Abstract
Retroviral capsid cores are proteinaceous containers that self-assemble to encase the viral genome and a handful of proteins that promote infection. Their function is to protect and aid in the delivery of viral genes to the host’s nucleus, and, in many cases, infection pathways are influenced by capsid-cellular interactions. From a mathematical perspective, capsid cores are polyhedral-cages and, as such, follow well-defined geometric rules. However, marked morphological differences in shapes exist, depending on virus type. Given capsid’s specific roles in the viral life cycle, the availability of detailed molecular structures, particularly at assembly interfaces, opens novel avenues for targeted drug development against these pathogens. Here, we summarize recent advances in the structure and understanding of retroviral capsid, with particular emphasis on assemblies and the capsid cores.
The functional role the retroviral capsid core
The majority of enveloped viruses, including those in the Hepadnaviridae, Retroviridae, and Flaviviridae families, contain spherical or ovoid protein shells, the capsid core, that enclose the viral genome [1]. While these viruses exhibit significantly different mechanisms of replication, they all face similar problems during particle assembly. Retroviruses exhibit diverse assembly mechanisms. B- and D-type assembly viruses, like mouse mammary tumor virus (MMTV) and Mason-Pfizer monkey virus (M-MPV), form immature particles in the cytoplasm, while C-type assembly viruses, like bovine leukemia virus (BLV), Rous sarcoma virus (RSV) and human immunodeficiency virus (HIV), assemble at the cytoplasmic face of the plasma membrane and bud directly from the cell surface [2]. The determinants for assembly are encoded in the respective Group-specific antigen protein (Gag) polyproteins. Gag is the only viral protein required for assembly and generation of immature virus particles [3]. In C-type assembly, myristoylated Gag precursor proteins associate with viral RNA in a patch at the inner leaflet of the plasma membrane of the host cell, growing laterally until budding off [4]. As a consequence, the immature virus particle contains a cell-derived lipid envelope that is wrapped around the nascent viral capsid core [4,5].
In immature virus particles, unprocessed Gag is radially arranged, with the matrix protein (MA) at one end, anchored with its N-terminal myristoyl tail to the inner membrane surface, and nucleocapsid protein (NC) at the other end, interacting with the viral RNA, each playing independent roles. During viral maturation, Gag is cleaved by the viral protease (PR), generating three major protein cleavage products, MA, capsid (CA) and NC, which are common to all retroviruses, while different numbers of spacer-peptides and subdomains exist in specific viruses. Proteolytic processing by PR results in a dramatic rearrangement of Gag, with only MA remaining associated with the viral lipid envelope, while CA and NC condense around the viral genome. NC forms a ribonucleoprotein (RNP) complex inside the virion, and CA assembles into a lattice that ultimately generates the spherical or ovoid protein shell of the capsid core.
The capsid core plays numerous roles during the viral life cycle, including interfering with innate immune sensing [6-10], regulating reverse transcription [11-13], and controlling the nuclear import pathway [8,9,11,13-15]. A large number of these functions are highly dependent on interactions between the capsid core and cellular host factors [11,14-16]. For instance, some proteins stabilize the capsid core (Cyclophilin A (CypA)) [17], others disrupt it (TRIpartite Motif or TRIM proteins) [18], and several assist in nuclear import (Transportin 3, Nucleoporin 153, Nucleoporin 358 (Nup358), and the Cleavage and polyadenylation specific factor 6) [16,19,20]. Furthermore, the capsid core also engages with the cytoskeleton of the cell [21], using dynein and kinesin motors for trafficking along microtubules to the nucleus [5]. Given the importance of the capsid core for infection and the life cycle of retroviruses, the last two decades have witnessed enormous efforts focused on determining atomic structures of CA, alone, in assemblies, and in complexes with cellular interactors. In this review, we present the current knowledge regarding the structure of the retroviral capsid core and its relationship to biological function.
The common building block of immature and mature capsid assemblies - the CA protein
All retroviruses, irrespective of whether they assemble in the cytoplasm or at the cellular membrane, initially form immature viral particles [3], which mature by proteolytic cleavage of Gag, generating polymorphic conical or spherical capsid cores [22]. Despite this heterogeneity in size and morphology, the unit building block, the capsid protein, CA, is essentially identical among retroviruses [23]. It contains about 230 amino acids and is composed of two α-helical domains that are connected by a flexible linker (Figure 1A) [24]. The N-terminal domain (NTD) [25] and the C-terminal domain (CTD) [26,27] can exist as independently folded entities in solution [28]. The NTD is monomeric, while the CTD and the full-length CA protein exist either as a monomer (human T-cell lymphotropic virus type 1 HTLV-1, RSV) [29,30] or in a concentration-dependent monomer-dimer equilibrium (HIV type 1, HIV-1), modulated by experimental conditions [31,32].
Figure 1.
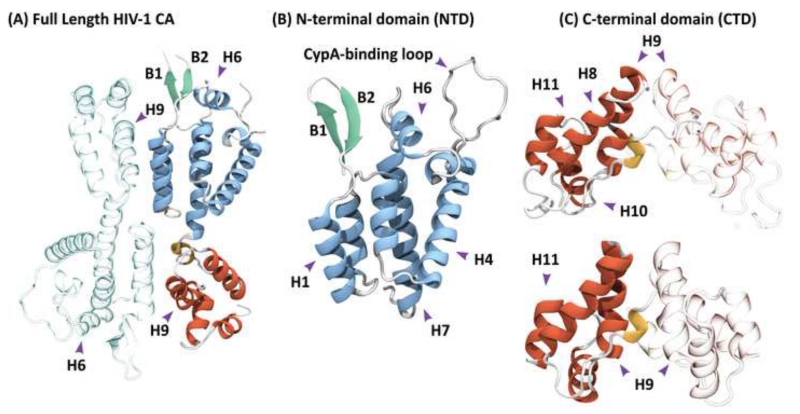
Structure of the full-length HIV-1 capsid (CA) protein. (A) The monomer is composed of two independent domains, which are arranged as an anti-parallel dimer in the X-ray structure (PDB accession code 3NTE) [24]. (B) Crystal structure of the N-terminal domain (PDB accession code 1GWP), comprising residues 1 to 146 arranged in seven α-helices. (C) Solution NMR (top) and X-ray (bottom) structures of the C-terminal domain (PDB accession codes 2KOD and 1A43), comprising residues 144 to 231 and 146 to 231, respectively. Both structures contain four alpha helices and a short 310 helix at the N-terminus. Residues 153-172 are commonly referred to as the major homology region (MHR). One of the monomeric units in the dimers (A) and (C) is shown in grey, with the other monomer in color. The NTD and CTD alpha helices are colored blue and red, respectively, and the beta-hairpin is colored green; in (A) and (C) the 310 helix is colored yellow. In (B) the location of the CypA-binding loop is indicated.
The NTD, comprising ca. 150 amino acids, consists of six or seven helices (Figure 1B) with a beta-hairpin at the N-terminus. In lentiviruses, the loop following helix 4 constitutes the binding interface for CypA, a human protein involved in immunological responses. The CypA-binding loop mediates interactions between the CA protein and this important host factor in a cell-type specific manner [9,33]. HIV-1 is unique among primate lentiviruses in its ability to interact with CypA and, therefore, is uniquely sensitive to cyclosporine A, compared to human immunodeficiency virus type 2 (HIV-2) or simian immunodeficiency virus of macaques (SIVmac) [34]. Intriguingly, in rhesus macaques, the cypA sequence has been incorporated by retrotransposition into the trim gene. The superfamily of TRIM proteins are associated with the innate immune response. TRIM5α restricts retroviral infection early after viral entry into the target cell [18,35], recognizing and binding to the surface of capsid assemblies using a SPRY domain [36]. Similarly, TRIMCyp recognizes the retroviral capsid core by using a CypA domain, rather than the SPRY domain [9]. The CypA-binding loop of CA is conserved in lentiviruses and is also present in feline immunodeficiency virus (FIV) [37] and endogenous lentiviruses in rabbits (rabbit endogenous lentivirus type K (RELIK)) and lemurs (prosimian immunodeficiency virus (PSIV)). Since the structure of the CA NTD-CypA complex is remarkably similar for RELIK/PSIV and HIV-1 [35], a conserved function of CypA in lentiviral infection is likely. Indeed, the equivalent loop in RSV’s CA protein is essential for infectivity [38], although no direct evidence of interactions between this loop and a CypA-type host factor has been shown.
The CTD contains four helices, numbered eight to eleven, as well as a 310 helix (Figure 1C). The region around residues ~150-170 is known as the major homology region (MHR) and is highly conserved among all retroviruses [1] as well as the Ty3 retrotransposon [39]. Intriguingly, in higher vertebrates, the fold of an endogenous synaptic protein, Arc, is remarkably similar to the retroviral CTD and has been proposed to be a remnant from a previous retroviral infection [40]. Together, helix 9 and the 310 helix constitute the core of the CTD-CTD dimerization interface (Figure 1C and Figure 2), irrespective of whether in an individual CA protein dimer or a multimeric assembly (see below). Located at the center of the dimer interface are hydrophobic amino acids, such as L151, W184 and M185 in HIV-1, W133, I168 and L172 in BLV, and W153, L180 and V188 in RSV. The variability in the relative arrangement of monomers at the CTD dimeric interface is remarkable, and several distinctly different conformations are seen for the HIV-1 and RSV dimer interfaces (Figure 2A, 2B, 2E and 2F). For both CA proteins, dimerization affinity is pH dependent, and the quaternary structure of the dimer is governed by the protonation state of E175 in HIV-1 [31] and by D191 in RSV [30] (Figure 2E, 2F).
Figure 2.
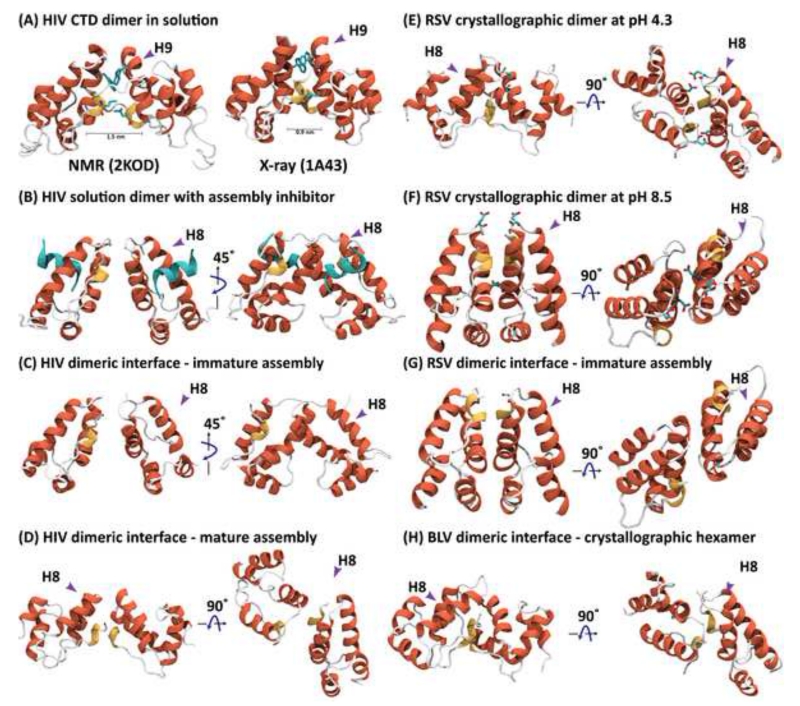
Structures of several individual retroviral CTD dimers (A, B, E, and F) and the CTD dimeric arrangement in CA assemblies (C, D, and G) or crystallographic hexamer (H). (A) NMR solution (PDB accession code 2KOD) [27] and crystal structure (PDB accession code 1A43) of the HIV-1 CTD [26]. (B) Crystal structure of the HIV-1 CTD dimer in complex with the inhibitory CAI peptide (PDB accession code 2BUO) [55]. (C) HIV CTD dimer arrangement in immature virus-like particles, obtained from single particle cryo-EM, using sub-tomogram averaging (PDB accession code 4USN) [47]. (D) HIV CTD arrangement in tubular assemblies of CA protein seen by cryo-EM (PDB accession code 3J34) [66]. (E) Crystal structure of the RSV CTD at pH 4.3 (PDB accession code 3G21) [56]. (F) Crystal structure of the RSV CTD at pH 8.4 (PDB accession code 3G1I) [56]. (G) RSV CTD dimer arrangement in immature virus-like particles, obtained from single particle cryo-EM, using sub-tomogram averaging (PDB accession code 5A9E) [49]. (H) CTD dimer interface in the X-ray structure of the BLV hexamer (PDB accession code 4PH0) [45]. For each structure, two views, rotated by 90°, are provided. Alpha helixes are colored in red and 310 helices in yellow. In (A) the W185 and L151 side chains and in (E) and (F) the D179 and D191 side chains are shown in stick representation. In (B), the inhibitor peptide is depicted in cyan.
Although structures and conformational details of the CA protein and its individual domains are informative, higher-order oligomeric arrangements constitute the units of extended CA lattices or helical assemblies. As we discuss below, the quaternary structures of the isolated CTD dimers and the relative orientations of the monomeric units in these dimers (Figures 2 A, B, E and F) exhibit striking similarities to their counterparts observed in assembled and crystallographic lattices (Figures 2 C, D, G and H), highlighting their biological relevance. In the following sections, we describe how the CA protein is put together in these oligomers.
Hexamer and Pentamers of the CA protein
Retroviral capsid cores exhibit different overall shapes (spherical, spherocylindrical, or conical), and their structures are created according to common design principles of convex polyhedral cages [41]. Like icosahedral viral capsids, all retroviral capsid cores are built from hexameric lattices, and the closure of the shell is accomplished by incorporating twelve pentamers. The pleiotropic shapes of capsid cores are a consequence of the distribution of the pentamers in the shells, simply governed by mathematical considerations: spherical capsid cores have twelve uniformly distributed pentamers, spherocylinders have six pentamers at each end of a tube, and cones have different numbers of pentamers at the ends, the smaller number at the narrow end and the larger number at the wide end [41]. Non-symmetric structures contain randomly and non-uniformly distributed pentamers. Intriguingly, the first retroviral pentamers were visualized in cryo-electron microscopy (cryo-EM) studies of in vitro assembled double-shelled RSV particles, which contained as the inner shell a sphere made-up from 12 pentamers, surrounded by an outer shell, containing 20 hexamers and 12 pentamers [42].
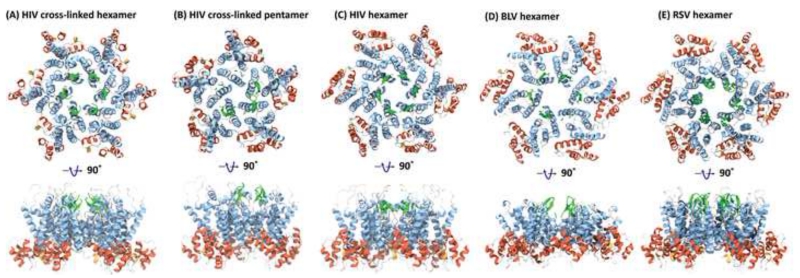
Given the importance of the hexameric and pentameric building blocks in retroviral capsid cores, high-resolution structures of hexamers and pentamers were pursued. Thus, HIV-1 CA proteins were engineered such that stable, cysteine cross-linked hexamers [43] or pentamers [44] were created for X-ray crystal structure determination of these oligomers. To that end, specific mutant CA proteins (A14C/E45C/W184A/M185A for the hexamer and N21C/A22C/W184A/M185A or P17C/R18L/T19C/W184A/M185A for the pentamer) were prepared, which permitted cysteine disulfide crosslinking between NTDs, while disrupting the CTD dimer (Figures 3A & 3B). The cross-linked CA hexamer is made up of two concentric rings, with NTDs forming the inner ring and CTDs the outer ring (Figure 3A). While extensive contacts exist between the NTDs, with helix 2 from one monomer packing against helices 1 and 3 from the neighboring monomer, no contacts are seen between CTDs. The cross-linked CA pentamer has a similar structure, except that the inner concentric ring contains five NTDs, with 15 helices forming the inter-monomer contacts (Figure 3B), compared to 18 in the hexamer (Figure 3A).
Figure 3.
Structures of retroviral hexamers and pentamers. (A) X-ray structure of a cross-linked HIV-1 mutant hexamer (PDB accession code 3H47). (B) X-ray structure of a cross-linked HIV-1 mutant pentamer (PDB accession code 3P05) [44]. (C) X-ray structure of a native HIV-1 hexamer (PDB accession code 4XFX) [46]. (D) X-ray structure of a native BLV hexamer (PDB accession code 4PH0) [45]. (E) Pseudo-atomic model of the RSV hexamer (PDB accession code 3TIR) [80]. Two views, rotated by 90°, are provided. Alpha helices are colored in blue and red for the NTD and CTD, respectively; the 310 helices are colored in yellow and the beta-hairpin in green.
Essentially identical hydrophobic contacts are seen between the helices at this interface for the hexamer and pentamer, and the alternative packing is achieved by a ~10° rotation of adjacent monomers around the center of the three helices. At the center of the cross-linked pentamer, the NTDs are in closer proximity than at the center of the cross-linked hexamer (cf. Figures 3A and 3B). Although it is clear that pentamers must be present in a capsid core (see below), compelling evidence for particular biophysical/biochemical effects that allow transition between hexamer and pentamer is lacking. Clearly, subtle changes in curvature can be introduced at the three interfaces, NTD-NTD, CTD-CTD and CTD-NTD, by conformational changes in the hinge between NTD and CTD, creating the rich polymorphism that is observed for retroviral cores [42].
Recently, X-ray structures of full-length native CA protein from HIV-1 and BLV (Figures 3C & 3D) [45,46] were solved. These crystal structures permitted the unambiguous identification of inter-hexameric interactions for the hexameric arrangement of the CA proteins without crosslinking. For HIV-1, these differ considerably from those derived from the cross-linked hexameric structure. Details of hydrophilic and water-mediated interactions at the dimer and trimer interfaces in the hexagonal lattice were determined [46]. A large number of structural water molecules form an extensive hydration layer, which are associated with packing differences at the two- and three fold interfaces in the hydrated and non-hydrated hexamer [46]. In addition, chloride and iodide ions mediate intra-hexameric interactions in the periphery and at the center of the hexamers. Whether these anion-mediated interactions play any biological role in assembly is unclear; they may simply aid ordering the protein and nucleate crystallization. The water-mediated packing differences have been proposed to permit isoenergetic structural rearrangements, important in conferring structural plasticity, necessary in the formation of an asymmetric conical capsid core.
In summary, the structural convergence between HIV-1, BLV and RSV hexamers suggests a conserved role for these oligomers in retroviral capsids (Figures 3A-E). Furthermore, there is strong evidence to support the existence of pentameric units in HIV-1 and RSV capsid cores, however, high resolution atomic structures of native pentamers are yet to be determined.
Higher order structures of the immature retroviral lattice
Inside immature virions, Gag is arranged in a quasi-spherical shell [47] with discernible boundary defects; the latter can be considered a sign of 3D spatial geometric frustration [48]. Because of the size and disorder in immature retroviral Gag lattices, atomic models are hard to derive. However, a combination of cryo-EM [49-52] and computational modeling [53,54], recently generated detailed models for the packing of the Gag protein in immature lattices. These models included structural specifics on the subdomains unique to the immature assembly, like the p10 peptide in RSV [49,54], the spacer peptide (SP) in RSV and spacer peptide 1 (SP1) in HIV [34,39,41].
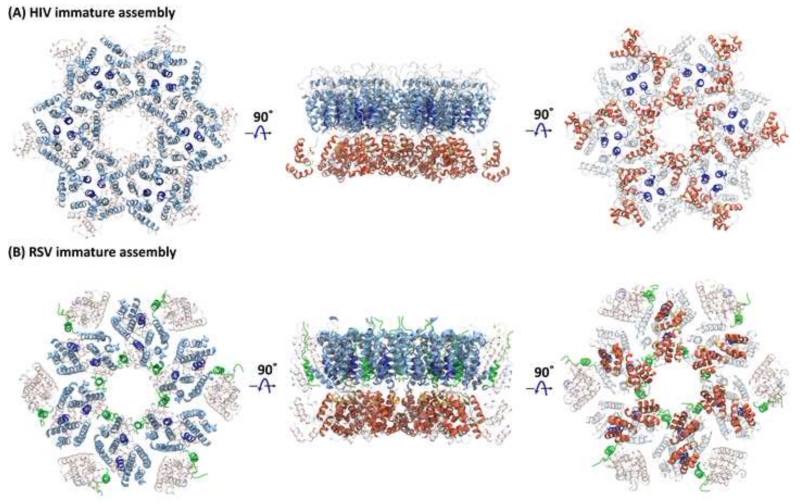
In the immature assembly, the packing arrangement in the CA-NTD layer is virus-specific. In HIV (Figure 4A), helix 1 resides at the NTD interfaces, while helix 2 forms the pseudo three-fold symmetry axis [47]. By comparison, in RSV, helices 2 and 7 reside at the NTD interfaces, while helix 4 is arranged around a pseudo three-fold symmetry axis (Figure 4B). A quaternary organization similar to RSV is also seen for M-MPV [34,39,41,53,54]. In addition, the short p10 peptide, present only in RSV (Figure 4B), propagates the lattice by connecting adjacent NTDs from different Gag monomers [49,54]. The causes or functional consequences of these structural differences between immature assemblies of different retroviruses are currently unknown, although it is possible that differences in the cellular mechanisms of assembly for D-type (M-MPV) versus C-type (RSV,HIV) retroviruses may play a role [49].
Figure 4.
Higher-order CA structures in immature retroviral particles. Three views of (A) the hexameric arrangement of HIV-1 CA (PDB accession code 4USN) [47] and of (B) the RSV hexameric assembly (PDB accession code 5A9E) [49,54]. Helix 2, which resides at the interface between adjacent NTDs in RSV, and at the center of a pseudo three-fold symmetry axis in HIV-1, is colored dark blue and the p10 peptide in RSV is shown in green. Note, a very different packing arrangement of hexamers is seen for HIV-1 and RSV, highlighted by the altered location of the NTD helix 2 (dark blue) in both.
In contrast to the varied arrangement of the CA-NTDs, the packing of the CA-CTDs in immature assemblies is conserved among retroviruses [49]. The dimer interface always involves helix 9, although marked differences in the relative helix 9-helix 9 orientations are seen in the different CTD domain dimer structures (described above and Figure 2). For the CTD arrangements in immature RSV particles, cryo-EM data suggest that hexameric CTD-rings are possibly present, given their close proximity (Figure 4B) [34,39,41]. In the case of HIV-1, the arrangement of the CTDs in the immature lattice (Figure 2C) is remarkably similar to the structure of the CTD dimer in complex with a CA assembly inhibitor (CAI) peptide (Figure 2B) [55], suggesting that this inhibitor functions by stabilizing the immature assembly, preventing maturation into the conical capsid cores after cleavage of the SP1 domain. In addition, the quaternary structure of the low pH 4.3 RSV CTD X-ray dimer (Figure 2E) [56] resembles the arrangement at the dimer interface in mature HIV capsid assemblies (Figure 2D) [50], while the high pH 8.5 RSV CTD dimer structure (Figure 2F) is similar to what is observed for the dimeric interface in immature RSV capsid assemblies (Figure 2G) [49].
The region between the CTD of CA and NC in Gag, namely SP in RSV, SP1 in HIV, a short, non-cleavable SP1-like junction in M-MPV and a long, ‘charged assembly helix’ at the C-terminus of CA in murine leukemia virus (MLV), play important roles for virus function, since mutations or deletions interfere with Gag-Gag assembly and affect virus infectivity [47,54,57].
At present, there is still uncertainty about the conformation of the region between CA and NC in immature assemblies of Gag. In individual SP/SP1 or CA-SP1 polypeptides, SP/SP1 is flexible and disordered [53,57], although a helical conformation of SP1 can be induced by appending a leucine zipper to the SP1 peptide [58]. In addition, it has been proposed that in immature assemblies the SP/SP1 region of RSV/HIV forms a six helix bundle, tethering together six Gag molecules [53,54]. Likewise, for MLV, cryo-electron tomography (cryo-ET) of in vitro assembled virus-like particles suggested the presence of helices between CA and NC [59]. For RSV and HIV, the SP/SP1 region has been evoked to function as a molecular switch, which, if present, stabilizes the immature assembly. Removal of SP/SP1 by proteolytic cleavage, therefore, may relieve constraints on Gag and would allow conversion to the mature capsid core [58,60]. The importance of the region between CA and NC is further supported by the observation of altered phenotypes when mutations are introduced in or near the SP/SP1 cleavage site [61] and by the effects of maturation inhibitors that target specific regions in SP1 [62]. In addition, differences between viruses in the linkage between Gag CA and NC may be related to virus-specific assembly pathways, since M-MPV, a virus lacking a cleavable SP/SP1 peptide, assembles in the cytoplasm.
Structure of the mature HIV-1 capsid core
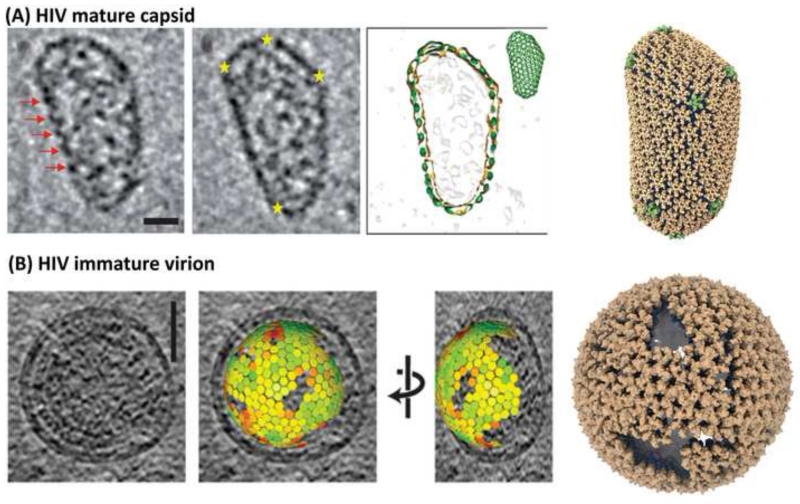
Atomic models for asymmetric conical HIV-1 capsid cores were determined by a combination of experimental and computational approaches. Employing molecular dynamics flexible fitting (MDFF) [63] and large-scale molecular dynamics [64], a hybrid model was created in which the cross-linked hexameric CA X-ray structure [65] and the dimeric CA CTD NMR structure [27] were fit into the cryo-EM density of in vitro generated tubular capsid assemblies. In the first step, a hexamer of hexamers (HOH) was created, in which CA hexamers were connected by CA CTD dimers. Assembling a large number of HOH permitted the generation of a curved hexameric lattice, recapitulating the arrangement in tubular capsid assemblies. Next, placing the cross-linked pentameric CA X-ray structure at the center, a pentamer of hexamers (POH) was modeled, creating units that permitted closure of the conical HIV-1 core. A POH constitutes an acutely curved unit, in contrast to the flat HOH [66]. Differences between the POH and HOH structures revealed key interactions, such as details at the three-fold axis that modulate the bite angle between neighboring oligomers. In the POH and HOH, the same three helices form the trimer interface, although it is more tightly packed in the POH than in the HOH. These interactions were probed and validated by mutagenesis. For example, the A204C variant, which upon oxidation cross-links the capsid cores at the trimer interfaces, results in a hyperstable capsid core and non-infectious virions [66]. Finally, an atomic model of a native HIV-1 capsid core was derived, based on the shape determined by cryo-ET and the POH and HOH building blocks This all-atom capsid core model contains 216 hexamers and 12 pentamers (Figure 5A).
Figure 5.
Mature and immature retroviral capsid cores. (A) Mature HIV-1 capsid core, composed of hexamers and pentamers. In the tomogram, locations of hexamers and pentamers on the periphery of the ovoid are indicated by red arrows and yellow stars, respectively. The pseudo-atomic model of the mature HIV-1 capsid core contains 286 hexamers (brown) and 12 pentamers (green) [66]. (B) Immature HIV-1 virus-like-particle, observed by cryo-EM [47] and the computational model of the capsid core derived from the cryo-EM data. The lattice in the model is composed entirely of hexamers and contains multiple gaps to permit the spherical morphology; the color gradient, from green (smaller) to red (larger), of the individual hexamers illustrate the magnitude of local structural deviations from a reference hexamer [54].
Suggestions about maturation derived from HIV capsid structures
The generation of infectious HIV virions from immature viral particles entails as the final step a global re-arrangement of the Gag assembly, known as maturation, which is triggered by sequential cleavages of Gag polyprotein by PR [67,68]. Starting from a quasi-spherical assembly of Gag (Figure 5B), cleavage between SP1 and NC releases the NC-SP2-p6 polypeptide, bound to the viral RNA genome. Subsequently, CA is cleaved off from the membrane-attached MA, followed by cleavage of SP1 from the CTD of CA, which leads to the formation of the conical capsid core (Figure 5A). Similarly, the NC-SP2-p6 polypeptide undergoes further cleavages, first releasing p6, followed by SP2 [70], resulting in the final NC containing RNP core. The ‘scars’ or defects seen in the immature lattice (Figure 5B) may relate to the mechanism of maturation, since gaps in the lattice may allow access of PR from the inside of the virion to the lamina between MA and CA. In turn, this may suggest that PR-mediated release of CA from MA in Gag begins at the edges of the immature lattice. However, in general, the molecular mechanism(s) of maturation remain elusive and two alternative pathways exist: a displacive-transition and a disassembly-reassembly maturation pathway. Some experimental evidence supports the former [69-71], although there is also biochemical data that support the latter [61,72]. Irrespective, given the large structural differences between mature and immature assemblies, the global re-arrangement upon maturation most likely requires the disruption of the CTD dimer, before self-assembly of the mature capsid core can happen.
Implications of the HIV capsid core structure for the interplay with cellular factors
The HIV capsid core is genetically fragile [73] and has to straddle between being stable enough to protect the HIV genome from cytosolic nucleic acid sensors, evading restriction factors, but still allow for reverse transcription and formation of the pre-integration complex. Efficient generation of pre-integration complexes, their association with the nuclear pore [11,15] and nuclear import are critically dependent on the presence of CypA in the target cell [9,11,74,75]. As described above, interactions between the CA protein and CypA involve the CypA-binding loop, which engages the catalytic site of CypA [76]. At the nuclear pore, the Nup358 protein also contains a CypA domain (Nup358Cyp), which has been shown to interact with the CA protein, albeit with a larger binding interface, compared to CypA [77]. Intriguingly, although structures of the CA protein with CypA are available, the precise biological role of CypA during lentiviral infection has still not been elucidated.
Some hints towards CypA’s role on capsid assemblies have recently emerged from magic-angle-spinning solid state NMR. These experiments permitted the evaluation of conformational dynamics at the atomic level and revealed exceptional mobility on the nano- to microsecond timescales for the CypA-binding loop [31,78]. Interestingly, these motions were significantly attenuated upon CypA binding, and the dynamics profiles of CypA escape mutants closely resembled those of CypA-bound capsid assemblies [51]. This resemblance in motional properties suggests that changes in the sequence-dependent conformational dynamics may be a critical factor in the escape mechanism of HIV-1 CA mutants from CypA dependence. In addition, a single-CypA molecule can simultaneously interact with two hexameric units of capsid assemblies using a non-canonical interface on CypA, with the second interaction interface possibly involved in the protective role played by CypA [79].
Concluding Remarks
In summary, viral capsid cores are proteinaceous containers, which exhibit highly plastic, yet robust, properties. In retroviruses, the CA protein and capsid assemblies play important roles in nearly every step of the life cycle, from the early, immediate post-entry step, via trafficking to the nucleus, to formation and budding and final maturation of the viral particle. Elucidating the relationship between structure, dynamics and function of capsid assemblies in virions requires deciphering the interfaces between subcomponents and building blocks and generating a detailed atomic overall model, which is only attainable by combining and integrating data obtained from a plethora of experimental techniques, supplemented by functional and computational studies. Such studies have yielded important insights already, opened new avenues for further research and, no doubt, will continue to do so in the future.
Box 1. Outstanding Questions.
Is capsid maturation a displacive-transition or a diffusive disassembly-reassembly reaction?
What are the most critical determinants for capsid stability?
Are the pentamers in a capsid fixed, defined locations, or could they be mobile and diffuse throughout the capsid?
Does the assembly of the mature capsid start from a pentamer or are pentamers incorporated last? Is it an ordered or stochastic process?
Where in the mature capsid resides the genomic RNA? How do nucleotides permeate through the capsid for reverse transcription.
Are differently shaped capsids different in their potential to traffic or become imported through the nuclear pore?
What is the role of the major homology region in the CA protein? Is it important for the structure of the mature capsid or for the interaction with a cellular co-factor?
Trends.
In retroviruses, the capsid plays a role in nearly every step of the life cycle, from early, immediate post-entry, via trafficking to the nucleus, to formation and budding and final maturation of the viral particle.
Recent structures suggest that quaternary arrangements in mature capsid are structurally conserved among retroviruses, while the quaternary arrangements in immature retroviral assemblies are poorly conserved.
Water and ions mediate interactions at several interfaces in the mature capsids, permitting subtle structural rearrangements between hexameric and pentameric building blocks, important for conferring structural plasticity.
Acknowledgments
We thank Boon Chong Goh for rendering the capsid assembly in an immature virion and helpful discussions, Klaus Schulten for continuous support and enlightening conversations, Chris Aiken, Peijun Zhang and all members of the Pittsburgh Center for HIV Protein Interactions for years of fruitful collaborations, and Teresa Brosenitsch for editorial support. This work is a contribution from the Pittsburgh Center for HIV Protein Interactions and was supported in part by National Institutes of Health grants P50GM082251 (A.M.G), R01GM067887 (J.R.P).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wills JW, Craven RC. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–54. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Coffin JM, et al. Retroviruses. Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 3.Gheysen D, et al. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 4.Dick RA, Vogt VM. Membrane interaction of retroviral Gag proteins. Front. Microbiol. 2014;5:187. doi: 10.3389/fmicb.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell EM, Hope TJ. HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol. 2015;13:471–483. doi: 10.1038/nrmicro3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kane M, et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goujon C, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, et al. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe. 2013;14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Rasaiyaah J, et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503:402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstone DC, et al. Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. Proc Natl Acad Sci U S A. 2014;111:9609–9614. doi: 10.1073/pnas.1402448111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaller T, et al. HIV-1 Capsid-Cyclophilin Interactions Determine Nuclear Import Pathway, Integration Targeting and Replication Efficiency. PLoS Pathog. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arhel N. Revisiting HIV-1 uncoating. Retrovirology. 2010;7:96. doi: 10.1186/1742-4690-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilditch L, Towers GJ. A model for cofactor use during HIV-1 reverse transcription and nuclear entry. Curr. Opin. Virol. 2014;4:32–36. doi: 10.1016/j.coviro.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price AJ, et al. CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog. 2012;8:e1002896. doi: 10.1371/journal.ppat.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matreyek KA, et al. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog. 2013;9:e1003693. doi: 10.1371/journal.ppat.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah VB, et al. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J. Virol. 2013;87:422–432. doi: 10.1128/JVI.07177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi M, et al. Cyclophilin A-dependent restriction of human immunodeficiency virus type 1 capsid mutants for infection of nondividing cells. J Virol. 2008;82:12001–12008. doi: 10.1128/JVI.01518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys.[see comment] Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 19.Wong RW, et al. Impact of Nucleoporin-Mediated Chromatin Localization and Nuclear Architecture on HIV Integration Site Selection. J. Virol. 2015;89:9702–5. doi: 10.1128/JVI.01669-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin CR, et al. Direct Visualization of HIV-1 Replication Intermediates Shows that Capsid and CPSF6 Modulate HIV-1 Intra-nuclear Invasion and Integration. Cell Rep. 2015;13:1717–31. doi: 10.1016/j.celrep.2015.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez J, et al. Microtubule-associated proteins 1 (MAP1) promote human immunodeficiency virus type I (HIV-1) intracytoplasmic routing to the nucleus. J. Biol. Chem. 2015;290:4631–46. doi: 10.1074/jbc.M114.613133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganser BK, et al. Assembly and analysis of conical models for the HIV-1 core. Science (80-.) 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 23.Gelderblom HR. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–37. [PubMed] [Google Scholar]
- 24.Du S, et al. Structure of the HIV-1 full-length capsid protein in a conformationally trapped unassembled state induced by small-molecule binding. J. Mol. Biol. 2011;406:371–386. doi: 10.1016/j.jmb.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang C, et al. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 26.Worthylake DK, et al. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr. Sect. D-Biological Crystallogr. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- 27.Byeon I-JJL, et al. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campos-Olivas R, et al. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J. Mol. Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- 29.Campos-Olivas R, Summers MF. Backbone dynamics of the N-terminal domain of the HIV-1 capsid protein and comparison with the G94D mutant conferring cyclosporin resistance/dependence. Biochemistry. 1999;38:10262–10271. doi: 10.1021/bi990991x. [DOI] [PubMed] [Google Scholar]
- 30.Khorasanizadeh S, et al. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J. Mol. Biol. 1999;291:491–505. doi: 10.1006/jmbi.1999.2986. [DOI] [PubMed] [Google Scholar]
- 31.Byeon I-JL, et al. Motions on the millisecond time scale and multiple conformations of HIV-1 capsid protein: implications for structural polymorphism of CA assemblies. J. Am. Chem. Soc. 2012;134:6455–66. doi: 10.1021/ja300937v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deshmukh L, et al. Structure and dynamics of full-length HIV-1 capsid protein in solution. J. Am. Chem. Soc. 2013;135:16133–47. doi: 10.1021/ja406246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherry B, et al. Role of cyclophilin A in the uptake of HIV-1 by macrophages and T lymphocytes. Proc Natl Acad Sci U S A. 1998;95:1758–1763. doi: 10.1073/pnas.95.4.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braaten D, et al. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIV(CPZ)GAB but not group O HIV-1 or other primate immunodeficiency viruses. J. Virol. 1996;70:4220–4227. doi: 10.1128/jvi.70.7.4220-4227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nisole S, et al. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 36.Yang H, et al. Structural insight into HIV-1 capsid recognition by rhesus TRIM5alpha. Proc Natl Acad Sci U S A. 2012;109:18372–18377. doi: 10.1073/pnas.1210903109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin T-Y, Emerman M. Cyclophilin A interacts with diverse lentiviral capsids. Retrovirology. 2006;3:70. doi: 10.1186/1742-4690-3-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spidel JL, et al. Genetic Studies of the beta-hairpin loop of Rous sarcoma virus capsid protein. J.Virol. 2007;81:1288–1296. doi: 10.1128/JVI.01551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orlinsky KJ, et al. Mutations in the Ty3 major homology region affect multiple steps in Ty3 retrotransposition. J.Virol. 1996;70:3440–3448. doi: 10.1128/jvi.70.6.3440-3448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, et al. Structural basis of arc binding to synaptic proteins: implications for cognitive disease. Neuron. 2015;86:490–500. doi: 10.1016/j.neuron.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen TT, et al. Continuum theory of retroviral capsids. Phys. Rev. Lett. 2006;96:078102. doi: 10.1103/PhysRevLett.96.078102. [DOI] [PubMed] [Google Scholar]
- 42.Cardone G, et al. Visualization of a missing link in retrovirus capsid assembly. Nature. 2009;457:694–698. doi: 10.1038/nature07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pornillos O, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–92. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pornillos O, et al. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–7. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obal G, et al. Conformational plasticity of a native retroviral capsid revealed by x-ray crystallography. Science. 2015;349:95–8. doi: 10.1126/science.aaa5182. [DOI] [PubMed] [Google Scholar]
- 46.Gres AT, et al. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science. 2015;349:99–103. doi: 10.1126/science.aaa5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schur FK, et al. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature. 2015;517:505–508. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 48.Manoharan VN. Colloidal matter: Packing, geometry, and entropy. Science (80-.) 2015;349:1253751–1253751. doi: 10.1126/science.1253751. [DOI] [PubMed] [Google Scholar]
- 49.Schur FKM, et al. The structure of immature-like Rous sarcoma virus Gag particles reveals a structural role for the p10 domain in assembly. J. Virol. 2015 doi: 10.1128/JVI.01502-15. DOI: 10.1128/JVI.01502-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schur FKM, et al. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature. 2015;517:505–8. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 51.Bharat TA, et al. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc Natl Acad Sci U S A. 2014;111:8233–8238. doi: 10.1073/pnas.1401455111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bharat TAM, et al. Structure of the immature retroviral capsid at 8 Å resolution by cryo-electron microscopy. Nature. 2012;487:385–389. doi: 10.1038/nature11169. [DOI] [PubMed] [Google Scholar]
- 53.Bush DL, et al. Higher Order Structure of the Rous Sarcoma Virus SP Assembly Domain. J. Virol. 2014;88:5617–5629. doi: 10.1128/JVI.02659-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goh BC, et al. Atomic Modeling of an Immature Retroviral Lattice using Molecular Dynamics and Mutagenesis. Structure. 2015;23:1414–1425. doi: 10.1016/j.str.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ternois F, et al. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol. 2005;12:678–682. doi: 10.1038/nsmb967. [DOI] [PubMed] [Google Scholar]
- 56.Bailey GD, et al. Proton-linked dimerization of a retroviral capsid protein initiates capsid assembly. Structure. 2009;17:737–748. doi: 10.1016/j.str.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 57.Han Y, et al. Magic angle spinning NMR reveals sequence-dependent structural plasticity, dynamics, and the spacer peptide 1 conformation in HIV-1 capsid protein assemblies. J. Am. Chem. Soc. 2013;135:17793–17803. doi: 10.1021/ja406907h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Datta SAK, et al. Dimerization of the SP-1 region of HIV-1 Gag induces a helical conformation and association into helical bundles: implications for particle assembly. J. Virol. 2015 doi: 10.1128/JVI.02061-15. DOI: 10.1128/JVI.02061-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hadravova R, et al. In vitro assembly of virus-like particles of a gammaretrovirus, the murine leukemia virus XMRV. J. Virol. 2012;86:1297–1306. doi: 10.1128/JVI.05564-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Datta SAK, et al. On the role of the SP1 domain in HIV-1 particle assembly: a molecular switch? J. Virol. 2011;85:4111–21. doi: 10.1128/JVI.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fontana J, et al. Identification of an HIV-1 mutation in spacer peptide 1 that stabilizes the immature CA-SP1 lattice. J. Virol. 2015 doi: 10.1128/JVI.02204-15. DOI: 10.1128/JVI.02204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Urano E, et al. Alkyl Amine Bevirimat Derivatives are Potent and Broadly Active HIV-1 Maturation Inhibitors. Antimicrob. Agents Chemother. 2015 doi: 10.1128/AAC.02121-15. DOI: 10.1128/AAC.02121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.G.Trabuco L, et al. Flexible Fitting of Atomic Structures into Electron Microscopy Maps Using Molecular Dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perilla JR, et al. Molecular dynamics simulations of large macromolecular complexes. 2015;31:64–74. doi: 10.1016/j.sbi.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pornillos O, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao G, et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiegers K, et al. Sequential Steps in Human Immunodeficiency Virus Particle Maturation Revealed by Alterations of Individual Gag Polyprotein Cleavage Sites. J. Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Freed EO. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015;13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu Z, et al. Unclosed HIV-1 capsids suggest a curled sheet model of assembly. J. Mol. Biol. 2013;425:112–123. doi: 10.1016/j.jmb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frank GA, et al. Maturation of the HIV-1 core by a non-diffusional phase transition. Nat. Commun. 2015;6:5854. doi: 10.1038/ncomms6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woodward CL, et al. Electron cryotomography studies of maturing HIV-1 particles reveal the assembly pathway of the viral core. J. Virol. 2015;89:1267–77. doi: 10.1128/JVI.02997-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keller PW, et al. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. J. Virol. 2013;87:13655–64. doi: 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rihn SJ, et al. Extreme Genetic Fragility of the HIV-1 Capsid. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sokolskaja E, et al. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J Virol. 2004;78:12800–12808. doi: 10.1128/JVI.78.23.12800-12808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hatziioannou T, et al. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 2005;79:176–183. doi: 10.1128/JVI.79.1.176-183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Howard BR, et al. Structural insights into the catalytic mechanism of cyclophilin A. Nat. Struct. Biol. 2003;10:475–481. doi: 10.1038/nsb927. [DOI] [PubMed] [Google Scholar]
- 77.Bichel K, et al. HIV-1 capsid undergoes coupled binding and isomerization by the nuclear pore protein NUP358. Retrovirology. 2013;10:81. doi: 10.1186/1742-4690-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu M, et al. Dynamic Allostery Governs Cyclophylin A-HIV Capsid Interplay. Proc. Natl. Acad. Sci. U. S. A. 2015;112:14617–22. doi: 10.1073/pnas.1516920112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu C, et al. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat. Commun. 2016;7:10714. doi: 10.1038/ncomms10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bailey GD, et al. A structural model for the generation of continuous curvature on the surface of a retroviral capsid. J. Mol. Biol. 2012;417:212–23. doi: 10.1016/j.jmb.2012.01.014. [DOI] [PubMed] [Google Scholar]