Abstract
A versatile strategy is reported for the multi-gram synthesis of discrete oligomers from commercially available monomer families, e.g., acrylates, styrenics, siloxanes. Central to this strategy is the identification of reproducible procedures for the separation of oligomer mixtures using automated flash chromatography systems with the effectiveness of this approach demonstrated through the multi-gram preparation of discrete oligomer libraries (Đ = 1.0). Synthetic availability, coupled with accurate structural control, allows these functional building blocks to be harnessed for both fundamental studies as well as targeted technological applications.
INTRODUCTION
The preparation of synthetic polymers with precise structures, compositions and functions is a grand challenge for polymer and material chemists. The advent of convenient and robust synthetic protocols for controlled polymerization (e.g., ATRP, RAFT, ROMP)1-4 has greatly increased the ease in which large quantities of well-defined polymers can be prepared. This increased availability has enabled the engineering of materials for a range of applications. Recent studies in this area of well-defined macromolecular design have been directed towards the development of polymers with added dimensions of control, such as the specific order in which monomers are arranged, in analogy to the sequence-defined polymers found in nature (e.g., DNA, RNA, proteins).5–12 In addition to sequence, another important characteristic of these biopolymers is their monodispersity (Đ = 1.0). Methods to prepare monodisperse synthetic polymers using direct polymerization techniques, however, do not exist due to the inherent statistical nature of polymer growth processes. Step-wise strategies are available, but limited in scope and utility.
As a result, the importance of monodispersity for synthetic polymers remains an open question, especially for low molecular weight-polymers (oligomers) where the impact of dispersity is more pronounced.13, 14 For example, a low dispersity oligomer (DP ~ 8, Đ = 1.2) contains a mixture of major species ranging from tetramer to dodecamer, with an actual octamer content of < 15 mol%. The properties of this mixture therefore result from the combination of individual oligomers, each with unique solubility and chemical reactivity. Since only a small subset of these oligomers may have desirable performance, the use of oligomer mixtures may not be optimal for many applications or fundamental studies. For example, recent work by Meijer and coworkers clearly demonstrates the impact of a single monomer addition in the self-assembly of oligomeric materials.15, 16 Unfortunately, a major challenge with existing step-wise approaches to discrete oligomers (Đ = 1.0) is their impractical nature for large-scale synthesis with many starting monomers not available.
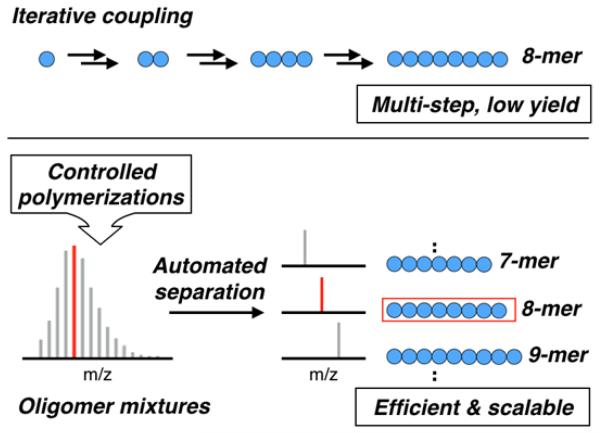
The construction of oligomers via iterative synthesis involves successive steps of monomer coupling and protecting group removal, resulting in control over the target structure and sequence (Scheme 1). While solid-phase oligopeptide17 and dendrimer18 synthesis are successful examples of this strategy, most iterative approaches are only suited for preparing small amounts of material due to its time-consuming, multi-step nature.19–24 These issues are further compounded with increasing oligomer length. Additionally, robust synthetic methods need to be established for coupling individual monomer units, preventing access to many important monomer classes, such as acrylates, styrenics, and functional siloxanes.25–32
Scheme 1.
Comparison of the traditional, iterative coupling approach to discrete oligomers with the separation of oligomeric mixtures prepared by controlled polymerization techniques.
In this manuscript, an alternative strategy involving the initial preparation of low dispersity oligomer mixtures by controlled polymerization techniques (i.e. ATRP, ROP) followed by the efficient, large scale separation of these mixtures into their constituent species is described. While separation of oligomers and polymers has been examined using a subset of chromatographic techniques, these techniques are generally only useful for small-scale analysis and not for the isolation of synthetically useful quantities. For example, thin layer (TLC),33–36 liquid (LC)37–39 and size exclusion (SEC)40, 41 chromatography have been employed to separate synthetic oligomers, but are limited by loading capacity and/or separation resolution. More specialized methods, such as aqueous displacement chromatography or thermal gradient fractionation, have been employed for gram-scale separations, but are highly oligomer specific, require specialized equipment, and do not represent general strategies.42–45
RESULTS AND DISCUSSION
The development of a versatile, scalable and straight-forward method for the synthesis and purification of discrete oligomers from commercially available monomer families would therefore be highly desirable. This approach is enabled by the combination of facile polymerization procedures (e.g., ATRP) and ubiquitous purification processes (e.g., flash chromatography). While relatively unexplored for oligomer separation, flash chromatography has the added advantage of being automated through various commercial chromatographic systems (e.g., Biotage®), leading to consistent and scalable protocols for separating oligomer mixtures. This allows large-scale preparation of libraries of discrete oligomers from various functional monomers in a single synthetic step.
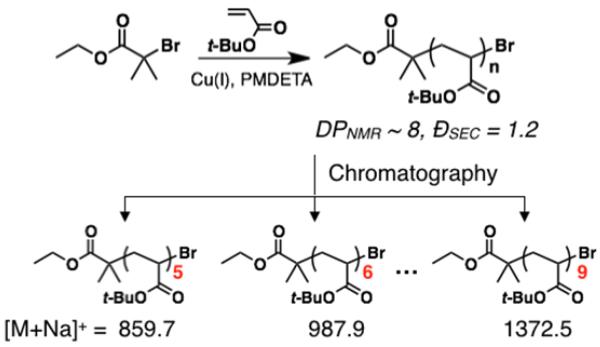
To demonstrate the feasibility of flash chromatography for large-scale oligomer separation, t-butyl acrylate oligomers (oligoTBA) were synthesized using ATRP on a 50-100 g scale (Scheme 2). As expected, a large distribution of species was observed by MALDI-MS analysis of the crude mixture, despite the low dispersity as measured by SEC analysis (ĐSEC = 1.2). Using automated flash chromatography, oligomer mixtures with a variety of molecular weights could be readily separated using an optimized hexane/ethyl acetate solvent gradient on standard columns. As an illustrative example, analysis of fractions from oligoTBA (DPNMR ~ 8) showed a regular elution of oligomers in a step-wise fashion with shorter oligomers being eluted first, implying the separation arises from adsorption rather than through a size exclusion mechanism. Combination of fractions then allowed discrete oligoTBAs from trimer to decamer to be isolated with each individual oligomer being fully characterized by a combination of spectroscopic and chromatographic techniques (Figure 1 and see the Supporting Information).
Scheme 2.
Synthesis of oligo(t-butyl acrylate) (oligoTBA) using ATRP and subsequent separation into discrete oligomers.
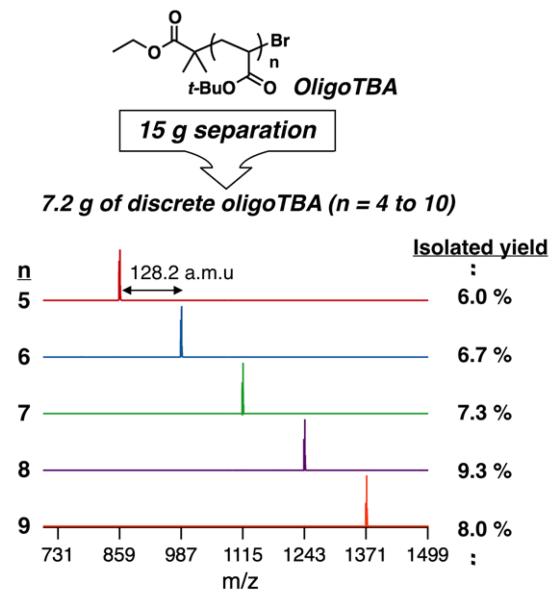
Figure 1.
MALDI-MS spectra of discrete oligoTBAs n = 5 to 9 isolated after chromatographic separation of a DPNMR ~ 8 mixture together with isolat-ed yields. Please note that trimer and n = 10+ fractions were also obtained.
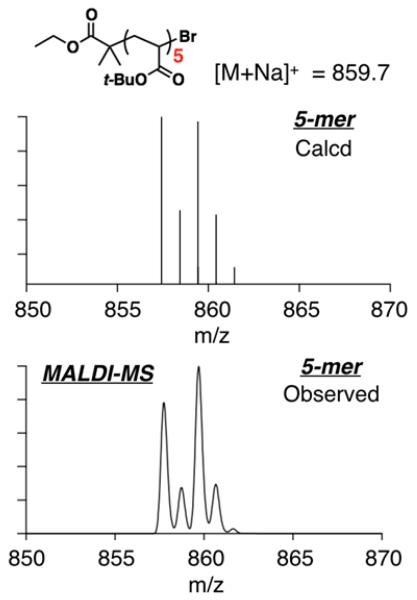
As shown in Figure 1, single molecular ions were observed by MALDI-MS analysis for each of the discrete oligomers with the molecular ion correlating with the expected structure (Scheme 2). Each discrete oligomer species is separated by 128 a.m.u. which corresponds to the t-butyl acrylate repeat unit and, as shown in Figure 2, the molecular ion corresponds to the sodium adduct with a single bromo- and a single ethyl isobutyrate end group. Of particular note is the high isolated yield (6-10%) for each discrete oligomer with gram quantities of oligomers from n = 4 to n = 10 being obtained from a typical 15 g batch of starting oligomer mixture. Using the automated chromatography system and the identified hexane/ethyl acetate solvent gradient, reproducible separation could be achieved for other oligomer mixtures starting from DP ~ 5 to DP ~ 12 with mixed samples only obtained above n = 14. Such a straightforward and reproducible strategy for isolating a library of discrete functional oligomers at a pre-parative scale (1–3 grams each) is synthetically powerful.
Figure 2.
The calculated and observed isotope patterns of oligoTBA pentamer.
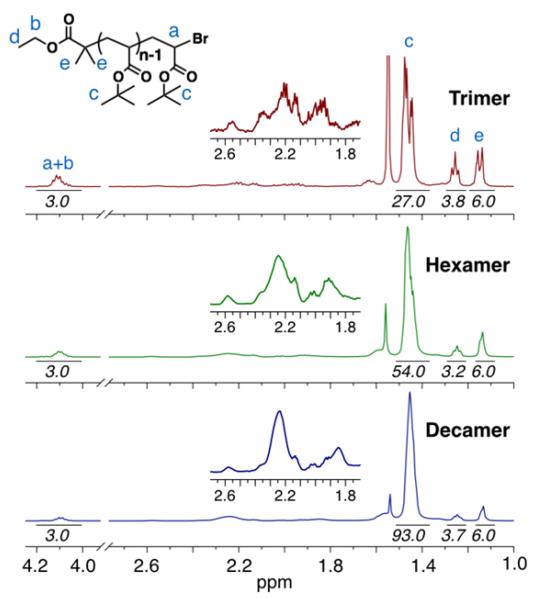
1H-NMR analysis of the discrete oligomers further reinforces the purity shown by the MALDI data while also providing insights to subtle structural changes as the size of the oligomers increases (Figure 3). Progressing from n = 3 (trimer) to n = 10 (decamer), the relative intensity of the chain-ends steadily decreases when compared to the backbone/t-butyl protons with the integration ratios closely correlating to the molecular structure. The overall appearance of the backbone protons (1.75-2.75 ppm region) also evolves as the oligomer size increases. The trimer has sharp peaks well dispersed in this region, presumably from the overlay of several distinct splitting patterns of individual diastereomers. As the series progresses towards the decamer, the resonances for these backbone protons broaden and become characteristic of higher molecular weight poly(t-butyl acrylate). This broadening reflects the increased number of stereochemical configurations, the minimized influence of chain-ends, and the gradual transition of oligomers to polymers as their chain length increases.
Figure 3.
1H-NMR spectra for the discrete trimer, hexamer and decamer oligo(t-butyl acrylate) derivatives. All spectra are normalized to the resonance for the t-butyl protons (c, 1.45 ppm). Integrations of the peaks are shown in italics.
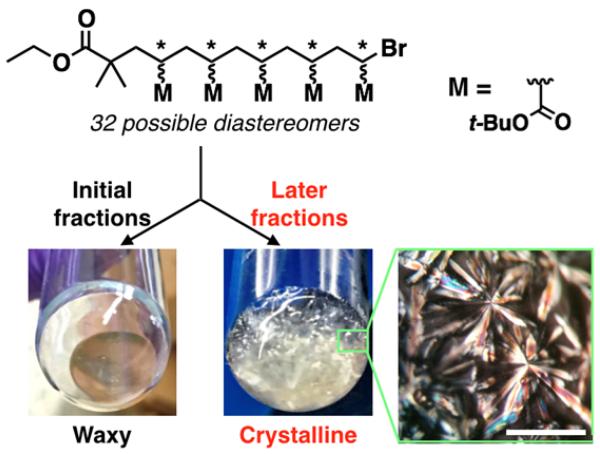
The physical properties of the discrete oligoTBA derivatives were also found to be dramatically different from each other and from the parent mixture. While the crude oligomers were viscous oils, discrete oligomers above n = 4 were obtained as solid waxes, with the glass transition temperature (Tg) of each oligomer increasing with degree of polymerization from −30 to 0 °C (Figure S6). Surprisingly, the purification strategy described above also allowed separation within individual oligomeric samples. For example, the initial fractions of the n = 5, oligoTBA pentamer derivative was found to give a waxy solid in direct contrast to later fractions which afforded a solid, spherulite-rich material (Figure 4). The spherulite-rich samples displayed Maltese-cross birefringence patterns when viewed under a polarized microscope, and gave a series of sharp diffraction peaks when measured using XRD, indicating the crystalline nature of the material (Figure S4). It should be noted that in both cases only a single molecular ion is observed at 859.7 a.m.u. corresponding to pure oligoTBA pentamer. NMR analysis also showed integration values consistent with a pentameric structure, though with differences in the backbone region corresponding with tacticity. These results strongly imply that the chromatography process is not only able to separate discrete oligomers by length but is also able to resolve the oligoTBA pentamers according to the stereochemical configuration along the backbone (32 possible stereoisomers). Similarly, reproducible separations within a discrete oligomer were also observed for the n = 6, hexamer and the n = 7 heptamer, leading to amorphous and crystalline fractions for each of these discrete oligomers.
Figure 4.
Variation in physical appearance for n = 5 penta-TBA5 derivatives. Initial fractions were obtained as a waxy solid with later factions being crystalline. Inset: polarized micrograph of crystalline penta-TBA5, scale bar is 1 mm.
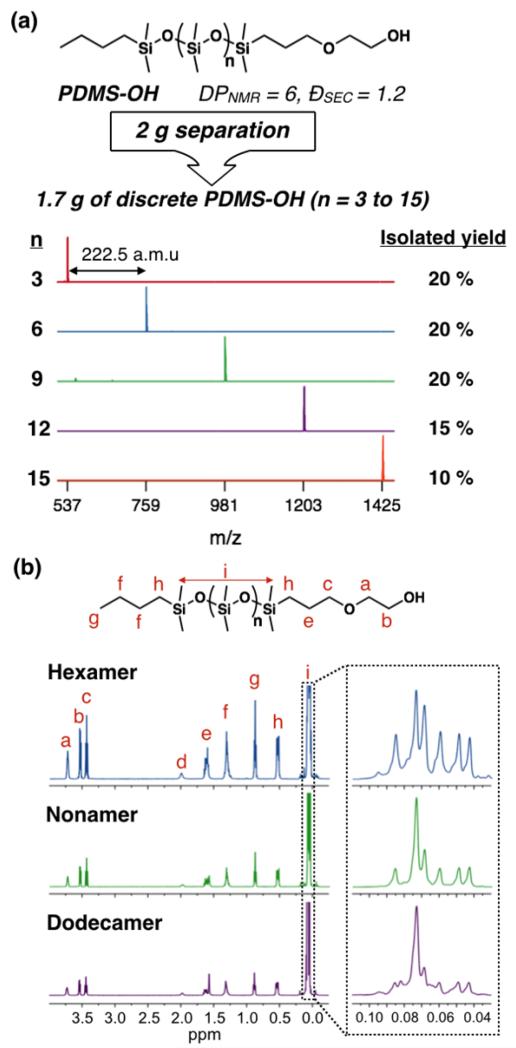
Having demonstrated efficient separation of oligoacrylates, this method was further extended to another important materials class, polysiloxanes. Siloxane-based oligomers, oligoDMS, present a distinct challenge because they are highly non-polar and therefore cannot be well resolved with a normal-phase silica column. While investigating the commercially available monohydroxyl-terminated polydimethylsiloxane oligomers (PDMS-OH, Gelest), we found that simply switching to a reverse-phase C18 column cartridge allows the isolation of a library of discrete species, as demonstrated by MALDI-MS analysis (Figure 5a). Once again, this separation could be performed on a significant scale (2.0 – 10.0 grams) to give the corresponding discrete oligomers in synthetically useful quantities with excellent mass recovery.
Figure 5.
(a) MALDI analysis of discrete PMDS-OH oligomers obtained from reversed-phase separation. (b) 1H NMR analysis of hexamer, nonamer and dodecamer PDMS-OH. Inset: NMR spectra of the dimethyl proton region.
In analogy with the acrylate libraries, 1H NMR analysis of the discrete species shows the relative intensity of the chain-ends steadily decreases when compared to the dimethyl protons of the backbone. The overall appearance of the backbone protons (0.04–0.10 ppm region) also evolves due to decreasing chain-end effects as the oligomer size increases (Figure 5b). The structure of the oligoDMS derivatives has a few notable differences from the polyacrylates that also assists in their separation. Since the PDMS-OH was prepared by anionic ring-opening polymerization of hexamethylcyclotrisiloxane, the isolated oligomer sizes are always separated by three siloxane units. Additionally, the oligosiloxanes lack stereocenters, thereby reducing the number of species present in the mixture and results in straightforward isolation of truly unimolecular materials. From a 2.0 gram mixture of oligomers, ca. 85% yield of discrete oligomers, n = 3, n = 6, n = 9, n = 12 and n = 15, was obtained which shows the efficiency of this process.
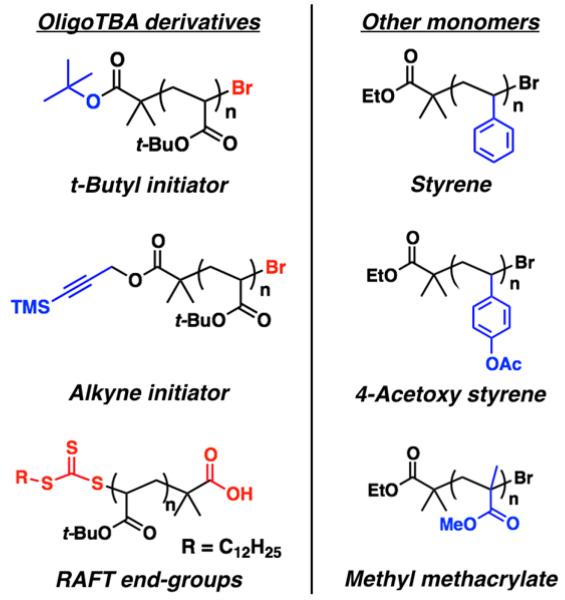
To illustrate the generality and scope of this strategy for the preparation of discrete oligomers, a wide range of oligomeric materials with different chain-ends and repeat units were then investigated (Figure 6). Without modification to the gradient established for ethyl isobutyrate terminated oligoTBA, oligomer mixtures derived from functional initiators containing alkyne or t-butyl ester groups could be efficiently separated on multi-gram scale. An analogous oligoTBA sample prepared using reversible-addition fragmentation chain-transfer (RAFT) also proved to be separable, which demonstrates compatibility of this approach with dramatically different chain ends - carboxylic acid and trithiocarbonate chain-ends for the RAFT derived oligomers.
Figure 6.
Scope of the controlled polymerization/chromatographic separation strategy highlighting the range of chain-ends and monomer-types accessible.
This versatility was further illustrated by extension to other repeat units based on commercially available monomers. By adjusting elution solvents, oligomer mixtures of methyl methacrylate, 4-acetoxystyrene, and styrene could be separated on multi-gram scale to give libraries of discrete oligomers (ca. n = 3 to n = 10), with purity comparable to the oligoTBA and oligoDMS derivatives described above. The wide scope of this separation protocol coupled with the chain-end fidelity of large-scale controlled polymerization techniques allows these discrete oligomers to be viewed as precision building blocks for the construction of well-defined materials with accurate physical and chemical properties.
CONCLUSION
In summary, we describe a general strategy for the large-scale synthesis and purification of discrete oligomer libraries from commercially available monomers. By combining high yielding controlled polymerization procedures with automated chromatography techniques, functional oligomers can be rapidly and reproducibly accessed with NMR, MALDI and SEC characterization demonstrating unique structural purity. The synthetic availability of these discrete oligomers libraries, coupled with accurate control over molecular weight and chain ends, suggests unprecedented opportunities as building blocks for the preparation of designer materials and macromolecular architectures.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the MRSEC program of the National Science Foundation (DMR 1121053, C.J.H.) and The Dow Chemical Company through the Dow Materials Institute at UCSB (J.L., S.L., C.F., A.S.K., W.R.G., A.J.M., B.V.K.J.S., C.J.H.) for financial support. W.R.G. thanks the NIH for a postdoctoral fellowship (F32GM108323). J.M.R. thanks the Victorian Endowment for Science, Knowledge and Innovation (VESKI) for a postdoctoral fellowship. B.V.K.J.S. was supported by a fellowship within the Postdoc-Program of the German Academic Exchange Service (DAAD).
Footnotes
Supporting Information. Experimental procedures and characterization data for all compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Boyer C, Corrigan NA, Jung K, Nguyen D, Nguyen T-K, Adnan NNM, Oliver S, Shanmugam S, Yeow J. Chem. Rev. 2016;116:1803–1949. doi: 10.1021/acs.chemrev.5b00396. [DOI] [PubMed] [Google Scholar]
- (2).Anastasaki A, Nikolaou V, Nurumbetov G, Wilson P, Kempe K, Quinn JF, Davis TP, Whittaker MR, Haddleton DM. Chem. Rev. 2016;116:835–877. doi: 10.1021/acs.chemrev.5b00191. [DOI] [PubMed] [Google Scholar]
- (3).Moad G, Rizzardo E, Thang SH. Chem. Asian J. 2013;8:1634–1644. doi: 10.1002/asia.201300262. [DOI] [PubMed] [Google Scholar]
- (4).Bielawski CW, Grubbs RH. Prog. Polym. Sci. 2007;32:1–29. [Google Scholar]
- (5).Lutz J-F, Ouchi M, Liu DR, Sawamoto M. Science. 2013;341:1238149. doi: 10.1126/science.1238149. [DOI] [PubMed] [Google Scholar]
- (6).Rosales AM, Segalman RA, Zuckermann RN. Soft Matter. 2013;9:8400–8414. [Google Scholar]
- (7).Soejima T, Satoh K, Kamigaito M. J. Am. Chem. Soc. 2016;138:944–954. doi: 10.1021/jacs.5b11631. [DOI] [PubMed] [Google Scholar]
- (8).Weiss RM, Short AL, Meyer TY. ACS Macro. Lett. 2015;4:1039–1043. doi: 10.1021/acsmacrolett.5b00528. [DOI] [PubMed] [Google Scholar]
- (9).Gutekunst WR, Hawker CJ. J. Am. Chem. Soc. 2015;137:8038–8041. doi: 10.1021/jacs.5b04940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Porel M, Alabi CA. J. Am. Chem. Soc. 2014;136:13162–13165. doi: 10.1021/ja507262t. [DOI] [PubMed] [Google Scholar]
- (11).Zhang J, Matta ME, Hillmyer MA. ACS Macro. Lett. 2012;1:1383–1387. doi: 10.1021/mz300535r. [DOI] [PubMed] [Google Scholar]
- (12).Yang J, Gitlin I, Krishnamurthy VM, Vazquez JA, Costello CE, Whitesides GM. J. Am. Chem. Soc. 2003;125:12392–12393. doi: 10.1021/ja035978l. [DOI] [PubMed] [Google Scholar]
- (13).Binauld S, Damiron D, Connal LA, Hawker CJ, Drockenmuller E. Macromol. Rapid Commun. 2011;32:147–168. doi: 10.1002/marc.201000548. [DOI] [PubMed] [Google Scholar]
- (14).Abe F, Einaga Y, Yamakawa H. Macromolecules. 1993;26:1891–1897. [Google Scholar]
- (15).Van Genabeek B, de Waal BFM, Gosens MMJ, Pitet LM, Palmans ARA, Meijer EW. J. Am. Chem. Soc. 2016;138:4210–4218. doi: 10.1021/jacs.6b00629. [DOI] [PubMed] [Google Scholar]
- (16).Zha RH, de Waal B, Lutz M, Teunissen AJP, Meijer EW. J. Am. Chem. Soc. 2016 doi: 10.1021/jacs.6b02172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Merrifield RB. J. Am. Chem. Soc. 1963;85:2149–2154. [Google Scholar]
- (18).Fréchet JMJ. Science. 1994;263:1710–1715. doi: 10.1126/science.8134834. [DOI] [PubMed] [Google Scholar]
- (19).Schaffert D, Wagner E. Gene Ther. 2008;15:1131–1138. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- (20).Gillies ER, Fréchet JMJ. Drug Discov. Today. 2005;10:35–43. doi: 10.1016/S1359-6446(04)03276-3. [DOI] [PubMed] [Google Scholar]
- (21).Tour JM. Chem. Rev. 1996;96:537–554. doi: 10.1021/cr9500287. [DOI] [PubMed] [Google Scholar]
- (22).Zhang L, Colella NS, Liu F, Trahan S, Baral JK, Winter HH, Mannsfeld SCB, Briseno AL. J. Am. Chem. Soc. 2013;135:844–854. doi: 10.1021/ja3104796. [DOI] [PubMed] [Google Scholar]
- (23).Burns M, Essafi S, Bame JR, Bull SP, Webster MP, Balieu S, Dale JW, Butts CP, Harvey JN, Aggarwal VK. Nature. 2014;513:183–188. doi: 10.1038/nature13711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kumarasamy E, Sanders SN, Pun AB, Vaselabadi SA, Low JZ, Sfeir MY, Steigerwald ML, Stein GE, Campos LM. Macromolecules. 2016;49:1279–1285. [Google Scholar]
- (25).Zhang H, Li X, Shi Q, Li Y, Xia G, Chen L, Yang Z, Jiang Z-X. Angew. Chem., Int. Ed. 2015;54:3763–3767. doi: 10.1002/anie.201410309. [DOI] [PubMed] [Google Scholar]
- (26).Brooke GM, Burnett S, Mohammed S, Proctor D, Whiting MC. J. Chem. Soc., Perkin Trans. 1. 1996:1635–1645. [Google Scholar]
- (27).Takizawa K, Tang C, Hawker CJ. J. Am. Chem. Soc. 2008;130:1718–1726. doi: 10.1021/ja077149w. [DOI] [PubMed] [Google Scholar]
- (28).Leibfarth FA, Johnson JA, Jamison TF. Proc. Nat. Acad. Sci. 2015;112:10617–10622. doi: 10.1073/pnas.1508599112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Barnes JC, Ehrlich DJC, Gao AX, Leibfarth FA, Jiang Y, Zhou E, Jamison TF, Johnson JA. Nature Chem. 2015;7:810–815. doi: 10.1038/nchem.2346. [DOI] [PubMed] [Google Scholar]
- (30).Vandenbergh J, Reekmans G, Adriaensens P, Junkers T. Chem. Sci. 2015;6:5753–5761. doi: 10.1039/c5sc02035b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Feldman KS, Bobo JS, Ensel SM, Lee YB, Weinreb PH. J. Org. Chem. 1990;55:474–481. [Google Scholar]
- (32).Klausen RS, Widawsky JR, Steigerwald ML, Venkataraman L, Nuckolls C. J. Am. Chem. Soc. 2012;134:4541–4544. doi: 10.1021/ja211677q. [DOI] [PubMed] [Google Scholar]
- (33).Litvinova LS, Bel’nikevich NG. Polym. Sci. Ser. A. 2010;52:1250–1256. [Google Scholar]
- (34).Litvinova L, Bel'nikevich N. J. Chromatogr. A. 2003;1005:165–176. doi: 10.1016/s0021-9673(03)00844-6. [DOI] [PubMed] [Google Scholar]
- (35).Iyengar DR, McCarthy TJ. Macromolecules. 1990;23:4344–4346. [Google Scholar]
- (36).Otocka EP, Hellman MY. Macromolecules. 1970;3:362–365. [Google Scholar]
- (37).Held D, Kilz P. Macromol. Symp. 2005;231:145–165. [Google Scholar]
- (38).Murgasova R, Hercules D. Anal. Bioanal. Chem. 2002;373:481–489. doi: 10.1007/s00216-002-1332-9. [DOI] [PubMed] [Google Scholar]
- (39).Mourey TH. Anal. Chem. 1984;56:1777–1781. [Google Scholar]
- (40).Trathnigg B. Size-Exclusion Chromatography of Polymers. In: Meyers RA, editor. Encyclopedia of Analytical Chemistry. J. Wiley and Sons Ltd.; 2000. pp. 8008–8034. [Google Scholar]
- (41).Liu Y, Rawlston J, Swann AT, Takatani T, Sherrill CD, Ludovice PJ, Weck M. Chem. Sci. 2011;2:429–438. [Google Scholar]
- (42).Hodges RS, Lorne Burke TW, Mant CT. J. Chromatogr. A. 1988;444:349–362. doi: 10.1016/s0021-9673(01)94036-1. [DOI] [PubMed] [Google Scholar]
- (43).Agner E. Method for displacement chromatography. 6,576,134 US Patent. 2003
- (44).Petro M, Svec F, Gitsov I, Fréchet JMJ. Anal. Chem. 1996;68:315–321. doi: 10.1021/ac950726r. [DOI] [PubMed] [Google Scholar]
- (45).Francuskiewicz F. Polymer Fractionation. Springer; Berlin: 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.