Summary
Emerging evidence indicates that precise regulation of iron (Fe) metabolism and maintenance of Fe homeostasis in Mycobacterium tuberculosis (Mtb) are essential for its survival and proliferation in the host. IdeR is a central transcriptional regulator of Mtb genes involved in Fe metabolism. While it is well understood how IdeR functions as a repressor, how it induces transcription of a subset of its targets is still unclear. We investigated the molecular mechanism of IdeR-mediated positive regulation of bfrB, the gene encoding the major Fe-storage protein of Mtb. We found that bfrB induction by Fe required direct interaction of IdeR with a DNA sequence containing four tandem IdeR-binding boxes located upstream of the bfrB promoter. Results of in vivo and in vitro transcription assays identified a direct repressor of bfrB, the histone-like protein Lsr2. IdeR counteracted Lsr2-mediated repression in vitro, suggesting that IdeR induces bfrB transcription by antagonizing the repressor activity of Lsr2. Together these results elucidate the main mechanism of bfrB positive regulation by IdeR and identify Lsr2 as a new factor contributing to Fe homeostasis in mycobacteria.
Introduction
Like most organisms, Mycobacterium tuberculosis (Mtb), the causative agent of human tuberculosis, needs iron (Fe) as a redox cofactor for essential cellular functions, ranging from respiration to DNA replication. Due to the insolubility of ferric ion, the concentration of free Fe available in the host is far below that required for normal growth of the pathogen. Mtb overcomes this Fe limitation by secreting siderophores (mycobactins) that sequester Fe(III) and deliver it to the bacterium via specialized Fe-siderophore transporters (Snow, 1970, Gobin et al., 1995, Rodriguez and Smith, 2006). Although essential, excess Fe can become toxic because it can catalyze the conversion of normal products of aerobic respiration to harmful free radicals (Keyer, 1996). To prevent Fe toxicity, all aerobic organisms tightly control free intracellular Fe. Failure to maintain this control due to mutation of factors involved in Fe sensing or Fe storage sensitizes Mtb to macrophage killing and antibiotics and renders it attenuated in mice (Pandey and Rodriguez, 2012, Pandey and Rodriguez, 2013) and guinea pigs (Reddy et al., 2011), indicating that precise regulation of Fe metabolism and maintenance of Fe homeostasis in Mtb are essential for Mtb pathogenicity.
In mycobacteria, the cellular Fe status is monitored by the Fe-dependent transcriptional regulator IdeR. Mtb lacking IdeR exhibits unrestricted Fe uptake and deficient Fe storage, resulting in toxic Fe overload, and attenuation of virulence (Pandey and Rodriguez, 2013). IdeR transcriptional activity is Fe-dependent: in complex with Fe(II), IdeR regulates the transcription of about 30 Mtb genes involved in Fe metabolism (Rodriguez, 2006). Among IdeR regulated genes in Mtb those involved in the synthesis, export and import of siderophores are repressed by Fe-IdeR, while genes involved in Fe storage (bfrA and bfrB) are positively regulated by Fe-IdeR (Rodriguez et al., 2002) (Pandey and Rodriguez, 2013). The mechanism of IdeR-mediated gene repression is reasonably well understood. In particular, it has been shown that IdeR, in complex with Fe, represses transcription by binding to a specific DNA sequence that overlaps the transcriptional start site and/or the −10 sequence of the target gene, thus impeding RNA polymerase access to the promoter (Gold et al., 2001). However, the molecular mechanism of IdeR-mediated induction of gene expression is still unknown.
Ferritins, Fe storage proteins that are widely distributed among living species and central to Fe homeostasis are upregulated by IdeR. Ferritin subunits associate with each other, forming a spherical shell within which up to 4500 atoms of Fe can be stored in a non-reactive state, thereby providing a non-toxic Fe reserve that can be used when external Fe is not available (Andrews, 1998). Mtb synthesizes a ferritin (BfrB) and a heme containing bacterioferritin (BfrA) encoded by Rv3841 and Rv1876 respectively, (Khare et al., 2011, Gupta et al., 2009) both of which are positively regulated by IdeR (Rodriguez et al., 2002). Our previous studies showed that while BfrA is needed under some stress conditions, BfrB is the major Fe storage protein in Mtb (Pandey and Rodriguez, 2012). Deletion of bfrB leads to Fe-mediated oxidative stress, increased sensitivity to antibiotics and decreased Mtb pathogenicity (Pandey and Rodriguez, 2012). Interestingly, ectopic expression of bfrB in an ideR mutant rescues its Fe toxicity and restores its survival in macrophages (Pandey and Rodriguez, 2013), underscoring the functional significance of IdeR-mediated induction of bfrB expression.
In this study, we investigated the mechanism by which IdeR induces bfrB transcription. We analyzed the effect of IdeR on bfrB transcription both in cells, using a transcriptional reporter in M. smegmatis, and by in vitro transcription assays, using purified components. We found that Fe-dependent induction of bfrB requires IdeR binding to multiple sites upstream of the bfrB promoter. Furthermore, we discovered that IdeR antagonizes Lsr2, a histone-like protein that directly represses bfrB in a Fe independent manner. Together, these results reveal the main mechanism of bfrB regulation by IdeR and uncover a novel role of Lsr2 in Fe homeostasis in Mtb.
Results
Characterization of the bfrB promoter
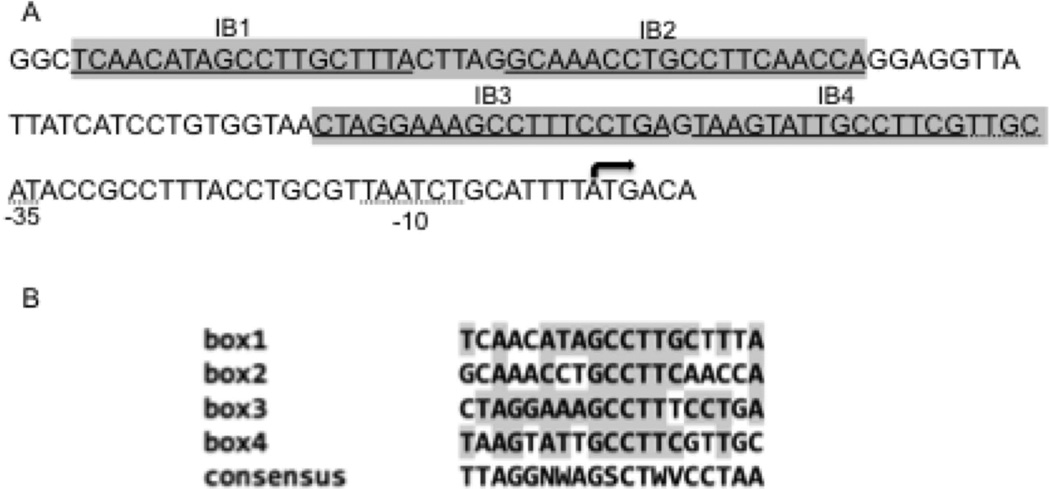
To begin deciphering the transcriptional regulation of bfrB we characterized the promoter of this gene (PbfrB). To identify the transcriptional start point (TSP) total RNA was isolated from Mtb grown in high Fe medium and primer extension was performed using three closely interspaced primers, as described in Experimental Procedures (Fig S1). We also determined the 5’ end of the bfrB transcript using 5’ RACE. Both approaches confirmed the location of the transcriptional start site of bfrB overlapping with the annotated start codon (Fig 1 and Fig S1). Thus, the bfrB transcript has no 5’ UTR. Leaderless transcripts are not unusual in actinomycetes. About one third of Mtb primary transcripts are leaderless (Cortes et al., 2013). Nevertheless, the molecular mechanisms for initiation of leaderless translation in Mtb is not understood. Sequences similar to the consensus −10 and −35 promoter elements (Gomez and Smith, 2000) were identified at the correct distance from the TSP suggesting that the bfrB promoter is recognized by σA-RNA polymerase (RNAp). Four sequences homologous to the consensus IdeR binding site (Rodriguez et al., 2002) are present upstream of bfrB (Fig 1).
Figure 1.
The bfrB promoter. The sequence of the regulatory region of bfrB is shown. The predicted IdeR recognition sequences are underlined (IB1-4) and the similarity to the consensus IdeR binding sequence is shown in the lower panel. Nucleotides that are identical to the consensus sequence are shaded. N= A,C,G or T; W= A or T; S= C or G; V= A,C or G. The regions protected by IdeR in footprinting analysis (according to Fig 2B) are shown in grey boxes. The transcriptional start site which coincides with the translational start site is indicated by the arrow and putative −10 and −35 sequences are underlined with a dotted line.
IdeR regulation of bfrB
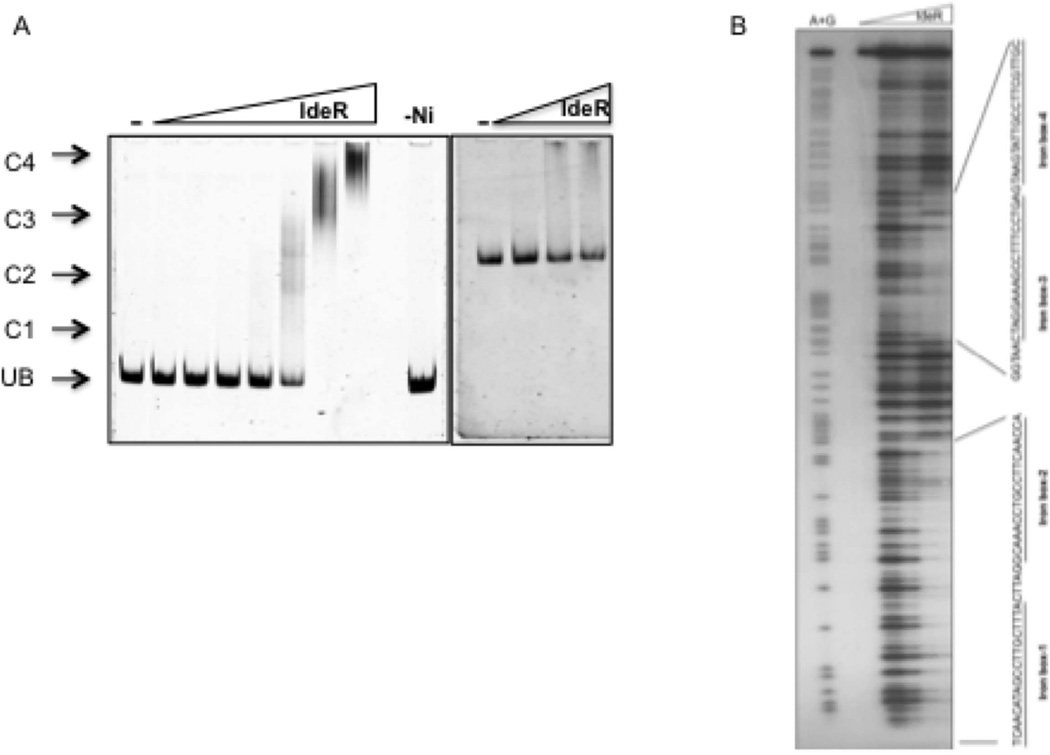
Gene expression analysis in a knock out and also in a conditional ideR mutant demonstrated that bfrB induction in high Fe was IdeR-dependent in Mtb (Rodriguez et al., 2002, Pandey and Rodriguez, 2013). However, whether IdeR directly or indirectly regulates bfrB is unknown. Previous in silico analyses predicted one IdeR binding sequence (iron box) at position −72 upstream of bfrB (Gold et al., 2001). Further examination of the upstream DNA sequence revealed three additional sequences similar to the consensus iron box (IB) (Fig 1) (Rodriguez et al., 2002, Wisedchaisri et al., 2007). To examine IdeR binding to this region a DNA fragment that included the four iron boxes (IB1–4) was PCR amplified and used as a probe in electrophoresis mobility shift assays (EMSA) using purified IdeR. Indeed, we found that IdeR retarded the mobility of the probe in a concentration and metal dependent manner (Fig. 2A). This binding was also specific. Under equivalent conditions IdeR did not retard the mobility of a non-specific DNA fragment used as control (Fig 2A).
Figure 2.
Direct interaction of IdeR with PbfrB. A. Gel shift assay of IdeR and a DNA fragment that includes −386 to +65 of bfrB. IdeR was added in increasing concentrations, from 0.03 µM to 2 µM to the double strand DNA probe, in the presence of 200 µM NiSO4 and DNA-protein complexes were resolved on a 6% Tris-acetate polyacrylamide gel. The right-most lane is a control reaction with 0.5 µM IdeR but without metal (−Ni). UB, unbound probe in the absence of IdeR; C1, C2, C3, C4 indicate IdeR-DNA complexes. The right panel shows that IdeR added at increasing concentrations (0.5 to 2 µM), in the presence of 200 µM NiSO4, does not shift a non specific DNA control. DNA is detected by SYBR Green staining. B. Footprinting of IdeR bound to the bfrB upstream region. A P32 labeled fragment was incubated with up to 1 µM purified IdeR in the presence of 200 µM NiSO4 in binding buffer and subjected to DNase digestion (as described in experimental procedures). Maxam and Gilbert A+G sequencing reactions were performed on the same probes to delineate the sequence protected by IdeR. Reaction products were separated on a 6% TBE polyacrylamide-urea sequencing gel. Protected sequences are indicated.
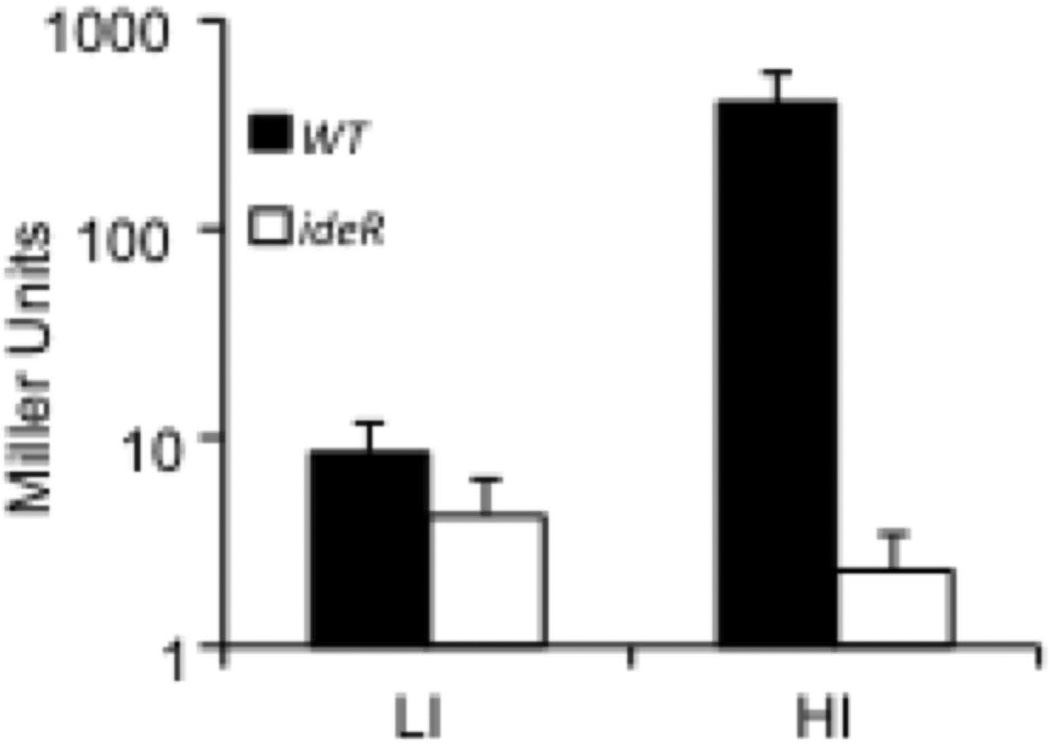
The precise sequences bound by IdeR were determined by DNase protection analysis. We identified two IdeR protected regions separated by 20 bp and including the predicted iron boxes (Fig. 2B). To investigate the molecular determinants of IdeR dependent regulation of bfrB we used the non-virulent, fast growing mycobacterium M. smegmatis (Msm) as Mtb surrogate. A DNA fragment containing PbfrB and the upstream IdeR binding region were fused to the lacZ reporter gene and integrated in the Msm chromosome. PbfrB-driven expression of lacZ was assessed by measuring β-galactosidase activity. Expression of bfrB was induced by 12 fold in wild type Msm cultured in high iron medium. This level of induction is similar to that previously observed in transcriptomic analysis in Mtb (Rodriguez et al., 2002). Importantly, inactivation of ideR abolished bfrB induction confirming that in Msm, as in Mtb, bfrB induction is IdeR dependent (Fig 3).
Figure 3.
Induction of PbfrB in high iron is IdeR dependent in Msm. A. PbfrB, lacZ was introduced into Msm wild type (filled bars) or SM3, an ideR knock out mutant (open bars) and β-galactosidase activity measured in cells grown in low (LI) or high (HI) iron medium as described in experimental procedures section. The data shows the means ± SD from biological triplicates.
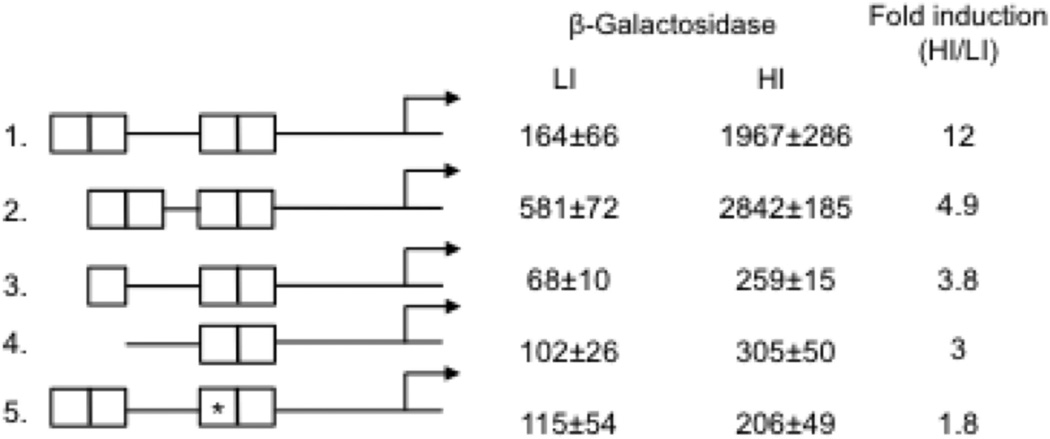
To begin to understand the mechanism of IdeR positive regulation we determined the functional significance of the multiple IdeR binding sites upstream bfrB. DNA fragments containing PbfrB and the upstream IdeR binding region deleted or mutated at the iron boxes (Fig 4) were also fused to lacZ and integrated in the Msm chromosome. PbfrB-driven expression of lacZ was again assessed by measuring β-galactosidase activity. Deletion of IB1 alone or together with IB2 (Fig. 4 constructs 3 and 4 respectively) resulted in decreased induction of PbfrB as did changing the IB3 central sequence AGCCTT to CTATGC (Fig. 4 construct 5). Interestingly, deleting 5 bp in the 20 bp spacer region between IB2 and IB3 (Fig. 4 construct 2) increased promoter expression constitutively suggesting that this short deletion altered control of basal and induced expression of the promoter (the relevance of this observation is discussed later). Collectively, these results show that modifications of the IdeR-binding region negatively impact Fe-dependent induction, underscoring the importance of the binding sites for IdeR mediated positive bfrB regulation.
Figure 4.
Altering the IdeR binding region decreases iron dependent induction of PbfrB. The intact or modified DNA sequences upstream of bfrB containing the four iron boxes (IB1-4) were fused to lacZ and the fusions were individually introduced into Msm. β-galactosidase activity in cells grown in low and high iron medium as described in experimental procedures is shown as well as the fold induction in high iron relative to low iron. 1 to 5 indicates the modifications in the IdeR binding region: 1. The intact IdeR binding region. 2. A 5 bp deletion was made in the spacer sequence between IB2 and IB3. 3. IB1 was deleted. 4. IB1 and IB2 were deleted. 5. *IB3 was mutagenized (AGCCTT changed to CTATGC). The background activity of strains transformed with the vector alone was subtracted from the total activity. The values represent means ± SD from biological triplicates.
Regulation of bfrB transcription in vitro
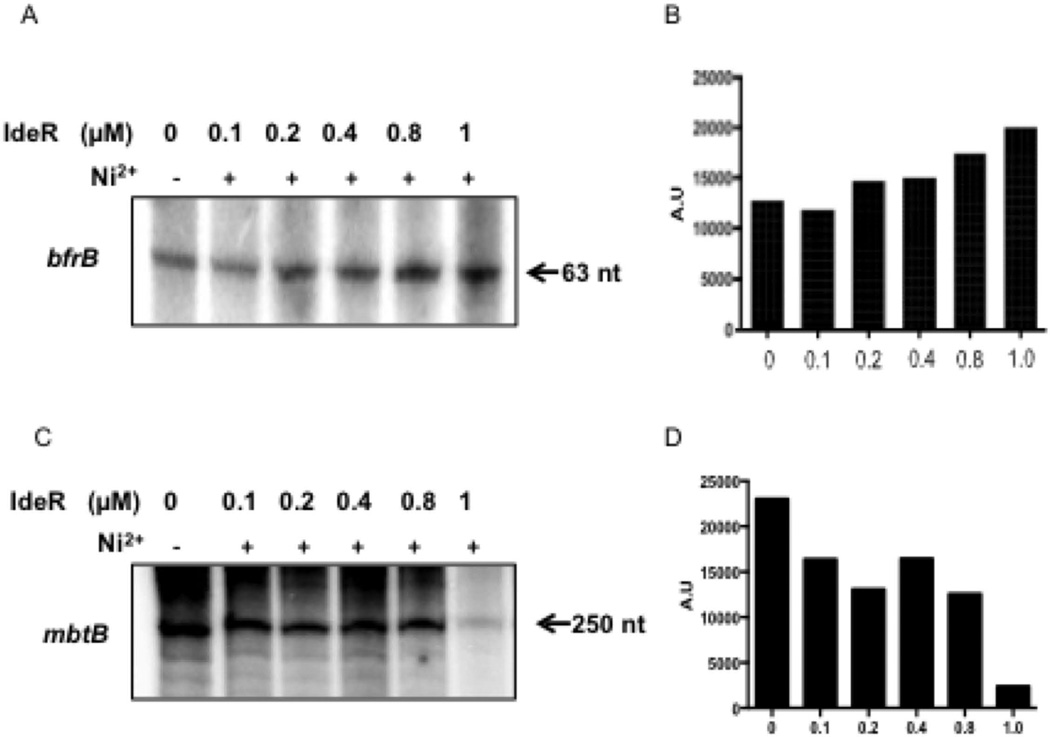
So far, the results demonstrate that IdeR binding to defined sequences upstream of bfrB is necessary for Fe-dependent bfrB induction. To determine if IdeR is also sufficient for induction we purified IdeR and mycobacterial σA-RNAp and conducted in vitro transcription assays. As template, a supercoiled DNA plasmid containing the sequence of a short bfrB transcript driven by PbfrB and flanked by transcriptional terminator sequences was used. A similar template was employed for transcription of the IdeR repressed gene, mbtB, as control. While in the presence of metal, IdeR repressed mbtB by about 10 fold (Fig. 5C–D), it had only a slight, although reproducible, positive effect on bfrB transcription: an average increase of 1.5 fold in the intensity of the bfrB transcript in the presence of IdeR was detected by densitometry in repeated experiments (Fig. 5A–B). This modest transcriptional activation by IdeR seems insufficient to account for the ten or more fold IdeR mediated induction of bfrB observed in vivo and suggests an additional effect of IdeR. One possibility is that IdeR bound to DNA upstream bfrB antagonizes the activity of a transcriptional repressor.
Figure 5.
In vitro transcription of Mtb iron regulated genes in the presence of IdeR. Transcript generated by Msm RNAP using a bfrB template (A) or a template of the iron and IdeR repressed gene mbtB (C). When indicated IdeR and Ni+2 were present in the reaction which was carried out as described in experimental procedures. Radiolabelled transcripts were separated by electrophoresis and detected by autoradiography. Shown are the results of one representative experiment that was repeated 4 times. Panels B and D show the densitometric quantification of the gels shown in A and C respectively.
Lsr-2 represses bfrB transcription
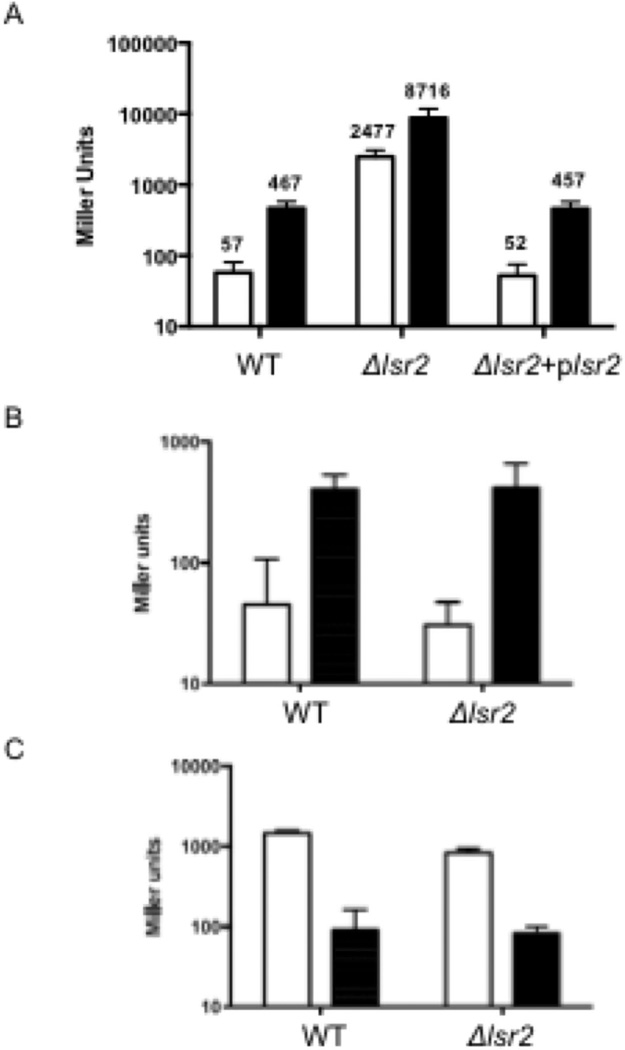
While searching for candidates for a bfrB repressor we noted that deletion of the nucleating histone-like protein Lsr2 in Msm resulted in elevated levels of bfrB transcript (Colangeli et al., 2007). Elevated bfrB transcript was also reported in an Mtb lsr2 mutant (Bartek et al., 2014). Lsr2 is a functional homolog of Escherichia coli H-NS and has been well characterized as a gene silencing protein in Mtb (Gordon et al., 2010). These observations led us to hypothesize that Lsr2 was a bfrB repressor. To test this hypothesis, we first validated Lsr2 repression of bfrB by introducing the PbfrB-lacZ construct into a Msm lsr2 deletion mutant (Δlsr2) (Colangeli et al., 2007), the parental and mutant-complemented strains and measuring β-galactosidase activity. As control we also generated, lacZ fusions of two other IdeR controlled genes PmbtB-lacZ and PbfrA-lacZ and introduced them in Δlsr2. In contrast to the wild type and complemented strains PbfrB-lacZ was highly expressed both in low and high iron in Δlsr2 confirming the role of Lsr2 in repression of bfrB (Fig 6A). The lsr2 deletion did not affect transcription of PbfrA or PmbtB (Fig. 6B and C) showing that repression of PbfrB is specific. Importantly, in the lsr2 mutant the β-galactosidase activity both in low and high iron was much higher than in the wild type strain even in high iron (Fig 6A) indicating that IdeR-Fe mediated induction of bfrB is incomplete and is limited by Lsr2. In addition, PbfrB was further induced (3–4 fold) in high iron relative to low iron in cells lacking Lsr2. This induction could reflect IdeR’s activity as transcriptional activator observed in vitro (Fig 5A) and/or it might be mediated by other factor(s). To investigate the possibility that IdeR contributed to this Lsr2 independent induction of bfrB we tried to generate a double Msm ideR-lsr2 mutant but despite repeated attempts a viable mutant was not recovered suggesting that the double mutation could be lethal. So, we approached this question by testing whether IdeR binding sequences were necessary for lsr2 independent, Fe induction of PbfrB. The PbfrB-lacZ fusions with mutated IBs were introduced into Δlsr2 and β-galactosidase activity was determined in cells grown in low and high iron. Deletion of IB1 and 2 did not affect PbfrB induction in high iron; however, altering the central sequence in IB3 reduced induction of PbfrB (Fig. S3), indicating that IB3 is necessary and suggesting that IdeR also contributes to Lsr2-independent induction of PbfrB. Other factors like change in the local DNA topology may also play a role. Generating a conditional double ideR-lsr2 mutant and additional studies are necessary to resolve the molecular events mediating Fe induction of PbfrB in cells lacking Lsr2.
Figure 6.
Deletion of lsr2 leads to derepression of PbfrB. β-galactosidase activity was determined from Msm (WT), Δlsr2 and Δlsr2 complemented (Δlsr2+plsr2 strains) carrying the PbfrB-lacZ (A), PbfrA-lacZ (B) or PmbtB-lacZ (C) reporter constructs and cultured in low (open bars) or high (filled bars) iron as described in experimental procedures. The background activity of strains transformed with the vector alone was subtracted from the total activity. For clarity, in panel A the plotted values are shown. The data represent means ± SD from biological triplicates.
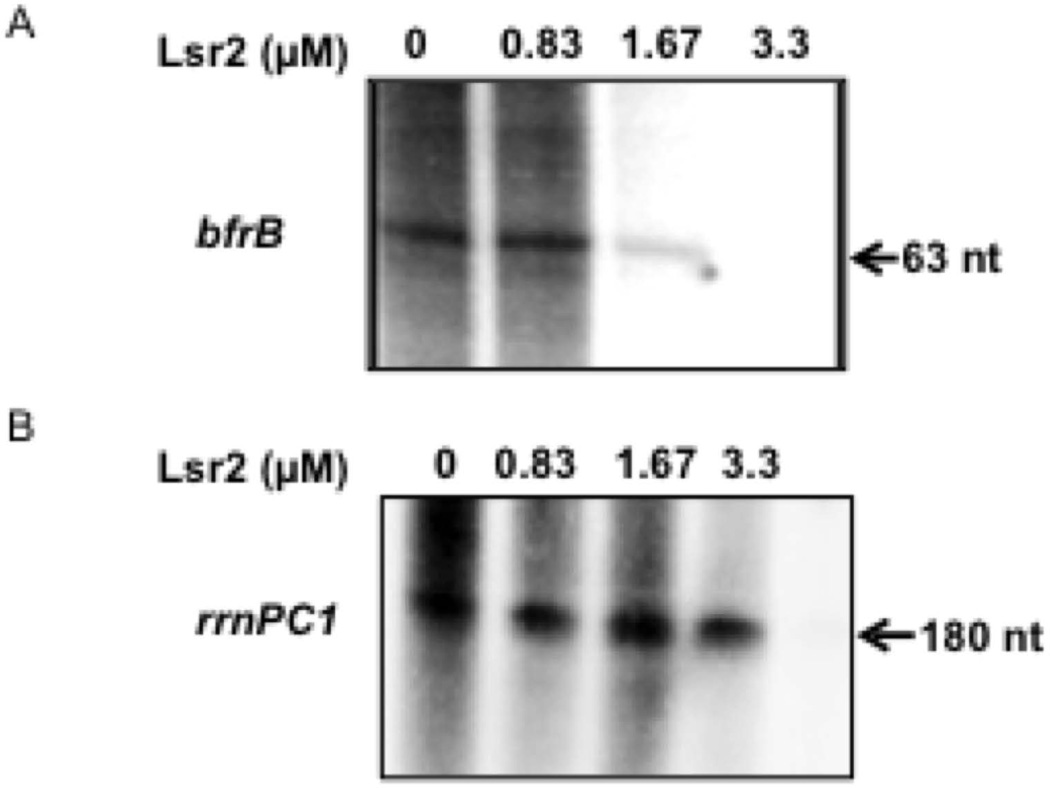
To determine whether Lsr2 represses bfrB directly or indirectly, we performed in vitro transcription assays in the presence of Lsr2. Addition of Lsr2 repressed transcription of PbfrB in vitro (Fig. 7A) at a concentration that did not inhibit transcription of a non-related promoter (Fig 7B).
Figure 7.
Lsr2 represses bfrB transcription in vitro. In vitro transcription of bfrB (A), and the non-related gene rrnPCl1 (B) by Msm RNAP was conducted in the absence or presence of increasing concentrations of purified Lsr2. Radiolabelled transcripts were separated by electrophoresis and detected by autoradiography as described in experimental procedures. Shown is one representative experiment repeated three times.
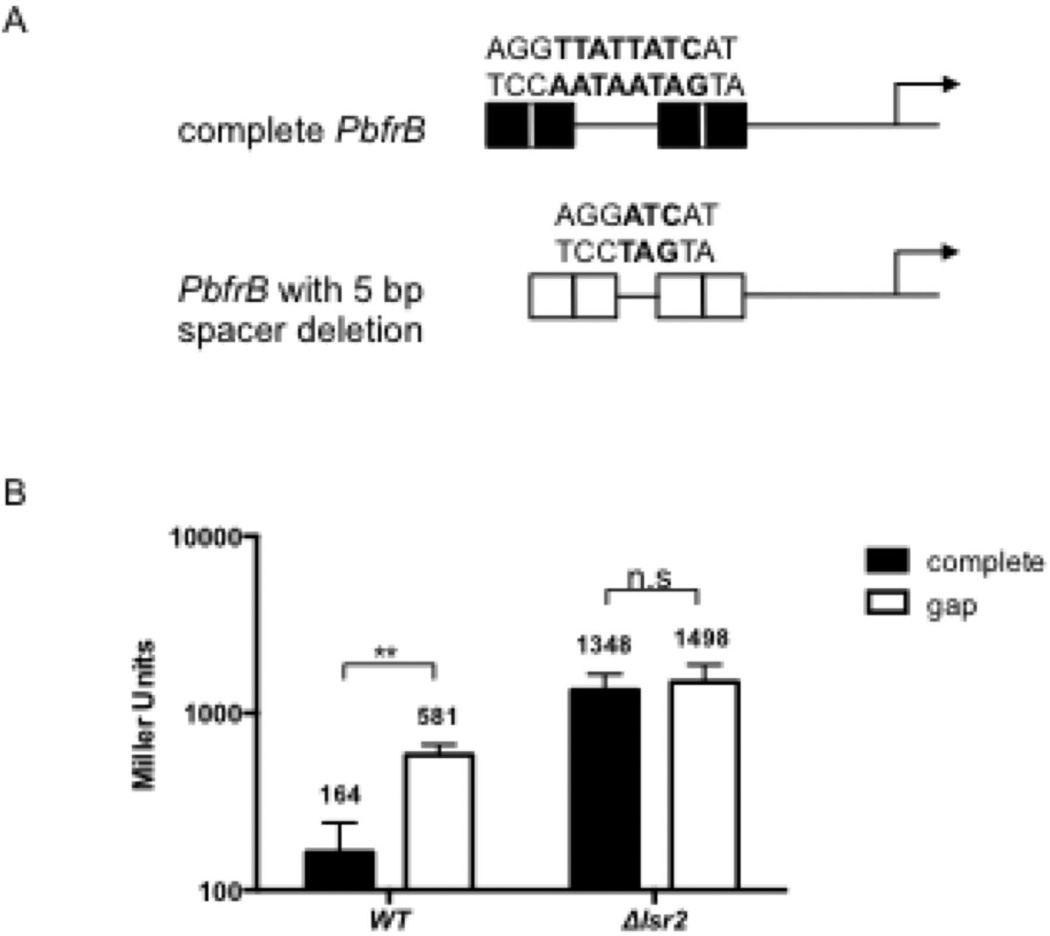
The derepression of PbfrB observed in the lsr2 mutant was mimicked in wild type Msm, although at a lower level, by the 5 bp deletion in the spacer between IB2 and IB3 (Fig 4). Hereafter the deleted promoter will be referred to as PbfrBΔ5. To investigate whether derepression of PbfrBΔ5 involved Lsr2, the level of expression of the PbfrBΔ5-lacZ fusion compared to the intact PbfrB-lacZ in wild type and Δlsr2 grown under repressive-low iron conditions was compared (Fig 8). PbfrBΔ5-lacZ was derepressed ~4 fold in the wild type strain compared to PbfrB and this effect was abolished in Δlsr2 indicating that repression of PbfrB is partially dependent on an intact IB2-IB3 spacer sequence and on Lsr2. Although in general, the upstream region of bfrB including the IdeR binding sites is AT-rich (57%) which makes it a possible binding region for Lsr2 the IB2-IB3 spacer sequence in particular includes an AT-rich, 8bp sequence AATAATAG identified previously in protein binding microarrays as an H-NS and Lsr2 preferred binding sequence (E-score 0.42 when maximum E-score was set at +0.5) (Gordon et al., 2011). The 5 bp deletion removed most of this Lsr2 binding site (Fig 8A). Thus, together the results suggest that Lsr2 binding to this sequence is needed for efficient PbfrB repression.
Figure 8.
An Lsr2 binding sequence located within the IdeR binding region is necessary for Lsr2-mediated repression of PbfrB. A. Schematic representation of the intact upstream sequence of bfrB harboring an Lsr2 binding sequence identified in protein binding microarrays (shown in bold), or a 5bp deletion (ΔTTATT) in the spacer region between IB2 and IB3 that destroys the Lsr2 binding site. lacZ fusions were generated with the intact and the deleted promoter and introduced into Msm WT and Δlsr2. B. β-galactosidase activity from the intact (filled bars) or deleted (open bars) promoter fusions in Msm (WT) and Δlsr2 strains grown under repressive (low iron) conditions. The deletion in the Lsr2 binding site resulted in (~4 fold) derepression of the promoter in the wild type Msm but did not lead to further derepression of the promoter in the Δlsr2 indicating that repression of PbfrB in low iron is partially dependent on an intact Lsr2 binding site present in the spacer sequence between IB2 and IB3. **, p<0.005, n.s. not significant.
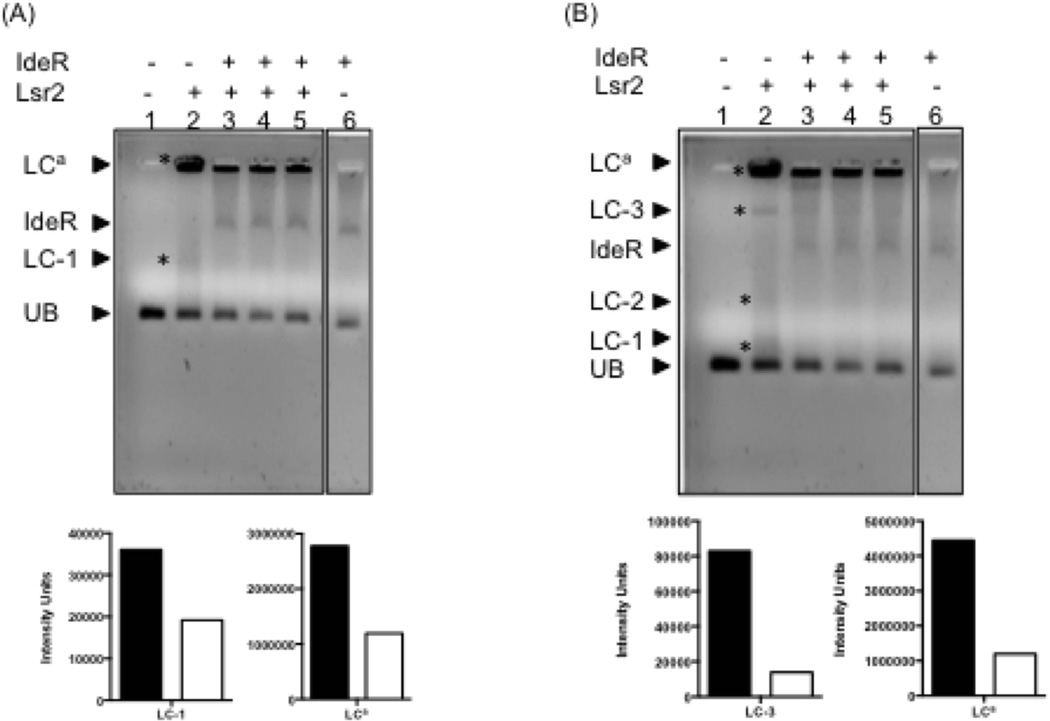
Having established that Lsr2 mediated bfrB repression involves binding to the region interacting with IdeR we next performed gel shift analysis that tested whether IdeR could displace Lsr2 from DNA. A DNA fragment encompassing the −155 to −34 region upstream of bfrB including the IdeR binding region was used. Individually both IdeR and Lsr2 bound to this DNA fragment and generated distinct protein-DNA complexes. Lsr2 binding generated large DNA protein aggregates that mostly remained in the well. IdeR added to pre-formed Lsr2-DNA complexes led to partial dissociation of these complexes. This was evidenced by the decrease in the amount of small Lsr2-DNA complexes and big aggregates coinciding with the formation of distinct IdeR-DNA complexes (Fig 9A). The same gel after a longer run (Fig 9B) shows higher molecular weight Lsr2-DNA complexes entering the gel. This is aided by adding a low concentration of IdeR. At higher concentrations IdeR leads to a further decrease in Lsr2-DNA aggregates including those that have entered the gel as well as those remaining on the well (Fig. 9B). This shows that IdeR can partially displace Lsr2 from DNA in vitro. This is consistent with the in vivo results, which indicate that IdeR relieves although does not eliminate Lsr2 control.
Figure 9.
IdeR can partially displace Lsr2 from PbfrB in vitro. A DNA fragment containing 121 bp upstream of bfrB including IB1-4 was incubated individually with Lsr2 (concentration 4.2 µM) (lane2), IdeR (concentration 6.7 µM) (lane 6) or first with Lsr2 followed by IdeR at increasing concentrations (4, 5 and 6.7 µM) (lanes 3–5), as described in experimental procedures. Protein-DNA complexes were resolved on a 0.9% TA agarose gel containing 1X SYBR green at 4°C. Digital images of the same gel were acquired after 45 or 60 min electrophoresis (panel A and B respectively ) to aid the visualization of low and high molecular weight Lr2-DNA complexes. The Lsr2 complexes in each gel are indicated by *. UB: PbfrB DNA probe (50 ng); IdeR: IdeR-PbfrB complex; LC-1: low molecular weight Lsr2-DNA complex; LC-2 and LC3: Intermediate weight Lsr2-DNA complexes; LCa: aggregates of Lsr2-DNA. The relative intensity of the bands corresponding to main Lsr2-DNA complexes is shown in the graphs under each gel image. The filled bars represent the intensity of the DNA shifts with Lsr2 alone (lane 2) and open bars represent the intensity of Lsr2 complexes in the presence of IdeR added at the highest concentration (lane 5).
IdeR reverses Lsr2 repression of bfrB
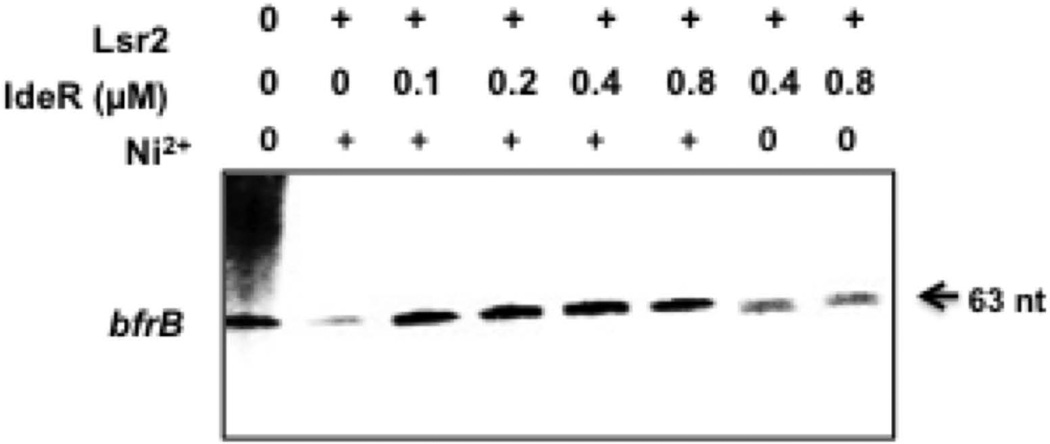
To validate the role of IdeR in relieving Lsr2 repression we tested whether IdeR could counteract Lsr2 mediated repression of bfrB in vitro. Purified Lsr2 was allowed to bind to the PbfrB DNA template and then increasing concentrations of metal activated or metal free IdeR were added, followed by σA-RNAp and ribonucleotides to initiate the reaction. At the lowest concentration added 0.1µM, IdeR relieved the repression of bfrB resulting from the addition of Lsr2 at 2.5 µM (Fig. 10). This result indicates that IdeR can indeed antagonize Lsr2 repression.
Figure 10.
IdeR reverses Lsr2 repression of PbfrB. The supercoiled bfrB template was first incubated with 2.5 µM of Lsr2 to allow DNA binding, then metal activated IdeR or apo-IdeR was added at the indicated concentrations and the reaction initiated by addition of RNAP and NTP mix. Radiolabelled transcripts were separated by electrophoresis and detected by autoradiography. Shown is one representative experiment repeated twice.
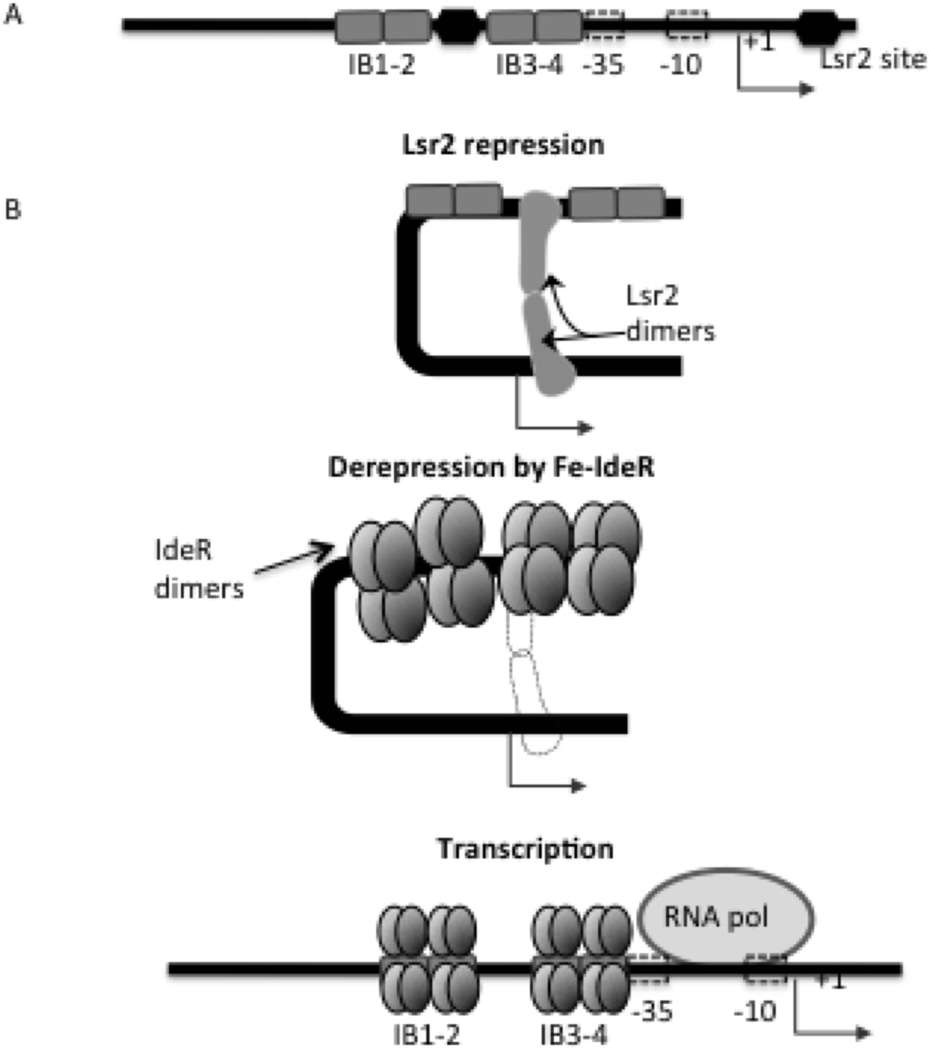
Based on the results we propose a regulatory model in which Lsr2 tightly represses bfrB. In high iron conditions IdeR binds Fe and as multiple dimers binds a DNA region that includes at least one confirmed Lsr2 binding site. IdeR partial displacement of Lsr2 relieves the DNA-distorting effects of Lsr2 allowing DNA accessibility for initiation of transcription by RNAP (Fig 11). In addition, as suggested by the results shown in Fig 5 and Fig S3, IdeR bound to the IB3–4 region may further promote PbfrB transcription independently of Lsr2.
Figure 11.
Model of IdeR-mediated induction of bfrB transcription. A. The regulatory region upstream of bfrB is shown locating the IdeR binding sites and the putative Lsr2 binding sites at −83 and at +24 bp (identified by ChiP). B. Lsr2 binding, oligomerization and DNA looping probably prevents transcription of bfrB. Under Fe sufficiency IdeR’s DNA binding activity is activated by Fe binding, multiple Fe-IdeR dimers bind to the iron boxes upstream bfrB partially displacing Lsr2 from the region thereby relieving bfrB repression. Additionally, IdeR binding to IB3-4 may also aid recruitment of the RNAP and promote transcription.
Discussion
To elucidate the mechanism of IdeR-mediated bfrB induction, we identified PbfrB sequence elements necessary for this induction and examined the effect of IdeR on bfrB transcription both in cells and in vitro. Like many other transcripts in Mtb, the bfrB transcript has no leader sequence (Cortes et al., 2013). Ongoing studies by several groups are expected to reveal the mechanism of ribosomal engagement and initiation of translation of this type of RNAs, which is currently unknown. IdeR directly interacted with tandem binding sequences upstream of PbfrB. Transcriptional reporter assays showed that the entire IdeR binding region was important for IdeR mediated induction of bfrB. Unexpectedly, while a transcriptional reporter controlled by the PbfrB upstream region was efficiently induced in a Fe-IdeR dependent manner in cells, in vitro bfrB transcription was independent of metal and enhanced only slightly by IdeR. These results indicated that although IdeR might activate transcription to a low level, transcriptional activation was not the main mechanism by which IdeR induced bfrB. We explored the possibility that in vivo, bfrB was subjected to transcriptional repression. Indeed, our subsequent analyses identified this repressor as the mycobacterial Lsr2 protein. Lsr2 is a homolog of the E. coli nucleoid-associated protein H-NS which plays a major role in silencing expression from sequences with high AT content (Gordon et al., 2010). Several lines of evidence, including analysis of expression of PbfrB reporter fusions, DNA binding assays, and in vitro transcription assays, demonstrated that Lsr2 directly repressed bfrB. Lsr2 mediated repression of bfrB was consistent with reported increases in bfrB transcript in a Msm and a Mtb lsr2 mutant (Colangeli et al., 2007), (Bartek et al., 2014). An Lsr2 binding site had been previously identified in the bfrB gene (+24 position) by ChIP studies (Gordon et al., 2010). Here, we identified a sequence previously reported as an H-NS/Lsr2 preferred binding sequence located in between two iron boxes that when disrupted led to partial derepression of PbfrB confirming the relevance of this site for Lsr2 mediated repression. In vitro transcription of bfrB in the presence of both Lsr2 and IdeR indicated that IdeR can relieve Lsr2-mediated transcriptional repression of bfrB. This finding and the ability of IdeR to partially displace Lsr2 bound to the bfrB upstream region, (Fig. 9) supports a model in which IdeR positively regulates bfrB by antagonizing Lsr2 repression. Interestingly, PbfrB was expressed at levels much higher in the lsr2 mutant than in the wild type in high iron, indicating that in vivo bfrB is subjected to Lsr2 repression even in high iron. This is consistent with IdeR relieving but not eliminating Lsr2 control. This tight Lsr2 mediated repression of bfrB might be critical for iron homeostasis. The cell must calibrate BfrB synthesis very precisely since excess apoferritin remaining after a spike in synthesis in response to high iron, might compete with metalloproteins for iron if suddenly, iron becomes limiting.
In the absence of Lsr2 an additional two to three fold induction of bfrB in high iron was observed. Reporter expression assays linked this induction to an intact IdeR binding site (IB3) (Fig. S3), suggesting that it may be mediated by IdeR. This is consistent with IdeR’s low activity as transcriptional activator in vitro (Fig 5). The DNase footprinting analysis did not show large hypersensitive regions indicative of DNA bending; however, IdeR might function as activator in other ways. For instance, the IB4 partially overlaps with the putative −35 element. Binding to DNA sites that overlap the −35 sequence is common to prokaryotic activators that interact with region 4 of σ70 (Busby and Ebright, 1994). In addition, preliminary studies indicate a direct interaction between IdeR and the beta subunit of RNA polymerase. Thus, IdeR may activate transcription by aiding to recruit the polymerase to the promoter. This possibility is currently under investigation. The impossibility of getting a surviving Msm ideR-lsr2 double mutant prevented us from conclusively determine the contribution of IdeR to Lsr2 independent induction of bfrB.
Lsr2 has been shown to form oligomers and multiple complexes with DNA, and to bridge distant DNA segments impeding transcription (Chen et al., 2008, Blair et al., 2010, Summers et al., 2012). Indeed multiple and high molecular weight complexes of Lsr2 and the bfrB DNA upstream region were visible in our gel shift assays. Accordingly, we propose that Lsr2 binding to at least two sites (−83 and +24) and subsequent oligomerization distorts DNA preventing initiation of transcription. When intracellular Fe levels increase, IdeR binds Fe, which activates its DNA binding activity. Multiple IdeR dimers bind to the iron boxes upstream of bfrB, partially displacing Lsr2 from this region and allowing transcription.
Interestingly, in E. coli a functional homolog of Lsr2, H-NS, also represses ferritin gene transcription, and this repression is reversed in high Fe by the ferric uptake regulator Fur, a functional homolog of IdeR (Nandal et al., 2010). Fur and IdeR belong to different protein families while Lsr2 is closely related to H-NS (Gordon et al., 2011). Thus, the same regulatory mechanism in E. coli and mycobacteria may be the result of convergent evolution.
Structural studies have shown that two IdeR dimers bind to one iron box on opposite sides of the DNA duplex (Wisedchaisri et al., 2004). It is unclear whether the presence of multiple iron boxes would have any effect on IdeR affinity. However, the existence of several IdeR binding sites upstream of IdeR-induced genes (bfrA and bfrB) is a main difference between IdeR repressed and induced genes, suggesting that binding of multiple IdeR dimers covering an extended region upstream of the promoter may be key to positive regulation by IdeR. IdeR positive regulation of bfrA was not dependent on Lsr2. Since IdeR directly interacts with the bfrA promoter (Gold et al., 2001) another repressor may be involved or IdeR may activate bfrA through interaction with RNAP.
The results shown here implicate Lsr2 as a new factor in Fe homeostasis and suggest that conditions that alter the concentration or activity of Lsr2 in the cell potentially could impact Fe metabolism via the effect on bfrB expression. For instance the upregulation of bfrB under low levels of oxygen, previously observed, correlates with down-regulation of lsr2 (Rustad et al., 2008). This and other conditions in which Lsr2 is decreased might lead to Fe-independent BfrB synthesis altering the normal pool of Fe in the cell. An Mtb lsr2 mutant is attenuated in mice (Bartek et al., 2014). It would be interesting to determine the contribution of derepressed bfrB expression to this phenotype. On the other hand, increased levels of Lsr2 under conditions like high temperature and nutrient starvation (Stewart et al., 2002, Betts et al., 2002) in which lsr2 is upregulated may also impact bfrB induction by favoring Lsr2 over IdeR occupancy of the bfrB upstream region. Lsr2 protein levels were reported to increase in Mtb cultured in high iron (Wong, 1999) and Lsr2 is annotated in the TB data base (http://www.tbdb.org) as an iron regulated protein. However, we found no evidence of Fe-mediated regulation of lsr2 at the level of transcription (data not shown). Thus, the mechanism and implications of augmented Lsr2 in high iron remain unclear.
We previously reported that enhanced intracellular free Fe, as a result of deficient Fe storage in the absence of BfrB, not only affects virulence but also potentiates the bactericidal effects of several clinically relevant antibiotics (Pandey and Rodriguez, 2012). The results of this study unveiled critical events in the regulation of bfrB and a connection between Lsr2 and iron regulation that could illuminate the design of new antitubercular strategies that target iron homeostasis in Mtb.
Experimental procedures
Bacterial strains, media and growth conditions
Escherichia coli strains GM161 or XL-10 (Stratagene) were used for cloning and BL21(DE3) rosetta strain (Novagen) was used for protein overexpression. E. coli strains were grown in Luria-Bertani (LB) broth. Mycobacterium smegmatis (Msm) mc2155 was grown in Middlebrook 7H9 broth or 7H10 agar medium supplemented with 0.2% glycerol and 0.05% Tween 80. M. tuberculosis H37Rv was grown in the same medium supplemented with bovine serum albumin (BSA), 0.2% dextrose and 0.85% NaCl (ADN). Minimal medium (MM) was used to culture strains under Fe defined conditions. MM contains 0.5% w/v asparagine, 0.5% w/v KH2P04, 2% glycerol, 0.05% Tween-80 and 10% ADN. The pH was adjusted to 6.8. To lower the trace metal contamination, the medium was treated with Chelex-100 (BioRad) according to the manufacturer instructions. Chelex was removed by filtration and then the medium was supplemented with 0.5 mg L −1 ZnCl2, 0.1 mg L−1 MnS04, and 40 mg L−1 MgS04. This medium contains less than 2 µM residual Fe as determined by atomic absorption spectroscopy and constitute the LI medium used in the experiments. The HI medium was generated by supplementing LI MM with 50 µM FeCl3.
Where indicated, antibiotics were used at the following concentrations: Kanamycin (Kan) 20 µg ml−1, streptomycin (Strep) 20 µg ml−1 and spectinomycin (Spec) 75 µg ml−1.
RNA isolation and transcriptional start point mapping
M. tuberculosis H37Rv was grown in 7H9 to logarithmic phase. Cells were collected by centrifugation and the pellet resuspended in 1 ml TRI reagent (Molecular Research Center). Suspensions were mixed with zirconia beads (0.1 mm diameter; Biospec Products, OH), and disrupted by two 1min pulses in a Bead-beater. RNA was purified as described previously (Rodriguez et al., 2002) using RNeasy (Qiagen) according to the manufacturer instructions. The bfrB transcriptional start site was determined by primer extension analysis as previously described (Gold et al., 2001). Briefly, 60 µg of RNA was hybridized to 1.5 pmol of γ-32P-labelled reverse primer and cDNA synthesized using C. thermo reverse transcriptase (Roche) according to the manufacturer instructions. At the end of a 60 min reaction, the cDNA-RNA mixture was ethanol precipitated and washed with 70% ethanol. Samples were resuspended in water and formamide loading buffer and separated on a 6% TBE polyacrylamide gel containing 8 M urea. A 10 bp ladder was used as a molecular weight standard to determine the length of the primer extension product. The transcriptional start point was verified using primers SP1, SP2 and bfrBL566 (Table S1) whose 3’ ends were 20 to 60 bp apart. Additionally, the transcriptional start site was confirmed by 5’ rapid amplification of c-DNA ends (RACE) using 5’/3’ RACE kit (Roche) and following the manufacturer instructions. Briefly, 2 µg of total RNA extracted from Mtb cultured in high iron was incubated with 12.5 µM of SP1 primer, 200 µM dNTP mix and 25 U of reverse transcriptase (Roche). The reaction was incubated at 55°C for 60 min. The first cDNA strand was column purified and eluted in 50 µl of nuclease free water. The purified cDNA was subjected to addition of homopolymeric A- tail at the 3’ end using terminal transferase and dATP (Roche 5’ RACE kit). Using the cDNA with the poly A tail as a template, 12.5 µM of oligo dT- Anchor primer and SP2 (internal to SP1) primer a PCR reaction was set up. The PCR product was separated on an agarose gel, excised from the gel, purified and sequenced using bfrBSP2 primer.
Construction of lacZ fusions
To generate the PbfrB, PmbtB and PbfrA-lacZ, transcriptional fusions 433 upstream of mbtB, the region encompassing −386 to +65 of bfrB and −378 to +52 of bfrA were PCR amplified using Mtb genomic DNA as template and cloned in front of a promoter-less lacZ gene in the integrative plasmid pSM128 (Gold et al., 2001) to generate plasmids pSM641, pSM475, and pSM393 respectively (Table 1) which were introduced into Msm by electroporation and transformants selected with Strep and Spec.
Table 1.
Strains and Plasmids
| Strains | Specification | Source |
|---|---|---|
| Msm strains | ||
| SM3 | mc2155 ideR null mutant | (Dussurget et al., 1996) |
| SM197 | mc2155 lsr2 deletion mutant. | (Colangeli et al., 2007) |
| SM17 | mc2155 carrying pSM128 | (Rodriguez et al., 1998) |
| SM154 | mc2155 carrying pSM393 | (Gold et al., 2001) |
| SM160 | mc2155 harbouring pSM475 | This study |
| SM163 | SM3 harbouring pSM475 | This study |
| SM173 | mc2155 harbouring pSM519 | This study |
| SM176 | mc2155 harbouring pSM526 | This study |
| SM177 | mc2155 harbouring pSM527 | This study |
| SM178 | mc2155 harbouring pSM529 | This study |
| SM193 | mc2155 harbouring pSM641 | This study |
| SM224 | SM197 harbouring pSM128 | This study |
| SM225 | SM197 harbouring pSM475 | This study |
| SM228 | SM225 transformed with pSM895 | This study |
| SM230 | SM197 harbouring pSM641 | This study |
| SM232 | SM197 harbouring pSM393 | This study |
| SM234 | SM197 harbouring pSM526 | This study |
| SM244 | SM197 harbouring pSM519 | This study |
| SM245 | SM197 harbouring pSM527 | This study |
| SM246 | SM197 harbouring pSM529 | This study |
| Plasmids | ||
| pSM128 | Mycobacterial integrative vector containing a promoter-less lacZ | (Rodriguez et al., 1998) |
| pSM207 | PSL1180 containing a T4 and a SinR T2T3 terminator flanking a unique Pst1 site | This study |
| pSM393 | transcriptional fusion of PbfrA lacZ in pSM128 | This study |
| pSM475 | transcriptional fusion of PbfrB lacZ in pSM128 | This study |
| pSM519 | pSM475 with IB3 mutated | This study |
| pSM526 | pSM475 with IB1 and IB2 deleted | This study |
| pSM527 | pSM475 with 5 bp deletion in the sequence between IB2 and IB3 | This study |
| pSM529 | pSM475 with IB1 deleted | This study |
| pSM641 | transcriptional fusion of PmbtB lacZ in pSM128 | This study |
| pMV261 | expression vector | (Stover et al., 1991) |
| pSM895 | lsr2 cloned into pMV261. Δlsr2 complementing plasmid | This study |
| pSM669 | pET30 derivative-Lsr2 expression plasmid,. | (Colangeli et al., 2007) |
| pSM918 | pET28TEV derivative-IdeR expression plasmid | This study |
| pSM495 | 200 bp upstream of Mtb mbtB and 250 bp downstream of the start codon cloned into Zero blunt Topo (Invitrogen). | This study |
| pSM749 | pSM207 carrying 387 bp upstream and 65 bp of Mtb bfrB orf | This study |
| pSM750 | pSM207 carrying 387 bp upstream and 65 bp of Mtb bfrB orf with IB1 deleted. | This study |
| pARN104 | pUC18 containing the Mtb rRNA promoter PCl1 and 180 bp downstream DNA followed by a transcriptional terminator from Rv1324 | Tare et al., 2012 |
Mutagenesis of IdeR binding sites
pSM475 carrying the wild type IdeR binding region was subjected to site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) following manufacturer instructions. Introduction of the desired changes was confirmed by DNA sequencing. The following mutant constructs were generated: pSM519 has sequence mutations in the IB3, pSM526 has IB1 and IB2 deleted, pSM527 has a 5 bp deletion of the spacer sequence between IB2 and IB3. pSM529 has deletion of IB1.
Complementation of the lsr2 mutant
The Msm lsr2 gene and its own promoter were PCR amplified using lsr2 Fp and lsr2 Rp primers and the PCR product was purified and cloned into BamHI site of pMV261 by infusion (Clonetech) following the manufacturer instructions. The resulting plasmid, pSM895, was verified by DNA sequencing and electroporated into Δlsr2 harboring the PbfrB-lacZ fusion to generate the complemented strain (SE225) (Table 1). Transformants were selected on 7H10-Kan.
IdeR purification
The DNA sequence encoding IdeR was PCR amplified using as template Mtb genomic DNA and the primers IdeRFp and IdeRRp (Table S2). The PCR product was purified and cloned at the NdeI-HindIII sites of pET28TEV (Ryndak et al., 2010) to create pSM918 having an in-frame fusion of IdeR with a His tag at the amino terminus that is cleavable by the TEV protease. E. coli BL21(DE3) rosetta harboring pSM918 was grown overnight in 50 ml of LB containing 50 µg/ml Kan at 37°C. Then 10 ml of this culture were inoculated into 1 L of LB-kan and cells were grown at 37°C with agitation (230 rpm) until reaching an optical density at 600 nm (OD600) of 0.6. Isopropyl-β-D-1-thiogalactopyranoside (IPTG) was added to the culture at a final concentration 0.5 mM and cells were allowed to continue growing for another 3.5 h. The cultures were centrifuged at 3,500 × g and the cell pellet was resuspended in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10% v/v glycerol (suspension buffer) supplemented with a protease inhibitor cocktail (Roche). Cells were lysed by sonication using a macro probe set at 50% duty cycle for 7 min at 4°C. The cell lysate was centrifuged at 20,000 × g for 30 min at 4°C. The supernatant was filtered through a 0.4 µm filter and loaded onto a His-Trap column (Amersham Biosiences). Unbound material was removed by washing with suspension buffer containing 50 mM imidazole. His-IdeR was eluted with a linear gradient of 50 to 400 mM imidazole. Eluted fractions were analyzed by SDS-PAGE and coomassie blue staining. The fractions containing IdeR which migrates as a 25 KDa (the calculated molecular mass of His-IdeR is 25,232) were pooled and dialyzed against the suspension buffer containing 20 mM EDTA and concentrated by filtration in a 10kDa cut-off Sartorius filter (Fig S2).
Lsr-2 purification
Mtb lsr2 was PCR amplified and cloned between the NdeI-HindIII sites of pET30 to create pSM669 having an in-frame fusion of Lsr2 with a His tag at the amino terminus. E. coli BL21 (DE3) rosetta strain containing pSM669 were pre cultured in 50 ml LB kan at 37°C overnight. The pre-culture was diluted (1:100) in fresh 1 L of LB-kan and cultured at 37°C at 230 rpm to OD600 0.8–1.0. The culture was then centrifuged at 3,500 × g and the cell pellet resuspended in suspension buffer. Cells were lysed in the presence of a protease inhibitor cocktail (Roche) in a sonicator using a macro probe set at 50% duty cycle for 7 min at 4°C. The cell lysate was centrifuged at 20,000 × g for 30 min at 4°C and the supernatant collected. The supernatant was filtered through a 0.4 µm filter and loaded onto a His-Trap column (Amersham Biosiences). Unbound material was removed by washing the column with suspension buffer containing 50 mM imidazole. His-Lsr2 was eluted from the column with a linear gradient of 50 to 400 mM imidazole. The eluted fractions were analyzed by SDS-PAGE and coomassie blue staining. Those fractions containing His-Lsr2 which migrates as a 12 KDa protein (the calculated molecular mass of Lsr2-His is 12,232) were pooled and dialyzed against suspension buffer to remove imidazole. The dialyzed protein was further purified by affinity to Talon matrix (Clonetech) (Fig S2).
β-galactosidase assays
Msm strains harboring lacZ fusions were Fe deprived by growing them in MM to early stationary phase (OD600 0.8–1.0). Confluent cultures were diluted 1:10 in MM (low Fe) or MM supplemented with 50 µM FeCl3 (high Fe), grown to logarithmic phase, collected by centrifugation, washed with phosphate buffered saline (PBS) and re-suspended in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, pH 7.0). Cells were lysed by sonication and β-galactosidase activity in the extracts was measured as previously (Pardee et al., 1959). Units of β-galactosidase were calculated by the following formula: 1,000X the OD420 per milligram of protein per min (Miller Units). The protein concentration in the bacterial extracts was measured by BioRad Dc protein assay (BioRad). Msm transformed with the vector plasmid pSM128 were used as negative control. Statistical significance was calculated using Student’s two tailed unpaired ‘t’-test.
Electrophoretic motility gel shift assays (EMSA)
DNA sequences encompassing −386 to +65 of bfrB wild type were excised from pSM475 and used as probe for EMSA. As a non-specific DNA control, a 402 bp fragment containing rv1397c was used. Binding reactions included 50 ng (11.6 nM) of DNA, purified IdeR, binding buffer [20 mM Tris-HCl (pH 8.0), 1mM DTT, 50 mM KCl, 5 mM MgCl2, 0.05 mg ml−1 BSA and 10% glycerol] in a final volume of 15 µl. When indicated 200 µM NiSO4 was added as Fe surrogate. The reactions were incubated 30 min at room temperature. At the end of the incubation the samples were mixed with 6X loading buffer containing 10 mM Tris-HCl (pH 7.6), 0.03% bromophenol blue dye and 60% v/v glycerol and loaded on a 6 % polyacrylamide gel containing 40 mM Tris-acetate (pH 8.0). The gel was run at 110 V at 4°C and then stained with SYBR Green [1X in Tris-acetate buffer (TAB)] (Invitrogen). The digital images were acquired using Molecular Imager GelDoc XR+ (Biorad) instrument.
EMSA conducted to demonstrate IdeR displacement of Lsr2 were conducted as follows: A 121 bp upstream region of bfrB encompassing 155 to −34 bp Including Ider binding regions IB1 to IB4 was generated by PCR. The DNA (42 nM) was incubated with either Lsr2 or IdeR alone at indicted concentrations for 30 min in binding buffer. For competition reactions, the DNA was first incubated with Lsr2 for 30 min followed by addition of IdeR and additional 30 min incubation at room temperature. At the end of this period the samples were mixed with loading dye and the complexes were separated on a 0.9% agarose gel in TAB containing 1X SYBR green at 75 V and 4°C. The digital images were acquired and quantified using Biorad XR+ gel documentation system.
DNase footprinting analysis
Footprinting analysis were performed as previously (Gold et al., 2001). The oligonucleotides primers bfrB339 and bfrB523 (Table S2) corresponding to either the top or bottom strand of the bfrB promoter region were labeled with [γ−32P]-ATP using T4 polynucleotide kinase and then used to individually PCR amplify a ~200 bp fragment containing the IdeR binding sites in the presence of the non-labeled cognate oligonucleotide. The PCR products were purified from a 2% agarose gel using QIAquick Gel Extraction Kit (Qiagen). Binding reactions with IdeR were performed using the same conditions as in the gel retardation experiments in 20 µl with 100000 cpm of labeled fragment and purified IdeR. After incubation for 30 min at room temperature reaction volumes were adjusted to 100 µl with a Ca2+/ Mg2 solution (2.5 mM CaCl2 and 5 mM MgCl2 final concentrations). Then 0.15 U of DNase I (Promega) were added, and mixtures were incubated for 1 min at room temperature. Reactions were terminated by addition of 90 µl of the stop solution (200 mM NaCl, 20 mM EDTA, 1% SDS and 100 µg ml−1 yeast RNA). Samples were subsequently extracted with phenol-chloroform, ethanol precipitated and resuspended in a formamide containing gel loading dye and resolved by electrophoresis on a 6% TBE poly acrylamide-urea sequencing gel, dried and analyzed by autoradiography. Maxam-Gilbert A+G sequencing reactions were performed to identify protected regions.
In vitro transcription
For these experiments RNA polymerase (RNAp) with stoichiometric σA content and high promoter-specific-activity was purified from Msm over-expressing σA as previously described (China et al., 2012). Reactions were carried out using supercoiled plasmid bearing the wild type bfrB, mbtB or PrrnPCl1 templates flanked by a transcriptional terminator (pSM749, pSM495 and pARN104 respectively). The reaction mixtures contained 300 ng of template DNA that were incubated at 37°C with different concentrations of Mtb IdeR or Lsr2, as specified, in the presence or absence of NiSO4. Transcription reactions were carried out in 50 mM Tris-HCl (pH 8.0), 3 mM magnesium acetate, 100 µM EDTA, 100 µM DTT, 30 mM KCl, 50 µg ml−1 BSA and 5% glycerol. RNA polymerase, (200 nM) was added and the reactions were incubated for 10 min at 37°C followed by addition of 100 µM NTPs, 1 µCi αP32 UTP. After additional 15 min of incubation at 37°C, the reactions were terminated by the addition of stop buffer (95% formamide, 0.025% w/v bromophenol blue, 0.025% w/v xylene cyanol, 5 mM EDTA, 0.025% SDS and 8 M urea) and inactivation at 90°C for 5 min. The reactions were snap chilled on ice for 5 min and resolved in a 12% urea-polyacrylamide gel and analyzed by autoradiography.
Supplementary Material
Acknowledgments
We thank Issar Smith for helpful discussions and critical reading of the manuscript. This work was supported by NIH grant AI044856 (GMR).
References
- Andrews SC. Advances in Microbial Physiology. Academic Press; 1998. Iron Storage in Bacteria; pp. 282–351. [DOI] [PubMed] [Google Scholar]
- Bartek IL, woolhiser LK, Baughn AD, Barasaba RJ, Jacobs WR, Jr, Lenaerts AJ, Voskuil MI. Mycobacterium tuberculosis Lsr-2 is a global transcriptional regulator required for adaptation to changing oxygen levels and virulence. mBio. 2014;5:e0116–e0114. doi: 10.1128/mBio.01106-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts J, Lukey P, Robb L, McAdam R, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis perssistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- Blair R, Gordon S, Li Y, Wang L, Sintsova A, van Bakel H, Tian S, Navarre WW, Xia B, Liu J. Lsr-2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis . PNAS. 2010;107:5154–5159. doi: 10.1073/pnas.0913551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby S, Ebright R. Promoter Structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Chen JM, Ren H, Shaw JE, Wang YJ, Li M, Leung AS, Tran V, Berbenetz NM, Kocincova D, Yip CM, Reyrat J-M, Liu J. Lsr-2 of Mycobacterium tuberculosis is a DNA-bridging protein. Nucleic Acid Research. 2008;36:2123–2135. doi: 10.1093/nar/gkm1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- China A, Mishra S, Tare P, Nagaraja V. inhibition of Mycobacterium tuberculosis RNA polymerase by binding of a Gre factor homolog to the secondary channel. J Bacteriol. 2012;194:1009–1017. doi: 10.1128/JB.06128-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangeli R, Helb d, Vilcheze C, Hazbon M, Lee C, Safi H, Sayers B, Sardone I, Jones M, Fleischmann R, Paterson S, Jacobs WR, Jr, Alland D. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr-2 in M. tuberculosis . PloS Phatog. 2007;3:e87. doi: 10.1371/journal.ppat.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes T, Schubert O, Rose G, Arnvig K, Comas I, Aebersold R, Young DC. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis . Cell Reports. 2013;5:1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussurget O, Rodriguez GM, Smith I. An ideR mutant of Mycobacterium smegmatis has a derepressed siderophore production and an altered oxidative-stress response. Mol Microbiol. 1996;22:535–544. doi: 10.1046/j.1365-2958.1996.1461511.x. [DOI] [PubMed] [Google Scholar]
- Gobin J, Moore CH, Reeve JR, Jr, Wong DK, Gibson BW, Horwitz MA. Iron acquisition by Mycobacterium tuberculosis : Isolation and characterization of a family of iron-binding exochelins. Proc. Natl. Acad. Sci. USA. 1995;92:5189–5193. doi: 10.1073/pnas.92.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Rodriguez GM, Marras MP, Pentecost M, Smith I. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Molecular Microbiology. 2001;42:851–865. doi: 10.1046/j.1365-2958.2001.02684.x. [DOI] [PubMed] [Google Scholar]
- Gomez M, Smith I. Determinants of Mycobacterial gene expression. In: Hatfull GF, Jacobs WRJ, editors. Molecular Genetics of Mycobacteria. Washington, DC: American Society for Microbiology Press; 2000. pp. 111–129. [Google Scholar]
- Gordon BRG, Li Y, Cote A, Weirauch MT, Ding P, hughes T, Navarre WW, xia B, Liu J. Stuctural basis for recognition of AT-rich DNA by unrelated xenogenic silencing proteins. PNAS. 2011;108:10690–10695. doi: 10.1073/pnas.1102544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon BRG, Li Y, Wang L, Sintsova A, van Bakel H, Tian S, Navarre WW, Xia B, Liu J. Lsr2 is a nucleoid-associated protein that targets AT-rich seqences and virulence genes in Mycobacterium tuberculosis . PNAS. 2010;107:5154–5159. doi: 10.1073/pnas.0913551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Gupta RK, Khare G, Salunke DM, Tyagi AK. Crystal Structure of BfrA from Mycobacterium tuberculosis: Incorporation of Selenomethionine Results in cleavage and demetallation of Haem. PLOSone. 2009:4. doi: 10.1371/journal.pone.0008028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free iron levels. Pro Natl Acad Sci USA. 1996;193:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare G, Gupta V, Nangpal P, Gupta RK, Sauter NK, Tyagi AK. Ferritin structure from Mycobacterium tuberculosis: Comparative study with Homologues Identifies Extended C-Terminus involved in ferroxidase activity. PLOSone. 2011:6. doi: 10.1371/journal.pone.0018570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandal A, Huggins CCO, Rodríguez-Quiňones F, Quail MA, Guest JR, Andrews SC. Inductin of the Ferritin gene [fntA] of Escherichia coli by Fe+2-Fur is mediated by reversal of H-NS silencing and is RhyB independent. Mol Microbiol. 2010;75:637–657. doi: 10.1111/j.1365-2958.2009.06977.x. [DOI] [PubMed] [Google Scholar]
- Pandey R, Rodriguez GM. A Ferritin Mutant of Mycobacterium tuberculosis is Highly suceptible to Killing by Antibiotics and Is Unable to Establish a chronic Infection in Mice. Infection and Immunity. 2012;80:3650–3659. doi: 10.1128/IAI.00229-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Rodriguez GM. IdeR is required for iron homeostasis and virulence in Mycobacterium tuberculosis. Mol Microbiol. 2013;91:98–109. doi: 10.1111/mmi.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee AP, F J, J M. The genetic control and cytoplasmic expression of “inductibility” in the synthesis of B-galactosidase and tryptophanase induction in E. coli. J. Mol. Biol. 1959;1:3331–3342. [Google Scholar]
- Reddy PV, Puri RV, Khera A, TyGI AK. Iron storage proteins are essential for the survival and pathogenesis of Mycobactierium tuberculosis in the THP-1 macrophages and guinea pig model of infection. J. of Bacteriology. 2011;194:567–575. doi: 10.1128/JB.05553-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez GM. Control of iron metabolism in Mycobacterium tuberculosis . TRENDS in Microbiology. 2006;14:320–327. doi: 10.1016/j.tim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Rodriguez GM, Gold B, Gomez M, Dussurget O, Smith I. Identification and characterization of two divergently transcribed iron regulated genes in Mycobacterium tuberculosis . Tuber Lung Dis. 1998;79:287–298. doi: 10.1054/tuld.1999.0219. [DOI] [PubMed] [Google Scholar]
- Rodriguez GM, Smith I. Identification of an ABC Transporter Required for Iron Acquisition and Virulence in Mycobacterium tuberculosis . J Bacteriol. 2006;188:424–430. doi: 10.1128/JB.188.2.424-430.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, An essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad TR, Harrell MI, Liao R, Sherman DR. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS ONE. 2008;3:e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryndak M, Wang S, I S, Rodriguez GM. The Mycobacterium tuberculosis high-affinity iron importer, IrtA, contains an FAD-binding domain. J. Bacteriology. 2010;192:861–869. doi: 10.1128/JB.00223-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow GA. Mycobactins: iron chelating growth factors from mycobacteria. Bacteriol. Rev. 1970;34:99–125. doi: 10.1128/br.34.2.99-125.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GR, Wernisch L, Stabler R, Mangan JA, Hinds J, Laing K, Young DB, Butcher PD. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology. 2002;148:3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, et al. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- Summers EL, Meindl K, Usón I, Mitra AK, Radjainia M, Colangeli R, Alland D, Arcus VL. The structure of the oligomirization domain of Lsr-2 from Mycobacterium tuberculosis reveals a mechanism for chromosome organization and protection. PLos ONE. 2012;7:e38542. doi: 10.1371/journal.pone.0038542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisedchaisri G, Chou CJ, Wu M, Roach C, Rice AE, Holmes RK, Beeson C, Hol WG. Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis. Biochemistry. 2007;46:436–447. doi: 10.1021/bi0609826. [DOI] [PubMed] [Google Scholar]
- Wisedchaisri G, Holmes RK, Hol WG. Crystal structure of an IdeR-DNA complex reveals a conformational change in activated IdeR for base-specific interactions. J Mol Biol. 2004;342:1155–1169. doi: 10.1016/j.jmb.2004.07.083. [DOI] [PubMed] [Google Scholar]
- Wong D, Lee BY, Horwitz MA, Gibson B. Identification of Fur, Aconitase and other Proteins Expressed by Mycobacterium tuberculosis under Conditions of Low and High Concentrations of Iron by Combined Two-Dimentional Gel Electrophoresis and Mass Spectrometry. Infect and Immun. 1999;67:327–336. doi: 10.1128/iai.67.1.327-336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.