SUMMARY
Most tumor cells take up more glucose than normal cells but metabolize glucose via glycolysis even in the presence of normal levels of oxygen, a phenomenon known as the Warburg effect. Tumor cells commonly express the embryonic M2 isoform of pyruvate kinase (PKM2) that may contribute to the metabolism shift from oxidative phosphorylation to aerobic glycolysis and tumorigenesis. Here we show that PKM2 is acetylated on lysine 305 and that this acetylation is stimulated by high glucose concentration. PKM2 K305 acetylation decreases PKM2 enzyme activity and promotes its lysosomal-dependent degradation via chaperone-mediated autophagy (CMA). Acetylation increases PKM2 interaction with HSC70, a chaperone for CMA, and association with lysosomes. Ectopic expression of an acetylation mimetic K305Q mutant accumulates glycolytic intermediates and promotes cell proliferation and tumor growth. These results reveal an acetylation regulation of pyruvate kinase and the link between lysine acetylation and CMA.
INTRODUCTION
It was first noted by Otto Warburg that cancer cells rely mainly on aerobic glycolysis to generate ATP instead of more efficient mitochondrial oxidative phosphorylation, resulting in the increased rate of glucose uptake and lactate production even in the presence of sufficient oxygen supply (Warburg, 1956). Based on the dramatically increased glucose consumption in cancer cells, positron emission tomography (PET) of 2-(18F)-fluoro-2-deoxy-D-glucose (FDG) has been developed as a diagnostic technique to detect cancer cells in clinics (Funes et al., 2007). Activation of oncogenes or loss of tumor suppressor genes, such as mutations in Ras (Dang and Semenza, 1999; Ramanathan et al., 2005), AKT (Manning and Cantley, 2007), Myc (Gordan et al., 2007a, 2007b), and p53 (Bensaad et al., 2006; Matoba et al., 2006) increase glucose uptake and lactate production. These observations rekindle attention to Warburg effect and cancer metabolism.
A key glycolytic enzyme consistently altered in expression during tumorigenesis is pyruvate kinase (E.C. 2.7.1.40) (Altenberg and Greulich, 2004; Majumder et al., 2004), which catalyzes the transfer of phosphate from phosphoenolpyruvate (PEP) to ADP, resulting in the formation of pyruvate and ATP. There are four pyruvate kinase isoforms in mammals: L, R, M1, and M2. The L and R isoforms are specifically expressed in liver and red blood cells, respectively (Mazurek et al., 2005). PKM1 is expressed in most adult tissues, while PKM2 is exclusively expressed during embryonic development. Notably, most tumor cells re-express PKM2 (Dombrauckas et al., 2005; Mazurek et al., 2005), suggesting that the switch from PKM1 to PKM2 expression may be beneficial to tumor cells. Indeed, switching from PKM2 to PKM1 reverses aerobic glycolysis, providing the selective growth advantage of PKM2 expression for tumor cells in vivo (Christofk et al., 2008a). Recently, the PKM1-to-PKM2 switch was found to be regulated by the Myc oncogene (David et al., 2009), providing further evidence linking the re-expression of the M2 isoform to the tumorigenesis.
The benefit of expressing PKM2 isoform to the rapidly growing embryonic and tumorigenic cells is believed to result from a decreased PK activity, which would lead to accumulation of various glycolytic metabolites for macromolecular biosynthesis to support cell growth. According to this notion, a regulation that decreases and increases PK activity could favor active dividing and quiescent cells, respectively. Unlike PKM1, full activity of PKM2 requires allosteric activation by fructose 1, 6-bisphosphate (F-1, 6-BP). One such regulation is the binding of PKM2, but not PKM1, to phosphotyrosine, and this binding releases the allosteric activator F-1,6-BP from PKM2, leading to a decreased PKM2 activity and shifting catabolism from energy production to anabolic processes, leading to increased cell proliferation and tumor growth (Christofk et al., 2008b).
Protein acetylation has recently emerged as a broadly used modification in the regulation of a wide range of cellular processes (Choudhary et al., 2009; Kim et al., 2006; Zhao et al., 2010). In particular, we have found that most of the intermediate metabolic enzymes are acetylated and that acetylation can directly affect enzyme function (Zhao et al., 2010). Notably, acetylation of metabolic enzymes is regulated by extracellular cues, such as the nutrient availability. These findings indicate a broad role of acetylation in the coordination between the extracellular nutrients and intracellular metabolic pathways. In this paper, we report that PKM2 activity and protein stability are regulated by lysine acetylation. Specifically, acetylation of lysine K305 inhibits PKM2 activity and promotes lysosome-dependent degradation of PKM2 via CMA. Our study reveals an acetylation regulation of pyruvate kinase and the link between acetylation and CMA.
RESULTS
PKM2 Is Acetylated at K305
Protein acetylation has long been known to play a key role in regulation of chromatin structure and gene transcription through modification of histones and nuclear transcription regulators (Soutoglou et al., 2000). We and others have recently discovered that a large number of nonnuclear proteins involved in a wide range of cellular pathways are also acetylated in human and mouse liver and leukemia cells (Choudhary et al., 2009; Kim et al., 2006; Zhao et al., 2010). To further explore acetylation of nonnuclear protein in other tissues, we fractioned cell extracts of both human prostate cancer cell line LnCAP and cancer tissues into nuclear, mitochondrial, and cytosolic fractions. Cytosolic fractions were digested with trypsin, and acetylated peptides were immunoprecipitated with an antibody specific to acetyllysine. The immunopurified peptides were analyzed by tandem liquid chromatography-tandem mass spectrometry (LC-MS/MS). These analyses identified 113 acetylated proteins (data not shown), including several enzymes involved in glycolysis pathway. PKM2 was identified to be acetylated by the mass spectrometric analyses in samples from both cultured LnCAP cells and primary prostate cancer tissues. Identified acetylated PKM2 peptides are shown in Table S1, available online. To confirm its acetylation, Flag-tagged PKM2 was ectopically expressed into HEK293T cells and immunoprecipitated. Western blotting with anti-acetyllysine antibody confirmed that PKM2 was indeed acetylated and its acetylation was enhanced approximately 3-fold after treatment with trichostatin A (TSA, an inhibitor of histone deacetylase HDAC I and II) (Ekwall et al., 1997; Furumai et al., 2001) and nicotinamide (NAM, an inhibitor of the SIRT family deacetylases) (Avalos et al., 2005) (Figure 1A).
Figure 1. Acetylation at K305 Decreases PKM2 Enzyme Activity.
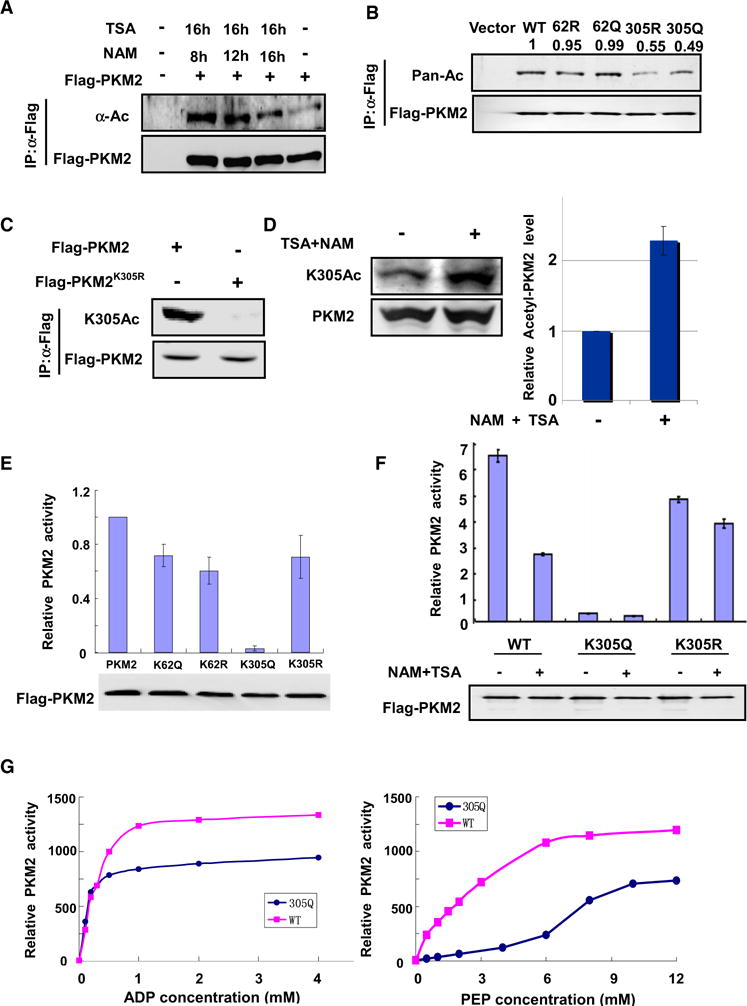
(A) PKM2 is acetylated. Flag-PKM2 was transfected into 293T cells followed by treatment with TSA and NAM for the indicated time, and PKM2 acetylation and protein levels were analyzed by western blot with indicated antibody, respectively.
(B) Mutation of K305 decreases PKM2 acetylation. The indicated plasmids were cotransfected into 293T cells, and protein was immunoprecipitated (IP) for acetylation analysis. Acetylation levels were normalized against β-actin.
(C) Characterization of acetyl-PKM2 (K305) antibody. The indicated plasmids were transfected into 293T cells, and acetylation level of IPed Flag-PKM2 was measured by the site-specific K305 acetylation antibody.
(D) Endogenous PKM2 is acetylated at K305. 293T cells were treated with TSA and NAM. Endogenous PKM2 was immunoprecipitated, and protein levels and acetylation of K305 were determined by western blot with indicated antibodies (left panel). Relative PKM2 K305 acetylation over protein level was quantified (right panel). Error bars represent ± SD for triplicate experiments.
(E) K305Q mutant decreases PKM2 enzyme activity. Flag-tagged wild-type and mutant PKM2 protein were expressed in 293T cells and purified by IP. The enzyme activity was measured and normalized against protein level. Mean values of relative enzyme activity of triplicate experiments with standard deviation (±SD) are presented.
(F) NAM and TSA treatment decreases PKM2 wild-type but not mutant enzyme activity. Flag-tagged wild-type and mutant PKM2 protein were expressed in 293T cells and treated with or without NAM and TSA, then purified by IP. The PKM2 enzyme activity was measured and normalized against protein level. Mean values of relative enzyme activity of triplicate experiments with standard deviation (±SD) are presented.
(G) K305Q mutation decreases the binding affinity toward PEP. The activities of wild-type and mutant PKM2 were assayed with increasing concentrations of ADP or PEP as indicated. Error bars represent ±SD for triplicate experiments.
Two acetylation sites were identified in PKM2, K62 and K305 (Table S1). K62 is not conserved, while K305 is conserved from C. elegans to mammals (Figure S1A). We generated Gln and Arg substitution mutants of both sites (K62Q, K62R, K305Q, and K305R) and transfected 293T cells with these mutants individually, followed by direct western blotting with anti-acetyllysine antibody. Mutation of K62 only slightly reduced the acetylation of PKM2, while mutation of K305 reduced the PKM2 acetylation by 45% (Figure 1B), and double mutant of K62 and K305 comparably reduced PKM2 acetylation level as K305 single mutant (Figure S1B), indicating that under this condition K305 is a major acetylation site of ectopically expressed PKM2. There is still residual acetylation in the double mutants, suggesting additional acetylation sites in PKM2. To determine if K305 is acetylated in vivo, we generated antibody specific to acetylated K305. To characterize the specificity of this antibody, we performed dot blot assay and found that PKM2 acetyl K305 antibody preferentially detected the acetylated peptide, but not the unmodified peptide (Figure S2A). Western blotting using this antibody detected strong signal of ectopically expressed wild-type PKM2, but not K305R mutant (Figure 1C). Moreover, PKM2 knockdown significantly decreased the signal detected by the acetyl K305 antibody (Figure S2B). We also found that the antibody recognition of PKM2 was competed by acetylation-modified peptide, but not by the unmodified peptide (Figure S2C), confirming the specificity of the antibody. Furthermore, western blotting of whole-cell extract from 293T cells detected a band that has a molecular weight of expected as PKM2 and whose intensity was substantially increased upon treatment of cells with TSA and NAM prior to the lysis (Figure 1D). Similarly, NAM and TSA treatment also increased PKM2 acetylation in LnCAP cells (Figure S3A). As shown in Figure S1C, both NAM and TSA increased endogenous PKM2 acetylation at K305. Moreover, IHC staining showed that PKM2 was indeed acetylated at K305 in human prostate cancer tissues (Figure S3B and Table S2). These results demonstrate an in vivo acetylation of PKM2 at Lys305.
Acetylation Mimetic Mutation at K305 Decreases PKM2 Activity
To determine the effect of acetylation at these two sites on PKM2 enzyme activity, we transfected 293T cells with individual PKM2 mutants and assayed for the PKM2 enzyme activity. Mutation of Lys62 to either Gln or Arg reduced the activity of PKM2 by 28% and 40%, respectively, while mutation of Lys305 to Arg reduces the PKM2 activity by 29%. Notably, mutation of Lys305 by Gln, which may mimic acetyl modification (Chen et al., 2008; Wang and Hayes, 2008; Zhang et al., 1998), dramatically decreased PKM2 activity by as much as 95% (Figure 1E). Furthermore, NAM and TSA treatment decreased PKM2 wild-type but not mutant enzyme activity (Figure 1F). Kinetic analyses of recombinant PKM2 proteins purified from E. coli revealed that the K305Q mutation reduced affinity toward its substrate PEP (Figure 1G). In contrast, the Km for ADP was not appreciably altered. Although the exact PEP concentration in prostate cancer cells is not known, the concentrations of PEP are reported to be between 4 and 140 μM in human and rat tissues (Liebermeister, 2005). These results suggest that the PKM2-K305Q would have a much lower activity under physiological PEP concentration, indicating an inhibitory role of K305 acetylation in PKM2 regulation. Taken together with the findings that K305Q has a much more dramatic reduction than K62Q in catalytic activity, we therefore focus the study on the molecular mechanism and physiological function of K305 acetylation in PKM2 regulation.
Glucose Increases K305 Acetylation and Decreases PKM2 Protein Level
To determine if the K305 acetylation of PKM2 is dynamically regulated in vivo, we cultured 293T cells with 25 mM glucose concentrations for different time points and examined the K305 acetylation. We found that K305 acetylation was increased in a time course-dependent manner (Figure 2A). More interestingly, the steady-state level of PKM2 decreased with the increase of PKM2 acetylation at K305 (Figure 2A). These results indicate a potential inverse correlation between PKM2 acetylation and protein levels in response to the change of glucose concentration.
Figure 2. Glucose and PCAF Regulate PKM2 Acetylation and Protein Levels.
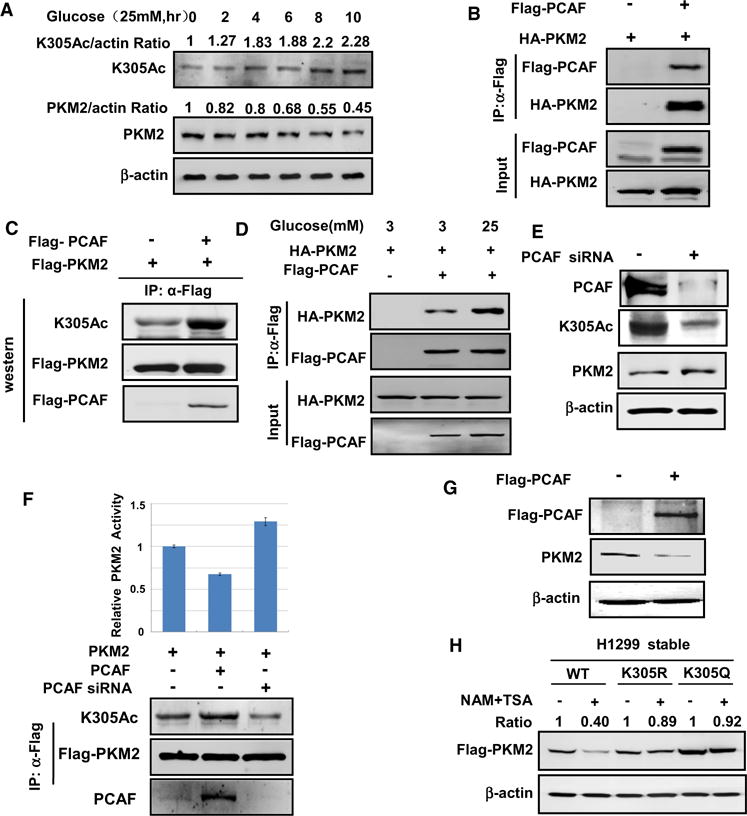
(A) High glucose increases PKM2 acetylation at K305 and decreases PKM2 protein level. HeLa cells were cultured with 25 mM glucose medium. The steady-state level and K305 acetylation of endogenous PKM2 were analyzed by western blot. PKM2 protein and acetylation levels were normalized against β-actin.
(B) PCAF binds with PKM2. 293T cells were transfected with indicated plasmids, and PCAF-PKM2 association was examined by IP and western blot.
(C) PCAF increases PKM2 acetylation at K305. 293T cells were transfected with indicated plasmids, and PKM2 acetylation at K305 was determined by western blot.
(D) High glucose promotes the interaction between PCAF and PKM2. The indicated plasmids were cotransfected into 293T cells, followed by indicated glucose concentration treatment, and the interaction between PKM2 and PCAF was examined by immunoprecipitation.
(E) Knocking down PCAF decreases endogenous K305 acetylation of PKM2. siRNA oligo targeting PCAF was transfected into 293T cells, and the levels of endogenous PCAF, PKM2 protein, and K305 acetylation were determined by western blot.
(F) PCAF negatively regulates PKM2 activity. PCAF expression plasmid or siRNA oligo targeting PCAF was cotransfected into 293T cells with a vector expressing Flag-PKM2. PKM2 was immunoprecipitated and activity was assayed. The mean value of triplicate experiments and standard deviation are presented.
(G) PCAF decreases endogenous PKM2 protein level. PCAF plasmid was transfected into 293T cells and endogenous PKM2 protein level was measured by western blot.
(H) TSA and NAM treatment decreases the level of wild-type PKM2 but not the PKM2K305Q and PKM2K305R mutants. wild-type, K305Q, and K305R mutants expressing stable cells were treated with or without TSA and NAM, followed by western blot to determine PKM2 protein levels. PKM2 protein levels were normalized against β-actin.
Acetyltransferase PCAF Promotes PKM2 Acetylation and Decreases PKM2 Protein Level
To identify the acetyltransferase responsible for PKM2 acetylation at K305, we examined four acetyltransferases, p300 (E1A binding protein, 300 Kda), CBP (CREB binding protein), PCAF (p300/CBP-associated factor, also known as K [lysine] acetyltransferase 2B, KAT2B), and GCN5 (KAT2A), and found that ectopically expressed PCAF bound to PKM2 (Figure 2B) and dramatically increased PKM2 acetylation at K305 (Figure 2C), whereas the other acetyltransferases had little effect (data not shown). Consistent with a role in cytoplasm, a fraction of PCAF was found in the cytoplasm based on subcellular fractionation (Figures S4A and S4B). Notably, glucose significantly increased the interaction between PKM2 and PCAF (Figure 2D), while it only slightly increased PCAF protein level (Figure S4C). Knocking down PCAF significantly reduced K305 acetylation (Figure 2E), indicating that PCAF is a potential K305 acetyltransferase in vivo. To determine if PCAF could affect PKM2 enzyme activity, we analyzed the effect of ectopically expressing PCAF as well as PCAF knockdown on PKM2 enzyme activity. As expected, PCAF ectopic expression decreased PKM2 activity by 33%, while PCAF knockdown increased PKM2 activity by 30% when the enzymatic activity was normalized against PKM2 protein (Figure 2F). These data are consistent with a model that acetylation inhibits PKM2 activity. Moreover, ectopically expressing PCAF decreased the steady-state level of endogenous PKM2 (Figure 2G). To further test the role of acetylation, particularly K305 acetylation, in regulation of PKM2 protein levels, Flag-PKM2-WT, K305Q, and K305R mutant were stably expressed in HEK293 cells, and the cells were treated with TSA and NAM. Inhibition of deacetylases by TSA and NAM treatment decreased the level of PKM2 wild-type (Figure 2H). In contrast, the same treatment had little effect on the protein levels of PKM2 K305Q and PKM2K305R, demonstrating that acetylation of K305 is important for PKM2 protein regulation (Figure 2H). Moreover, NAM and TSA treatment decreased the protein level of endogenous PKM2 but not PKM2K305R (Figure S4D). Together, our results indicate that PCAF decreases PKM2 protein level, possibly via acetylating PKM2 at K305.
Glucose-Induced PKM2 Degradation Requires CMA
The decrease of PKM2 protein level in cells grown in high glucose media led us to determine if it occurred at transcription level or protein stability. We found that the levels of PKM2 mRNA were moderately increased, but not decreased, with the increasing concentration of glucose (Figure 3A). Thus, the reduction of PKM2 protein by glucose is unlikely to be due to a transcriptional regulation. Similarly, inhibition of endogenous deacetylases by NAM and TSA treatment had no significant effect on PKM2 mRNA levels (Figure 3B). These results prompted us to test whether proteasome is involved in PKM2 protein degradation. We treated 293T cells with a proteasome inhibitor MG132 and observed that it had no effect on the steady-state level of PKM2 (Figure 3C). In contrast, a similar MG132 treatment increased the level of phosphoenolpyruvate carboxykinase (PEPCK), a gluconeogenic enzyme known to be degraded by the proteasome pathway in response to high glucose (Zhao et al., 2010). This result indicates that high glucose-induced decrease of PKM2 is due neither to downregulation of PKM2 transcription nor to proteasome-dependent degradation.
Figure 3. Glucose-Induced PKM2 Degradation Requires CMA.
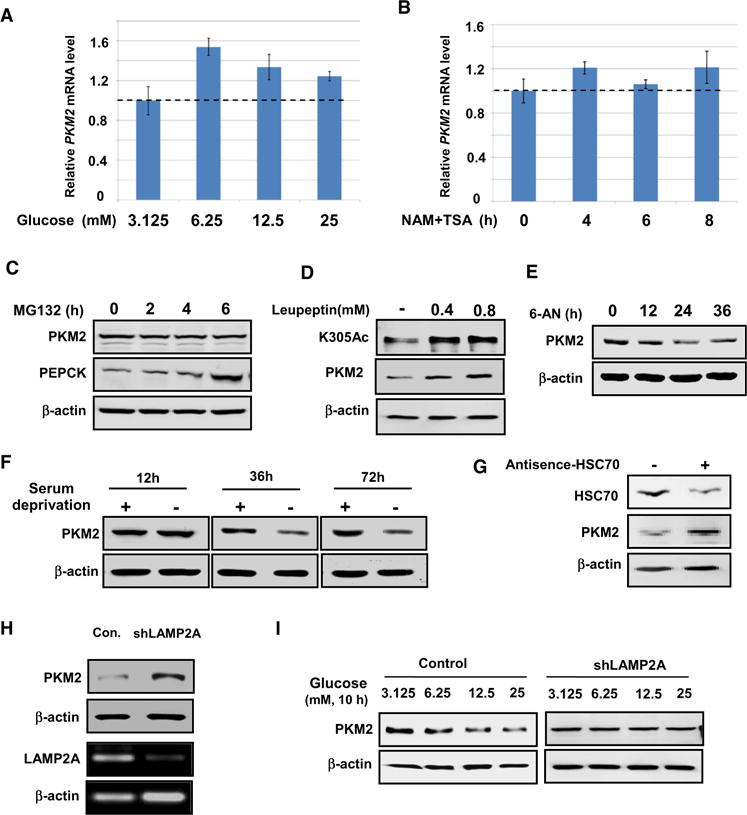
(A) High glucose induces moderate increase of PKM2 mRNA. HeLa cells were cultured with the increasing concentrations of glucose. The level of PKM2 mRNA was determined by qPCR and normalized against GAPDH. Error bars represent ±SD for triplicate experiments.
(B) Inhibition of deacetylases does not significantly affect PKM2 mRNA level. HeLa cells cultured in 25 mM glucose were treated with or without NAM and TSA. The levels of PKM2 mRNA were determined by qPCR using GAPDH as an internal control. The level of PKM2 mRNA in the cells treated with or without NAT and TSA are shown. Error bars represent ±SD for triplicate experiments.
(C) PKM2 is not degraded by the 26S proteasome pathway. 293T cells were treated with proteasome inhibitor MG132, and the PKM2 protein level was analyzed by western blotting. PEPCK, a known proteasome substrate, was included as a control.
(D) Leupeptin accumulates K305 acetylation and PKM2 protein. 293T cells were treated with or without Leupeptin. The levels of total and acetylated PKM2 were determined by western blot. PKM2 level was normalized against β-actin.
(E) CMA activator 6-aminonicotinamide decreases PKM2 protein levels. 293T cells were treated with or without 6-aminonicotinamide (6-AN), and PKM2 level was determined by western blot.
(F) Serum withdrawal decreases PKM2 protein. PKM2 level was determined by western blot after serum withdrawal as indicated in 293T cells.
(G) HSC70 knockdown accumulates PKM2. Plasmid expressing HSC70 antisense was transfected into 293T cells. HSC70 knockdown efficiency and PKM2 level were determined by western blot.
(H) LAMP2A knockdown accumulates PKM2. LAMP2A was stably knocked down in HeLa cells by shRNA. LAMP2A knockdown efficiency was determined by Q-PCR while PKM2 level was determined by western blot. b-actin protein (second bottom panel) and mRNA (bottom panel, determined by qPCR) are shown as controls.
(I) LAMP2A knockdown blocks the glucose effect on PKM2 protein levels. HeLa cell pools stably expressing LAMP2A shRNA were cultured in the presence of increasing concentrations of glucose. PKM2 expression level was determined by western blot.
To elucidate the mechanism underlying PKM2 degradation, we considered the other major pathway of protein degradation, the lysosome system or autophagy. Three different autophagy mechanisms, macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA), have been characterized. Both macroautophagy and microautophagy have the capacity to engulf large structures through both selective and nonselective mechanisms, whereas CMA degrades only soluble proteins, mostly in a selective manner (Mizushima et al., 2008). In CMA, heat shock protein Hsc70 binds to and recruits the target proteins to lysosomes through the lysosomal membrane receptor LAMP2A for degradation (Majeski and Dice, 2004).
We treated cells with Leupeptin, an inhibitor of lysosomal proteases (Jeong et al., 2009), and observed obvious accumulation of PKM2 protein and acetylation (Figure 3D) while treatment of cells with 6-aminonicotinamide, an ATP blocker and an activator of lysosomes (Yang et al., 2009), decreased PKM2 level (Figure 3E). Thus, these results indicate that lysosome is involved in high glucose-induced PKM2 degradation.
Prolonged serum deprivation can activate CMA (Cuervo et al., 1995; Wing et al., 1991). We therefore determined the effect of serum deprivation on the steady-state level of PKM2 and observed that serum starvation decreased PKM2 protein levels (Figure 3F), providing further support for a role of CMA in PKM2 degradation.
Both HSC70 and LAMP2A are critical components in the CMA pathway (Cuervo, 2009). To provide additional evidence to support the role of CMA in PKM2 degradation, we knocked down either HSC70 or LAMP2A and measured PKM2 protein levels. Knockdown of either HSC70 or LAMP2A significantly accumulated PKM2 (Figures 3G and 3H). Moreover, glucose-induced PKM2 degradation was blocked in LAMP2A knockdown cells (Figure 3I). We conclude that high glucose-induced PKM2 reduction is achieved through CMA-mediated protein degradation.
PKM2 Acetylation Promotes Its Binding with HSC70 and Uptake by Lysosomes
The apparent inverse correlation between PKM2 acetylation and protein levels led us to explore a potential role of K305 acetylation in PKM2 degradation. In CMA, HSC70 functions to recruit target proteins to lysosomes for degradation. We first employed acetyl mimetic mutant to examine if PKM2K305Q exhibits a binding with HSC70. Coimmunoprecipitation showed that K305Q mutant displayed a much stronger interaction with HSC70 than wild-type PKM2 (Figure 4A). Consistently, treatment of cells with deacetylase inhibitors TSA and NAM significantly increased the binding between PKM2 and HSC70 (Figure 4B), suggesting that PKM2 acetylation may increase its interaction with HSC70. Furthermore, coexpression of HSC70 dramatically decreased the level of K305-acetylated PKM2 (Figure 4C), consistent with a notion that HSC70 destabilizes the acetylated PKM2. The PKM2 immunoprecipitated by HSC70 had enriched K305-acetylated PKM2 (Figure 4D). The interaction between endogenous PKM2 and HSC70 was confirmed by coimmunoprecipitation (Figure 4E). These data demonstrate that HSC70 preferentially associates with the K305-acetylated PKM2. Importantly, the PKM2-HSC70 association was substantially increased with the increase of glucose concentrations (Figure 4F). Similarly, glucose treatment increased the interaction between endogenous PKM2 and HSC70 (Figure S4E). These results indicate that K305 acetylation promotes PKM2 degradation by enhancing its interaction with HSC70 in response to glucose.
Figure 4. PKM2 Acetylation Promotes Its Binding with HSC70 and Uptake by Lysosome.
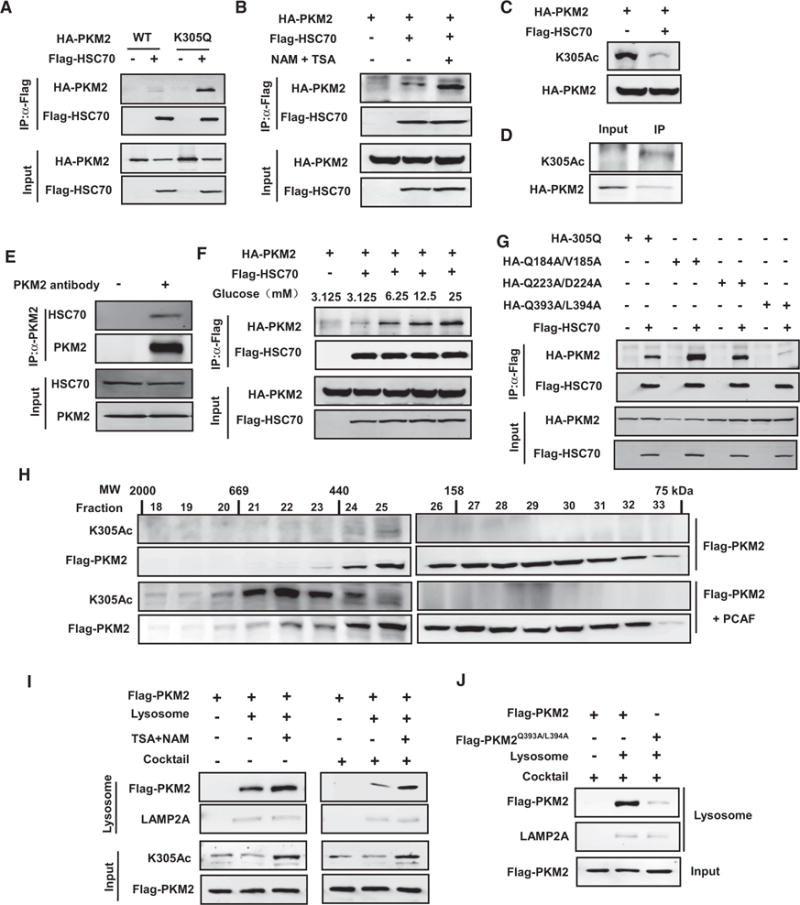
(A) Acetyl mimetic K305Q mutation increases the binding between PKM2 and HSC70. The plasmids were cotransfected into 293T cells as indicated, the binding between PKM2 and HSC70 were examined by IP-western analysis.
(B) Inhibition of deacetylases increases PKM2-HSC70 binding. The plasmids were cotransfected into 293T cells as indicated, followed by NAM+TSA treatment. PKM2-HSC70 binding was determined by IP-western analysis, and the levels of total and acetylated PKM2 were determined by western blot (lower panels).
(C) HSC70 reduces acetylated PKM2. The plasmids were cotransfected into 293T cells as indicated and followed by NAM+TSA treatment, and PKM2 acetylation at K305 was determined by western blot.
(D) The HCS70 preferentially interacts with K305-acetylated PKM2. HA-PKM2-transfected cells were immunoprecipitated with HSC70 antibody (indicated as IP). Relative acetylation of total cellular PKM2 (Input) and the HSC70-associated pKM2 (IP) were determined by western blot with indicated antibodies.
(E) Endogenous PKM2 associates with HSC70. Cell lysate from H1299 cells was immunoprecipated with PKM2 antibody and followed by western blot analysis as indicated.
(F) Glucose promotes the PKM2-HSC70 association. Indicated plasmids were cotransfected into 293T cells followed by treatment with media with different glucose concentrations. The binding between PKM2 and HSC70 was analyzed by IP-western.
(G) Identification of a HSC70 binding motif in PKM2. The binding between PKM2 mutants (generated on K305Q plasmid) and HSC70 was analyzed by IP-western.
(H) PCAF increases PKM2 acetylation and shifts it to higher molecular weight fractions. The indicated plasmids were cotransfected into 293T cells, and cell lysates were separated by gel filtration, followed by western analysis. Fraction numbers and elution of molecular weight markers (MW) are indicated. The acetylated PKM2 is exclusively present in the high molecular weight fractions.
(I) Inhibition of deacetylases promotes lysosomal uptake of PKM2. Flag-tagged wild-type or HSC70-binding deficient mutant PKM2 was immunopurified from 293T cells treated with or without TSA and NAM, and incubated with the lysosomes isolated from rat liver. Lysosomes were reisolated and the associated PKM2 (either inside or binding to the surface) was determined by western blot. The presence of protease inhibitors (cocktail) should block lysosomal degradation, thus such conditions should measure both PKM2 binding to lysosomes and uptake, whereas in the absence of cocktail this assay only measures PKM2 binding to lysosomes.
(J) The HSC70 binding-defective PKM2-393L/394A mutant has weak lysosomal binding. Experiments are similar to those in (I), except PKM2 mutant was used.
Proteins undergoing CMA-mediated lysosomal degradation often contain a loosely defined KFERQ motif important for HSC70 binding (Cuervo, 2009). The KFERQ motif usually contains five residues, including a critical glutamine (Q) residue that is preceded or followed by four amino acids consisting of a basic (R or K), an acidic (E or D), or a bulky hydrophobic residue (I, L, V, or F) (Dice, 1990). Inspection of PKM2 amino sequence reveals three potential HSC70 binding motifs, 184QVKQK188, 223EKDIQDLKF231, and 393QLFEE398. We replaced Q184V185, Q223D224, and Q393L394 by alanines and examined the interaction of each mutant with HSC70. While neither Q184A/V185A nor Q223A/D224A mutation decreased the interaction with HSC70, Q393A/L394A mutation significantly reduced its interaction with HSC70 (Figure 4G), providing an additional support for a CMA-mediated degradation of PKM2. Finally, gel filtration analysis showed that coexpression of PCAF led to a shift of a fraction of PKM2 to higher molecular complex and the acetylated PKM2 was exclusively present in the higher molecular weight fractions (Figure 4H). These observations are consistent with an idea that acetylation promotes the interaction of PKM2 and HSC70.
To determine if PKM2 can be uptaken by lysosomes, we incubated the immunopurified PKM2 with isolated lysosomes in vitro. The lysosomes were repurified and the associated PKM2 was detected by western blotting to determine PKM2 binding. When the above experiment was performed in the presence of lysosomal protease inhibitors, this assay would show the total PKM2 that was bound to and taken up by the lysososomes. We found that PKM2 bound to lysosomes. Moreover, the PKM2 isolated from TSA and NAM-treated cells showed more lysosomal binding and uptake (Figure 4I), suggesting that the acetylated PKM2 has higher affinity to lysosomes. Furthermore, the PKM2-Q393A/L394A mutant, which has mutations of the putative HSC70 recognition motif, significantly decreased lysosomal binding and uptake (Figure 4J). These results demonstrate that PKM2 can bind to and be uptaken by the lysosomes and that acetylation enhances these processes.
Acetylation Mimetic Mutant K305Q Accumulates Glycolytic Intermediates
Accumulation of glycolytic intermediates resulting from decreased PKM2 activity by cellular signals, such as binding with phosphotyrosine or phosphorylation by FGFR1 (Christofk et al., 2008a; Hitosugi et al., 2009), has been suggested to promote cell and tumor growth. We wanted to determine the physiological function of PKM2 acetylation. To this end, we generated stable H1299 cell lines in which the endogenous PKM2 was knocked down by short hairpin RNA (shRNA) and either wild-type (referred to as H1299/PKM2), K305Q mutant (referred to as H1299/PKM2K305Q), or K305R mutant (referred to as H1299/PKM2K305R) was stably expressed. Various glycolytic metabolites were measured by LC-MS/MS analysis. We found that the H1299/PKM2K305Q cells accumulated more fructose-6-phosphate (F-6P), glucose-6-phosphate (G-6P), and F-1,6-BP, but produced less lactate and pyruvate than H1299/PKM2 cells when these cells were cultured in the high glucose media (Figure 5A). The increase of F-6P, G-6P and F-1, 6-BP and decrease of lactate were statistically significant. In contrast, the H1299/PKM2K305R cells accumulated less F-6P and G-6P, but produced more lactate than the H1299/PKM2 cells under the same culture conditions (Figure S5A). The decrease of F-6P and G-6P and increase of lactate were statistically significant. Interestingly, we found that the glycolysis rate in K305R cells was indeed higher compared to wild-type and K305Q cells, suggesting that acetylation inhibits PKM2, and thus accumulates glycolytic intermediates for biosynthesis (Figure S5B). Furthermore, we also observed a significant increase and decrease of NADPH in H1299 cells expressing PKM2K305Q (Figure 5B) and PKM2K305R (Figure S5C), respectively. Together, these data show that expression of PKM2K305Q mutant accumulates glycolytic intermediates and NADPH.
Figure 5. Accumulation of Glycolytic Intermediates and NADPH in Cells Expressing Acetylation Mimetic K305Q Mutant.
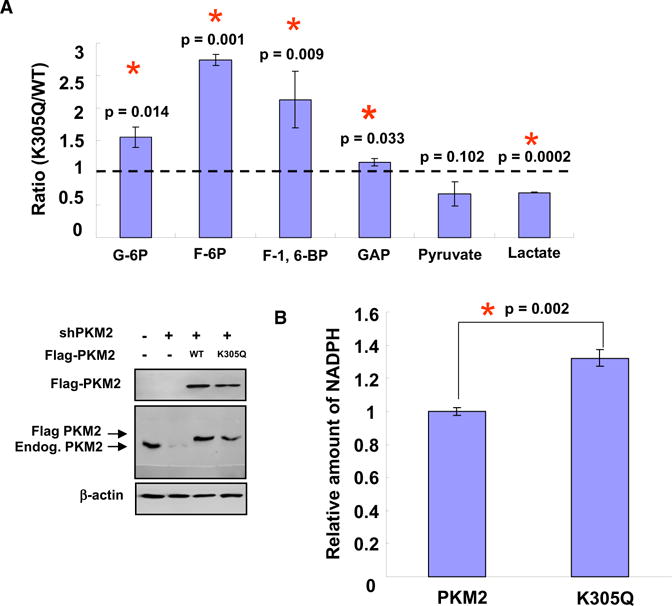
(A) Cells expressing acetylation mimetic K305Q mutant of PKM2 accumulate higher levels of glycolytic intermediates. Metabolites in H1299 cells wild-type (H1299/PKM2) or K305Q mutant (H1299/PKM2K305Q) PKM2 were analyzed by LC-MSMS. The ratios of individual metabolites in H1299/PKM2 expressing over H1299/PKM2K305Q-expressing cells are shown from three independent samples. PKM2 knockdown efficiency and re-expression were determined by western blot. Error bars represent ±SD for triplicate experiments.
(B) H1299/PKM2K305Q cells accumulate NADPH. Cells used in the assay were the same as in (A). NADPH was measured using a NADPH kit according to the manufacturer’s protocol. Error bars represent ±SD for triplicate experiments.
PKM2 Mutant Promotes Cell Proliferation and Tumor Growth
Given the high frequency of PKM2 expression in tumor cells, we next examined the effect of mutation at K305 of PKM2 on cell proliferation and tumor growth. We first verified the knocking down of endogenous PKM2 and stable ectopic expression of both wild-type and K305Q mutant PKM2 (Figure 6A). We found that H1299/PKM2K305Q cells proliferated twice as fast as the H1299/PKM2 cells (Figure 6B), indicating a growth advantage conferred by the acetyl mimetic substitution at K305.
Figure 6. PKM2 K305Q Promotes Cell Proliferation and Tumor Growth.
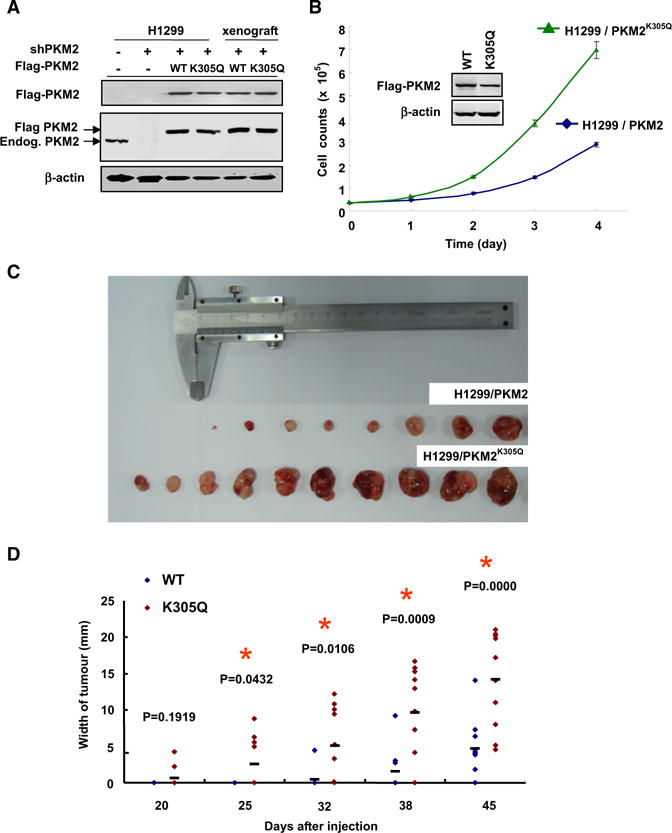
(A) The expression of PKM2 and PKM2K305Q in xenograft. Whole cell tracts were prepared from either original stable H1299/PKM2 and H1299/PKM2K305Q pools or xenograft tumors, followed by western blot.
(B) PKM2K305Q promotes cell proliferation. 3 × 104 H1299/PKM2 or H1299/PKM2K305Q cells were seeded in each well. Cell numbers were counted every 24 hr. Error bars represent ±SD for triplicate experiments.
(C and D) PKM2K305Q mutant promotes xenograft tumor growth. Nude mice injected on the left side with H1299/PKM2K305Q cells and the right side with H1299/PKM2 cells. The xenograft tumors were measured over time and dissected at the endpoint and shown as H1299/PKM2 (upper row) and H1299/PKM2K305Q (lower row) in (C). Quantification of average width of tumors over time is shown in (D). Error bars represent ±SD for ten tumors.
To determine whether PKM2K305Q mutant also rendered growth advantage to tumor cells in vivo, we performed xenograft studies. Three million H1299/PKM2 or H1299/PKM2K305Q cells were injected into nude mice, and tumor cell growth was monitored over a period of around 7 weeks. Sustained expression of Flag-tagged wild-type and K305Q mutant PKM2 in xenograft tumors was verified by western blot analysis using the antibody specific to Flag and PKM2 (Figure 6A). As shown in Figure 6C, larger tumors were developed in mice injected with H1299/PKM2K305Q than in those injected with H1299/PKM2 cells. Measurement of the width of tumor demonstrates that H1299/PKM2K305Q cells give rise to significantly larger tumors than the H1299/PKM2 cells (Figure 6D). Taken together, these results suggested that substitution of K305 with an acetylation mimetic residue confers tumors cell growth advantage both in vitro and in vivo.
DISCUSSION
Metabolic alteration in cancer cells has emerged as an exciting topic in cancer research. It is particularly noteworthy that cancer cells display a robust increase in glucose uptake and a higher demand of metabolic intermediates for biosynthesis to support rapid cell growth. Among the glycolytic enzymes, pyruvate kinase is unique in that it shows a cancer-specific switch of expression from PKM1 to PKM2; the latter is normally reserved for expression in rapidly proliferating embryonic cells. We have observed that PKM2 is acetylated in cancer cells and that the K305 acetylation inhibits PKM2 by two mechanisms. Acetylation reduces enzymatic activity, likely due to its decreased affinity toward the substrate PEP, and destabilizes PKM2 via HCS70-mediated CMA.
The eukaryotic cell uses two main degradation systems, the proteasome and the lysosomes, to regulate protein stability (Ciechanover, 2005). The proteasome usually degrades short-lived proteins, while the lysosome targets long-lived proteins. Among the three types of lysosome-dependent degradation (macroautophagy, microautophaqgy, and CMA), CMA is unique that it shows a selective degradation of target proteins (Levine and Kroemer, 2007; Massey et al., 2004). The selectivity is achieved through the binding of HSC70 to a KFERQ-like motif present in target proteins. It is estimated that approximately 30% of cytosolic soluble proteins contain KFERQ-like motifs (Dice et al., 1990), indicating a potential broad role of CMA in regulating protein stability.
A key step in CMA regulation is the interaction between chaperone and target proteins. For PKM2, it appears that acetylation plays a key role in regulating the interaction between HSC70 and PKM2. The acetylated PKM2 displays a stronger interaction with HSC70. For example, treatment with deacetylase inhibitors, NAM and TSA, significantly increases the interaction between HCS70 and PKM2. Moreover, HSC70 selectively coprecipitates the acetylated PKM2. Furthermore, acetylated PKM2 displays an enhanced interaction with lysosomes. Although the precise mechanism of acetylation in promoting the interaction between PKM2 and HSC70 is unclear, we have identified that the 393QLFEE398 is apparently essential for the interaction with HSC70. We speculate that acetylation of K305 may cause conformational changes in PKM2, thus the 393QLFEE398 sequence in the acetylated PKM2 becomes accessible for recognition by HSC70. Our study provides the first example that acetylation regulates CMA by promoting the interaction between chaperone and target proteins.
Christofk et al. have suggested that PKM2 is important for cancer cell growth. A unique feature of the pyruvate kinase is that it acts at the end of the glycolytic pathway and that its product, pyruvate, can be metabolized into two major directions. Pyruvate can be reduced to produce lactate, which is often accumulated in cancer cells, or oxidized to acetyl-coA, which can be fed into the TCA cycle for energy production. An inhibition of pyruvate kinase will result in accumulation of glycolytic intermediates, which could be used for biosynthesis for cell growth, if glucose is available. In contrast, an inhibition of enzymes in earlier steps, such as hexokinase, would not accumulate glycolytic intermediates, thus the cell would not accumulate building materials for biosynthesis. This may potentially explain why cancer cells specifically express PKM2, which is activated by F-1, 6-BP and inhibited by tyrosine phosphorylated proteins (Christofk et al., 2008b; Mazurek et al., 2005). In this study, we present another mechanism of PKM2 inhibition by acetylation. We also demonstrate that expression of acetylation mimetic mutant PKM2 enhanced glycolytic intermediates and NADPH. As a result, the acetylation mimetic mutant PKM2 increases cell proliferation in vitro and tumor growth in vivo.
Glucose regulates PKM2 acetylation and protein levels. What is the physiological significance of this glucose-dependent regulation in cancer cells? PKM2, but not PKM1, has a low activity that can be activated allosterically by FBP (Jurica et al., 1998; Mazurek et al., 2005). When glucose concentration is low, tumor cells would use the limited glucose for oxidative phosphorylation to generate energy for survival. So, under this condition, PKM2 is active due to activation by F-1, 6-BP and lacks acetylation-induced inhibition. When glucose concentration is high, tumor cells need to decrease PKM2 activity in order to accumulate glycolytic intermediates for cell growth. Although high glucose results in a high F-1, 6-BP, PKM2 activity is in check because high glucose induces PKM2 acetylation, which directly decreases PKM2 activity and indirectly stimulates the CMA-dependent degradation. Therefore, together with activation by F-1, 6-BP, PKM2 regulation by glucose-induced acetylation would provide an advantage for tumor cell growth under glucose sufficiency. One may speculate that drugs modulating PKM2 activity or acetylation could have potential value for cancer treatment.
EXPERIMENTAL PROCEDURES
Cell Lysis and Immunological Procedures
Cells were lysed in a NP-40 buffer, and western blot analysis was carried out according to standard methods. Antibodies specific to Flag (Sigma), HSC70 (Abcam), Lamp2A (Abcam), HA (Santa Cruz), actin (Sigma), and PSA (Bio-world) were purchased commercially. Antibodies to pyruvate kinase and acetyl-PKM2(K305) were generated by immunizing rabbits with antigen peptides at Shanghai Genomic Inc.
Measurement of Pyruvate Kinase Activity
Pyruvate kinase activity was measured by a continuous assay coupled to lactate dehydrogenase (LDH). The change in absorbance resulting from NADH oxidation was measured using a F-4600 Fluorescence Spectrophotometer (HITACHI). Assays for PK activity were carried out as previously described (Christofk et al., 2008a).
Lysosome Binding and Uptake Assay
Lysosomes were isolated by following previously described procedures (Cuervo et al., 1995). Both lysosome binding and uptake assays were carried out as described previously (Cuervo et al., 2004).
Short Hairpin RNA Constructs and Retroviral Production
A shRNA retrovirus targeting pyruvate kinase was constructed and retroviruses were produced using a two-plasmid packaging system as previously described (Christofk et al., 2008a).
Putting Back
Flag-tagged human PKM2 and PKM2K305Q containing two silent nucleotide substitutions in the sequence corresponding to the shRNA-targeted region were cloned into the retroviral vector (pQCXIH) and were cotransfected into 293T cells together with vectors expressing the gag and vsvg genes. Retroviral supernatant was harvested 36 hr after initial plasmid transfection and mixed with polybrene (8 μg/ml) to increase the infection efficiency. H1299 cells were infected with retrovirus and selected in hygromycin (350 μg/ml) for 2 weeks.
Cell Proliferation Analysis
3 × 104 cells were seeded in triplicate in 6-well plates, and cell numbers were counted every 24 hr over a 5 day period.
Measurement of Metabolites
To extract metabolites, 2 × 107 cells was directly chilled into 2 ml ice-cold 80% ethanol containing 0.1% formic acid, followed by a centrifugation at 10,000 rpm for 20 min at 4°C. The supernatant was collected and dried in Concentrator 5301 (Eppendorf). The dried extract was dissolved in 200 μl 20% acetonitrile, and the insoluble material was removed by centrifugation (10,000 rpm for 20 min at 4 C). Of the supernatant, 20 μl was used to for LC-MS analysis.
NADPH Assay
The assays were carried out according to the protocol of NADP/NADPH Assay Kit (BioVision).
Xenograft Studies
Nude mice (nu/nu, male 6- to 8-week-old) were injected subcutaneously with 3 × 106 H1299 cells. Around 7 weeks after injection, the tumors were dissected and weighed.
Immunohistochemistry
Prostate samples were acquired from Affiliated Renji Hospital of Jiaotong University. A physician obtained informed consent from the patients. The procedures related to human subjects were approved by Ethic Committee of the Institutes of Biomedical Sciences (IBS), Fudan University. IHC was performed as previously described (Lei et al., 2006).
Measurement of Glycolysis Rate
Glycolysis rate was measured by monitoring the conversion of 5-3H-glucose to 3H2O, as described previously (Liang et al., 1997).
Supplementary Material
Acknowledgments
We thank the members of the Fudan MCB laboratory for discussions throughout this study. PcDNA/Zeo(−)-antisense-Hsc70 was kindly provided by Drs. Janice S. Blum and Zixu Mao. This work was supported by 973 (grant numbers 2009CB918401, 2011CB910600), NCET (grant number 09-0315), and NSFC (grant numbers 30600112, 30871255, 31071192). This work was also supported by the 985 Program and Shanghai key project (grant number 09JC1402300); 100 Talents Programme of Shanghai Health; the Shanghai Leading Academic Discipline Project, project number B110; and National Institutes of Health (NIH) grants (Y. Xiong and K.-L.G.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and two tables and can be found with this article online at doi:10.1016/j.molcel.2011.04.025.
References
- Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol Cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008a;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008b;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab. 2009;21:142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2009;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Dice JF, Terlecky SR, Chiang HL, Olson TS, Isenman LD, Short-Russell SR, Freundlieb S, Terlecky LJ. A selective pathway for degradation of cytosolic proteins by lysosomes. Semin Cell Biol. 1990;1:449–455. [PubMed] [Google Scholar]
- Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- Funes JM, Quintero M, Henderson S, Martinez D, Qureshi U, Westwood C, Clements MO, Bourboulia D, Pedley RB, Moncada S, Boshoff C. Transformation of human mesenchymal stem cells increases their dependency on oxidative phosphorylation for energy production. Proc Natl Acad Sci USA. 2007;104:6223–6228. doi: 10.1073/pnas.0700690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumai R, Komatsu Y, Nishino N, Khochbin S, Yoshida M, Horinouchi S. Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc Natl Acad Sci USA. 2001;98:87–92. doi: 10.1073/pnas.011405598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007a;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007b;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Then F, Melia TJ, Jr, Mazzulli JR, Cui L, Savas JN, Voisine C, Paganetti P, Tanese N, Hart AC, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Mesecar A, Heath PJ, Shi W, Nowak T, Stoddard BL. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure. 1998;6:195–210. doi: 10.1016/s0969-2126(98)00021-5. [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Lei Q, Jiao J, Xin L, Chang CJ, Wang S, Gao J, Gleave ME, Witte ON, Liu X, Wu H. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 2006;9:367–378. doi: 10.1016/j.ccr.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2007;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Buettger C, Berner DK, Matschinsky FM. Chronic effect of fatty acids on insulin release is not through the alteration of glucose metabolism in a pancreatic beta-cell line (beta HC9) Diabetologia. 1997;40:1018–1027. doi: 10.1007/s001250050783. [DOI] [PubMed] [Google Scholar]
- Liebermeister W. Predicting physiological concentrations of metabolites from their molecular structure. J Comput Biol. 2005;12:1307–1315. doi: 10.1089/cmb.2005.12.1307. [DOI] [PubMed] [Google Scholar]
- Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, Manola J, Brugarolas J, McDonnell TJ, Golub TR, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A, Kiffin R, Cuervo AM. Pathophysiology of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2420–2434. doi: 10.1016/j.biocel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A, Wang C, Schreiber SL. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci USA. 2005;102:5992–5997. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol Cell Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wing S, Chiang HL, Goldberg AL, Dice JF. Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but notskeletal muscle, of fasted rats. Biochem J. 1991;275:165–169. doi: 10.1042/bj2750165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, Mao Z. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


