidoust M, Farshad S, Ziyaeyan M, Alborzi A, Japoni S, et al. Characterization of SCCmec types and antibacterial susceptibility patterns of methicillin-resistant Staphylococcus aureus in Southern Iran. Jpn J Infect Dis. 2011;64:28–33. [PubMed] [Google Scholar]
- 32.Bhutia KO, Singh T, Adhikari L, Biswas S. Molecular characterization of community-& hospital-acquired methicillin-resistant & methicillin-sensitive Staphylococcus aureus isolates in Sikkim. Indian J Med Res. 2015;142:330–5. doi: 10.4103/0971-5916.166600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arakere G, Nadig S, Ito T, Ma XX, Hiramatsu K. A novel type-III staphylococcal cassette chromosome mec (SCCmec) variant among Indian isolates of methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 2009;292:141–8. doi: 10.1111/j.1574-6968.2008.01482.x. [DOI] [PubMed] [Google Scholar]
- 34.Shabir S, Hardy KJ, Abbasi WS, McMurray CL, Malik SA, Wattal C, et al. Epidemiological typing of meticillin-resistant Staphylococcus aureus isolates from Pakistan and India. J Med Microbiol. 2010;59:330–7. doi: 10.1099/jmm.0.014910-0. [DOI] [PubMed] [Google Scholar]
- 35.Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46(Suppl 5):S350–9. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kahl BC, Becker K, Friedrich AW, Clasen J, Sinha B, Von Eiff C, et al. agr-dependent bacterial interference has no impact on long-term colonization of Staphylococcus aureus during persistent airway infection of cystic fibrosis patients. J Clin Microbiol. 2003;41:5199–201. doi: 10.1128/JCM.41.11.5199-5201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho CM, Hsueh PR, Liu CY, Lee SY, Chiueh TS, Shyr JM, et al. Prevalence and accessory gene regulator (agr) analysis of vancomycin-intermediate Staphylococcus aureus among methicillin-resistant isolates in Taiwan – SMART program, 2003. Eur J Clin Microbiol Infect Dis. 2010;29:383–9. doi: 10.1007/s10096-009-0868-4. [DOI] [PubMed] [Google Scholar]
- 38.Azimian A, Havaei SA, Fazeli H, Naderi M, Ghazvini K, Samiee SM, et al. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolate from the respiratory tract of a patient in a university hospital in northeastern Iran. J Clin Microbiol. 2012;50:3581–5. doi: 10.1128/JCM.01727-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Indrawattana N, Sungkhachat O, Sookrung N, Chongsa-nguan M, Tungtrongchitr A, Voravuthikunchai SP, et al. Staphylococcus aureus clinical isolates: Antibiotic susceptibility, molecular characteristics, and ability to form biofilm. Biomed Res Int 2013. 2013:314654. doi: 10.1155/2013/314654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolawole DO, Adeyanju A, Schaumburg F, Akinyoola AL, Lawal OO, Amusa YB, et al. Characterization of colonizing Staphylococcus aureus isolated from surgical wards’ patients in a Nigerian university hospital. PLoS One. 2013;8:e68721. doi: 10.1371/journal.pone.0068721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feil EJ, Nickerson EK, Chantratita N, Wuthiekanun V, Srisomang P, Cousins R, et al. Rapid detection of the pandemic methicillin-resistant Staphylococcus aureus clone ST 239, a dominant strain in Asian hospitals. J Clin Microbiol. 2008;46:1520–2. doi: 10.1128/JCM.02238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song Y, Du X, Li T, Zhu Y, Li M. Phenotypic and molecular characterization of Staphylococcus aureus recovered from different clinical specimens of inpatients at a teaching hospital in Shanghai between 2005 and 2010. J Med Microbiol. 2013;62:274–82. doi: 10.1099/jmm.0.050971-0. [DOI] [PubMed] [Google Scholar]
- 43.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323–36. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zafar A, Stone M, Ibrahim S, Parveen Z, Hasan Z, Khan E, et al. Prevalent genotypes of meticillin-resistant Staphylococcus aureus: Report from Pakistan. J Med Microbiol. 2011;60:56–62. doi: 10.1099/jmm.0.022707-0. [DOI] [PubMed] [Google Scholar]
- 45.Nagarajan A, Saravanan M, Padma K. Emergence of panton-valentine leucocidin among community-and hospital-associated meticillin-resistant Staphylococcus aureus in Chennai, South India. J Hosp Infect. 2010;76:269–71. doi: 10.1016/j.jhin.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 46.Mangili A, Bica I, Snydman DR, Hamer DH. Daptomycin-resistant, methicillin-resistant Staphylococcus aureus bacteremia. Clin Infect Dis. 2005;40:1058–60. doi: 10.1086/428616. [DOI] [PubMed] [Google Scholar]
- 47.Murthy MH, Olson ME, Wickert RW, Fey PD, Jalali Z. Daptomycin non-susceptible meticillin-resistant Staphylococcus aureus USA 300 isolate. J Med Microbiol. 2008;57(Pt 8):1036–8. doi: 10.1099/jmm.0.2008/000588-0. [DOI] [PubMed] [Google Scholar]
- 48.Biswas S, Brunel JM, Dubus JC, Reynaud-Gaubert M, Rolain JM. olistin: An update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther. 2012;10:917–34. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 49.Jain N, Mathur P, Misra MC. Trend of vancomycin susceptibility of staphylococci at a level 1 trauma centre of India. Indian J Med Res. 2013;138:1022–4. [PMC free article] [PubMed] [Google Scholar]
- 50.Vamsimohan A, Gupta S, Muralidharan S. Daptomycin resistance in methicillin-resistant Staphylococcus aureus: A report from Southern India. Germs. 2014;4:70–2. doi: 10.11599/germs.2014.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]


