SUMMARY
Many DNA and RNA regulatory proteins contain polypeptide domains that are unstructured when analyzed in cell lysates. These domains are typified by an over-representation of a limited number of amino acids and have been termed prion-like, intrinsically disordered or low complexity (LC) domains. When incubated at high concentration, certain of these LC domains polymerize into labile, amyloid-like fibers. Here we report methods allowing the generation of a molecular footprint of the polymeric state of the LC domain of hnRNPA2. By deploying this footprinting technique to probe the structure of the native hnRNPA2 protein present in isolated nuclei, we offer evidence that its LC domain exists in a similar conformation as that described for recombinant polymers of the protein. These observations favor biologic utility to the polymerization of LC domains in the pathway of information transfer from gene to message to protein.
INTRODUCTION
DNA and RNA regulatory proteins are composed of two functional domains. Gene specific transcription factors contain DNA binding domains that recognize specific sequences via structurally ordered states including zinc fingers, homeoboxes, helix-loop-helix domains and leucine zipper domains (Pabo and Sauer, 1992). Likewise, RNA binding proteins are able to bind RNA via structurally ordered KH domains, RNA recognition motifs and pumilio domains (Lunde et al., 2007).
Most DNA and RNA regulatory proteins also contain polypeptide domains that lack structural order when purified from cellular lysates. The unstructured activation domains of certain transcription factors contain an over-representation of acidic amino acids (Hope et al., 1988). In the context of gene specific transcription factors, these structurally disordered domains have been termed “acid blobs” or “negative noodles” (Sigler, 1988), and other conceptualizations invoking biological function in the absence of folded protein structure.
Not all activation domains associated with gene specific transcription factors are acidic. Some are enriched in glutamine residues, others in proline residues (Triezenberg, 1995). Common, however, among the majority of activation domains is the over-representation of one or a small grouping of amino acids. Instead of utilizing a balanced proportion of all 20 amino acids, these domains are of low complexity in nature. Nucleic acids deploy a four-lettered code, proteins a 20 letter code. LC domains operate via the deployment of a highly skewed distribution of amino acids, and would appear to be much more DNA- and RNA-like in the nature of their code.
RNA binding proteins also contain LC domains, including repetitive polymers of serine and arginine (SR) in many proteins that regulate pre-mRNA splicing (Manley and Tacke, 1996), and G/S-Y-G/S repeats in the LC domains associated with the FET, CIRBP/RBM3 and hnRNP families of RNA binding proteins (Kato et al., 2012). Compared with gene-specific transcription factors and their activation domains, less attention has been paid to the LC domains associated with RNA regulatory proteins. Some degree of attention has been focused on the LC domains associated with the FET family of RNA binding proteins, including fused in sarcoma (FUS), Ewing’ sarcoma (EWS) and TAF15. The amino terminal LC domains of these three proteins can be translocated onto DNA binding domains as the causative event in many forms of human cancer (Riggi et al., 2007). In the context of these fusion proteins, the LC domains of the FET proteins are understood to function as potent transcriptional activation domains.
Several years ago we inadvertently observed polymerization of the LC domains of the FET proteins as well as certain hnRNP proteins (Kato et al., 2012). When incubated at high concentration, the LC domains of these proteins polymerize into amyloid-like fibers. A combination of X-ray diffraction and electron microscopy gave evidence that the fibers were of the prototypic cross-β structure first described 50–60 years ago by Astbury (Astbury et al., 1959). Unlike irreversible, pathogenic amyloids, the fibers polymerized from LC domains present in FUS, TAF15 and hnRNPA2 are readily disassembled upon dilution. By comparing the effects of mutations in the LC domains of FUS and TAF15 on both transcription activation capacity and polymerization, a strong correlative relationship gave evidence that polymerization might be of critical importance to the function of these domains in living cells (Kwon et al., 2013).
Heretofore missing from this line of research was any evidence indicative of the structure of LC domains in their native state. In attempts to address this shortcoming, we developed a chemical probing strategy that allows generation of a footprint indicative of ordered structure. After having validated the utility of the approach using two enzymes of known structure, we deployed the footprinting strategy on fibrous polymers of the LC domain of hnRNPA2. Our observations give evidence that the LC domain of hnRNPA2 exists in the same structural state in both recombinant polymers of the protein and native hnRNPA2 within the nuclear compartment of mammalian cells.
RESULTS
Development of a Chemical Footprinting Method
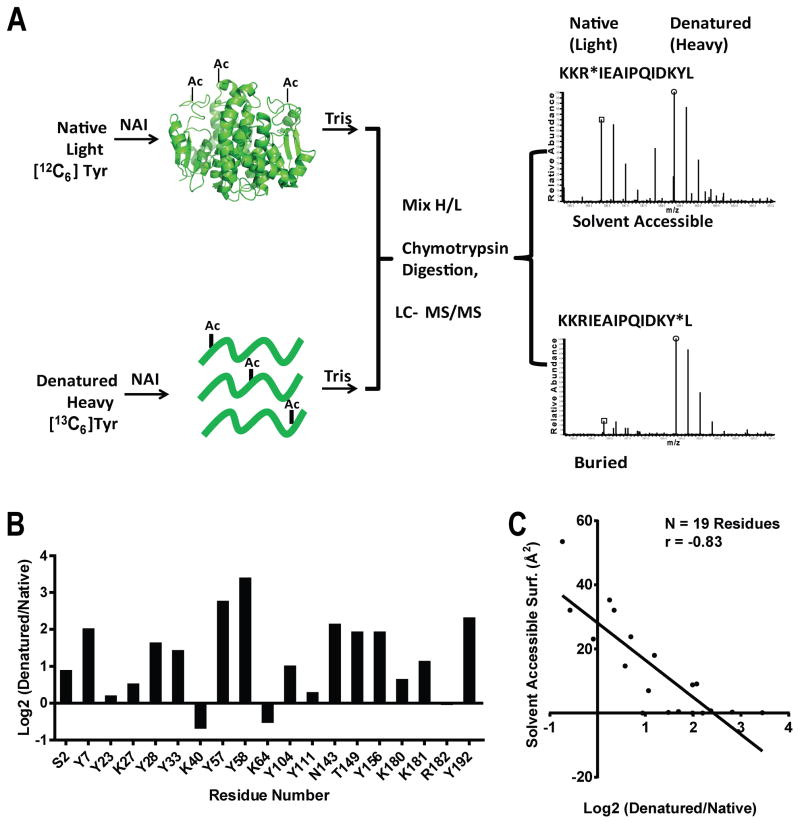
N-acetylimidazole (NAI) is a reactive chemical that is capable of acetylating certain amino acid side chains in proteins (Riordan et al., 1965; Timasheff and Gorbunoff, 1967). Under conditions of neutral pH the chemical can donate an acetyl group to serine, tyrosine, lysine, threonine, arginine and asparagine side chains. Reasoning that the ability of NAI to modify amino acids might be influenced by the structural state of a protein, we compared modification of glutathione-S-transferase (GST) as a function of its folded versus unfolded state. GST enzyme was prepared under conditions of isotopic labeling with 13C-labeled tyrosine to produce a “heavy” protein sample. This sample was denatured with a chaotropic reagent and exposed to NAI under conditions leading to roughly one modification per polypeptide. The reaction was quenched with 0.8M Tris, pH 8.8 which inactivates the NAI reagent. A corresponding “light” sample of GST was exposed to NAI in its folded state, again under conditions resulting in roughly one modification per polypeptide, and again quenched with Tris to terminate the reaction. The heavy and light samples were mixed at a 1:1 ratio, digested with chymotrypsin, then evaluated by SILAC mass spectrometry (Figure 1A; Experimental Procedures).
Figure 1. Differing Patterns of Acetylation of Folded and Denatured Samples of Glutathione-S-transferase Mediated by N-acetylimidazole.
A. Folded glutathione-S-transferase (GST) was exposed to N-acetylimidazole (NAI) under conditions leading to roughly one modification per polypeptide chain, with the reaction quenched by the addition of 0.8 M Tris. A separate batch of GST grown in bacterial cells supplemented with 13C-labeled tyrosine was denatured in 5 M guanidine thiocyanate prior to NAI treatment. Following quenching with Tris, the two samples were mixed, digested with chymotrypsin and subjected to SILAC mass spectrometry.
B. Nineteen acetylated side chains were scored for abundance in the two samples, yielding an NAI footprint. The degree of residue protection from NAI modification in the folded state, relative to the denatured state, is measured on the Y axis as log2 values.
C. Plot showing the correlative relationship between the degree of protection from NAI in the folded state, relative to the denatured state (X-axis), and the measured level of solvent accessibility determined from the X-ray crystal structure of GST (Y-axis).
See also Figure S1 and Table S1, S2.
The patterns of NAI reactivity with the denatured and folded states of GST were different. Certain amino acid side chains reacted similarly in the two protein samples (Y23, K27, K40, K64 and Y111), whereas others were acetylated to a lesser extent in the folded sample compared with denatured GST (Y7, Y57, Y58 and Y192). The NAI “footprint” of GST is shown in Figure 1B. We then compared this footprint with the degree of surface exposure of NAI-modified side chains as deduced from the X-ray crystal structure of the enzyme (Rufer et al., 2005) (PDP ID: 1Y6E). A strong correlative relationship was observed between NAI accessibility and solvent exposure in the structure (Figure 1C and Table S1). Surface-exposed residues tended to be NAI-accessible, whereas residues buried within the core of the enzyme tended to be NAI-inaccessible. We conclude that the correlative match between NAI-accessibility and protein structure gives evidence that the NAI footprint is properly reflective of protein structure.
Proceeding from a recombinant protein sample to a native protein within mammalian nuclei, we evaluated the difference in NAI modification of the poly-ADP-ribose polymerase (PARP) enzyme as a function of its folded versus denatured state. Nuclei were prepared from 293T cells that had been grown in either normal tissue culture medium (light) or medium deprived of tyrosine and supplemented with an isotopically labeled form of the amino acid (heavy). The light sample of nuclei was exposed to a 30mM level of NAI for 15 minutes before quenching with Tris. The heavy sample was denatured in 5 M guanidine thiocyanate prior to exposure to the same level of NAI, then also quenched with Tris. The samples were combined, digested with chymotrypsin overnight, and processed by mass spectrometry (Experimental Procedures).
NAI modification was monitored on 14 amino acid side chains in the native and denatured forms of PARP (Figure S1A). Six residues were modified by NAI far more extensively in the denatured sample than the intact enzyme (K621, T799, K802, Y817, S902 and S904), five residues were modified slightly more extensively in the denatured sample relative to the intact enzyme (K571, S782, S783, S808 and K816), and three residues were modified equally in the two samples (K616, K903 and Y907). We again observed a correlation between NAI accessibility and protein structure (PDB ID: 3GJW) (Figure S1B and Table S2). The three side chains that were modified equally in the two samples show a high level of predicted solvent accessibility in the X-ray crystal structure of PARP (Miyashiro et al., 2009). Likewise, five of the six residues observed to be highly protected from NAI modification are predicted to be solvent inaccessible by the crystal structure of the enzyme.
Analysis of three consecutive residues in the polypeptide chain of PARP is particularly revealing. Serine residue 902 is protected from NAI modification in nuclear PARP and buried beneath the surface of the enzyme. Lysine residue 903 is surface exposed and NAI accessible in the folded form of PARP. Finally, serine residue 904 is NAI inaccessible in the folded enzyme, and buried beneath the surface of the PARP crystal structure. We offer that the correlative relationship between NAI accessibility and the predicted level of surface exposure of a given amino acid side chain validates this means of probing protein structure both in a recombinant protein and a native enzyme present in nuclei of mammalian cells.
Determination of the NAI Footprint of Recombinant hnRNPA2 Fibers
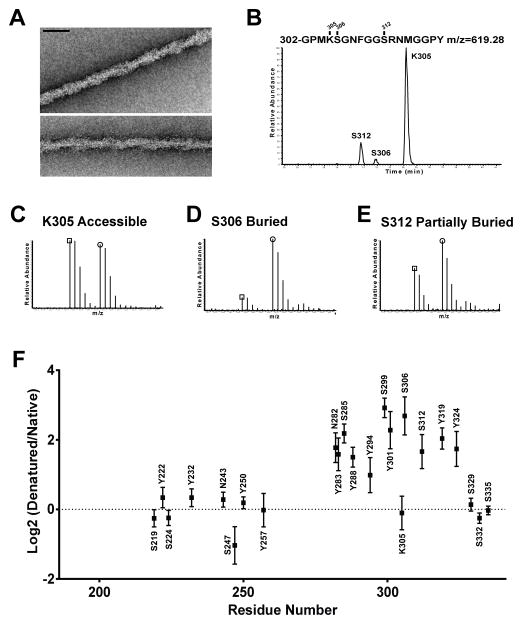
Hydrogel droplets were formed using a fusion protein linking mCherry to the LC domain of hnRNPA2 (Kato et al., 2012). This protein sample was exposed to NAI under conditions resulting in roughly one modification per polypeptide chain, then quenched with Tris (Experimental Procedures). Similarly prepared hnRNPA2 polymers were formed using protein isotopically labeled with heavy tyrosine. The latter sample was denatured with guanidine thiocyanate prior to NAI-mediated modification, followed by quenching with Tris. The two samples were combined at a 1:1 ratio, digested with chymotrypsin, then analyzed by mass spectrometry (Experimental Procedures).
Twenty-three amino acid side chains were evaluated for NAI accessibility. Twelve amino acids appeared to be equally accessible to NAI-mediated modification in the two samples, and eleven appeared to be less accessible in the native fibers relative to the denatured protein sample (Figure 2). Three of these acetylated amino acid residues could be identified in the same peptide spanning amino acids 302–319 of the hnRNPA2 polypeptide. HPLC chromatography was successful in separating variants of this peptide acetylated at lysine 305, serine 306, or serine 312 (Figure 2B). The peptide variant acetylated at K305 was found at equal abundance in both light and heavy samples, indicative of the ability of NAI to modify this residue irrespective of whether the protein was in the fibrous or denatured state. The variant acetylated at S306 was considerably less abundant in the light sample than the heavy sample, giving evidence of its protection from NAI modification in the fibrous state. Finally, the variant acetylated at S312 was slightly less abundant in the light sample relative to the heavy sample, consistent with partial protection from NAI modification when the LC domain of hnRNPA2 existed in the polymeric state.
Figure 2. Footprint of NAI-mediated Acetylation of Recombinant hnRNPA2 Polymeric Fibers.
A. Electron micrographs of negatively stained polymeric fibers formed from an mCherry:hnRNPA2 fusion protein (Experimental Procedures). Scale bar: 70 nm.
B. HPLC separation of chymotryptic digestion products of the LC domain of hnRNPA2 corresponding to residues 302–319. The S312 acetylated peptide eluted earlier from the column than the S306 acetylated peptide, which – in turn – eluted earlier than the K305 acetylated peptide (Experimental Procedures).
C. Relative abundances of the K305 acetylated peptides in folded versus denatured samples.
D. Relative abundances of the S306 acetylated peptides in folded versus denatured samples.
E. Relative abundances of the S312 acetylated peptides in folded versus denatured samples.
F. NAI footprint of the LC domain of hnRNPA2 (All data are presented as means ± SD).
The pattern of protection from NAI modification in polymeric fibers of the hnRNPA2 LC domain, or lack thereof, can be described in the following way. An extensive, N-terminal region of the protein was equally acetylated by the chemical probe irrespective of the fibrous or denatured state. An equally extensive segment corresponding to a more C-terminal region of the LC domain was protected in the polymeric state, relative to the denatured state, at 11 out of 12 acetylated residues. Right within the middle of this apparently ordered region of the LC domain, lysine residue 305 was found to be equally accessible in both the polymeric and denatured states of the protein. Finally, the three most C-terminal residues scored in the assay were all equally accessible under both fibrous and denatured states.
Relationship of hnRNPA2 Footprints Between Recombinant and Nuclear Forms of the Protein
Using the same methods described for determining an NAI footprint for the nuclear form of the PARP enzyme (Figure S1), we probed the structure of native hnRNPA2 present in nuclei freshly prepared from 293T cells (Experimental Procedures). Isotopically labeled heavy protein was probed under the denaturing conditions of 5 M guanidine thiocyanate. Light protein was probed via the exposure of nuclei to the NAI chemical reagent. Following quenching with Tris the samples were mixed, digested with chymotrypsin and evaluated by mass spectrometry.
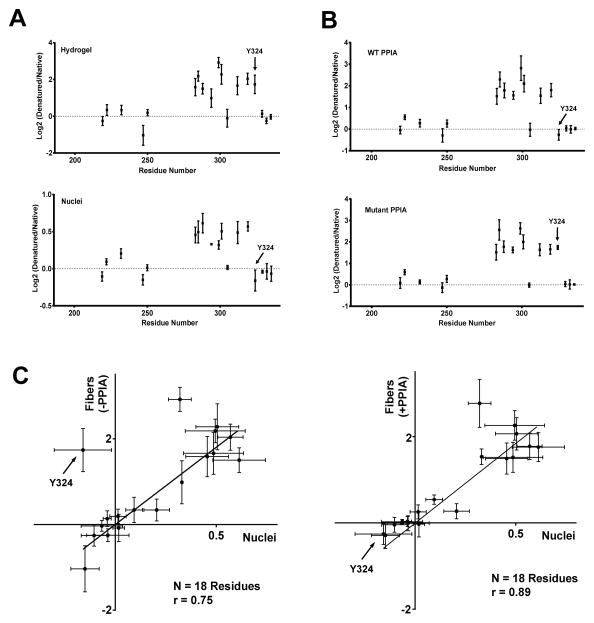
The NAI footprint observed for native, nuclear hnRNPA2 could be scored for 18 of the 23 acetylated residues observed in the footprint derived from recombinant hnRNPA2, and the two footprints were qualitatively similar (Figure 3A). Of the acetylation events detected in both footprints, all nine residues that were equally accessible to NAI-mediated acetylation in both polymeric and denatured samples of recombinant hnRNPA2 protein were also acetylated equally in the native hnRNPA2 irrespective of structural state. Seven of the eight residues that were preferentially protected from acetylation as a function of the fibrous state of recombinant hnRNPA2 protein were also preferentially protected in the native hnRNPA2 protein relative to nuclear protein that had been denatured with 5M guanidine thiocyanate. The single qualitative difference between the two footprints was tyrosine residue 324. This residue was preferentially protected from NAI-mediated acetylation in the fibrous form of recombinant hnRNPA2, yet was equally accessible to the chemical probe in native hnRNPA2 irrespective of whether nuclei were left intact or denatured.
Figure 3. NAI Footprints of the LC Domain of hnRNPA2 Deduced from Recombinant Protein, Native Nuclear hnRNPA2, and Recombinant Protein Co-expressed with Peptidyl-prolyl Cis-trans Isomerase (PPIA).
A. NAI footprint of recombinant hnRNPA2 fibers as described in Figure 2 (upper footprint) compared with NAI footprint deduced from native, nuclear hnRNPA2 (lower footprint). Note that tyrosine 324 is protected from NAI modification in the folded form of hnRNPA2 in the recombinant form of hnRNPA2, but not in the footprint deduced from the native, nuclear protein.
B. NAI footprint of recombinant hnRNPA2 co-expressed with active PPIA enzyme (upper footprint) compared with footprint of hnRNPA2 co-expressed with a catalytically inactive form of the enzyme (lower footprint). Note that co-expression of hnRNPA2 with the active form of PPIA causes tyrosine 324 to become exposed to NAI modification in the polymeric state.
C. Plots showing the correlative relationship of the NAI footprint of recombinant hnRNPA2 to that of the native, nuclear form of the protein. Correlation plot on left compares the footprint of recombinant hnRNPA2 not exposed to the PPAI enzyme with the nuclear hnRNPA2 footprint. Correlation plot on right compares the footprint of recombinant hnRNPA2 co-expressed with the active PPIA enzyme with the nuclear hnRNPA2 footprint.
See also Figure S3.
Despite displaying qualitative similarities, the NAI-generated footprints for recombinant and native hnRNPA2 differed quantitatively in a consistent manner. The NAI protected residues observed in recombinant hnRNPA2 yielded an average of roughly 3-fold (log2 ~1.8) difference when comparing peptide abundance in the light (fibrous) and heavy (denatured) samples. Turning to the native hnRNPA2 assayed in either intact or denatured nuclei, the average difference in peptides revealing NAI protected residues was roughly 1.5-fold (log2 ~0.5). Interpreted most simply, this difference gives indication that a smaller fraction of the native hnRNPA2 present in nuclei may exist in the structurally ordered state than the fraction deduced by studies of recombinant hnRNPA2 polymeric fibers.
Co-expression of hnRNPA2 with Peptidyl-prolyl Cis-trans Isomerase Causes Tyrosine 324 to Become NAI-accessible in Recombinant Polymers
The NAI footprint observed in recombinant hnRNPA2 polymeric fibers was qualitatively similar to that observed for native hnRNPA2 in intact nuclei. Among 18 residues defining the footprint, tyrosine 324 was the single amino acid that was clearly different in the two samples. This residue was protected from NAI-mediated acetylation in fibrous preparations of recombinant hnRNPA2, but not in the native hnRNPA2 present in intact nuclei.
Proline residues are found six positions on the amino terminal side of tyrosine 324, and two positions on its carboxyl terminal side (Figure S2). Proteomic studies of cellular proteins that bind to hydrogel droplets formed from the LC domains of both hnRNPA2 and FUS revealed retention of PPIA, the most abundant isoform of a family of peptidyl-prolyl cis-trans isomerase enzymes. PPIA has been reported to interact with RNA granule proteins upon biochemical fractionation (Lauranzano et al., 2015), and antibodies to the enzyme revealed co-localization with stress granules (Figure S3). We thus reasoned that the PPIA enzyme might affect the structure of hnRNPA2 fibers by facilitating cis-trans interconversion of the peptide bonds of proline residue 319 or 326 of the hnRNPA2 polypeptide.
To test this hypothesis mCherry:hnRNPA2 was co-expressed with either the native form of PPIA or a catalytically inactive mutant (Zydowsky et al., 1992). Following purification of the mCherry:hnRNPA2 protein, polymeric fibers were formed and exposed to the NAI probe under either the polymeric or denatured state. Co-expression of hnRNPA2 with the active form of PPIA yielded an NAI footprint wherein tyrosine residue 324 was equally accessible to acetylation irrespective of fibrous or denatured state (Figure 3B, top). By contrast, co-expression with the catalytically inactive form of PPIA yielded a footprint indistinguishable from that seen on recombinant hnRNPA2 never exposed to the enzyme (Figure 3B, bottom).
The bottom panel of Figure 3 (Figure 3C) correlatively compares the NAI footprints of hnRNPA2 observed in native protein within intact nuclei with that of recombinant protein expressed in either the absence or presence of PPIA. The r value of correlation of the native and recombinant footprints was 0.76, which increased to 0.89 when the recombinant hnRNPA2 had been co-expressed with PPIA.
Mutations in the NAI-protected Region of the hnRNPA2 LC Domain Impede Hydrogel Binding
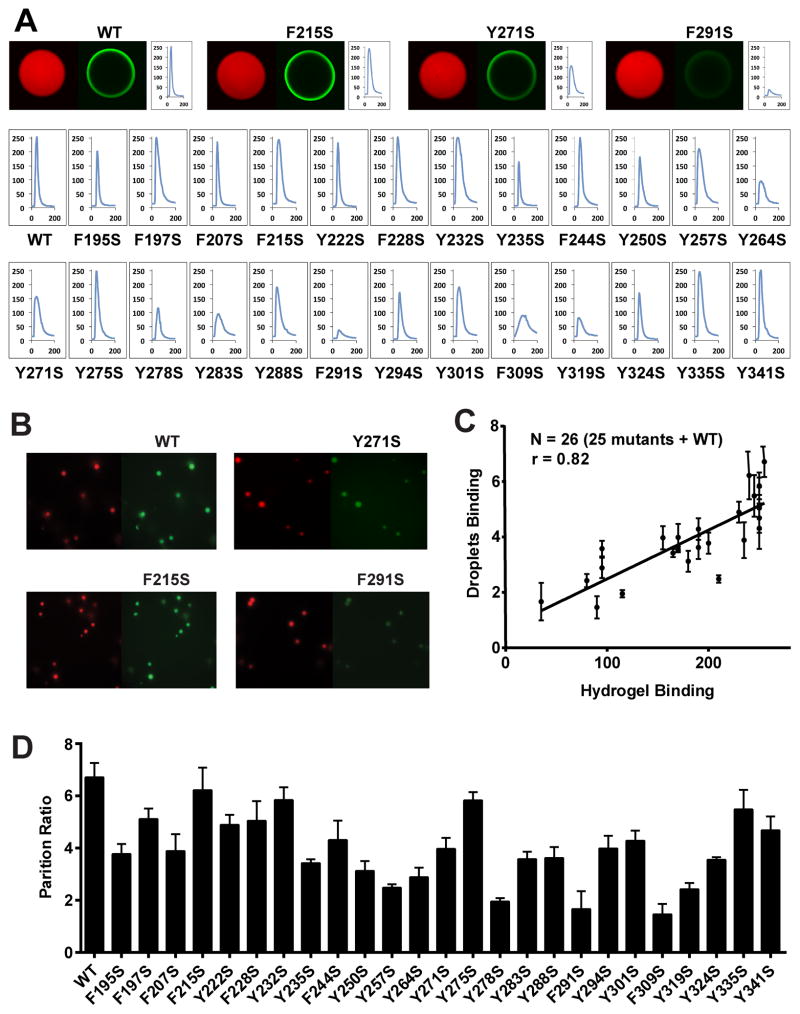
Is the NAI footprint telling us anything of functional relevance to the LC domain of hnRNPA2? To address this question we prepared mutated variants of the LC domain of hnRNPA2 wherein all 25 phenylalanine and tyrosine residues were individually mutated to serine (Figure S2). GFP fusion proteins representing wild type hnRNPA2 and all of the individual mutants were expressed in bacterial cells, purified and assayed for the ability to adhere to mCherry:hnRNPA2 hydrogel droplets (Figure 4A).
Figure 4. Correlative Relationship Between Binding of Mutated Variants of the LC Domain of hnRNPA2 to Hydrogels Relative to Their Partitioning into Liquid-like Droplets.
A. All phenylalanine and tyrosine residues within the LC domain of hnRNPA2 were individually mutated to serine, expressed as GFP fusion proteins, purified and tested for binding to mCherry:hnRNPA2 hydrogel droplets (Experimental Procedures). Top figures show images of hydrogel binding by GFP linked to the native LC domain of hnRNPA2 (WT), the F215S mutant, the Y271S mutant and the F291S mutant. Confocal images were scanned to yield the signal intensity of bound GFP (Experimental Procedures), yielding the 26 scans in the lower part of the figure. X-axis indicates the scanned distance in μm, and Y-axis indicates the GFP signal intensity in arbitrary units.
B. Liquid-like droplets formed upon binding of a PTB:hnRNPA2 fusion protein to a synthetic RNA containing five copies of the PTB recognition sequence (Experimental Procedures. See also Figure S4). The presence of a SNAP tag allowed the PTB:hnRNPA2 fusion protein to be appended with a red dye. When exposed to GFP alone, no partitioning into liquid-like droplets was observed (data not shown). When exposed to GFP fused to the native LC domain of hnRNPA2 (WT), clear evidence of partitioning was observed within minutes. Certain phenylalanine- or tyrosine-to-serine mutants partitioned well into liquid-like droplets (F215S), whereas others did not (Y271 and F291S).
D. Partitioning into liquid-like droplets was quantified for all phenylalanine- and tyrosine-to-serine mutants that had been constructed and assayed for binding to mCherry:hnRNPA2 hydrogel droplets (A). Histogram shows relative levels of partitioning of GFP linked to the native (WT) LC domain of hnRNPA2 as compared with the 25 individual mutants.
C. Plot showing the correlative relationship between hydrogel binding and partitioning into liquid-like droplets for GFP linked to the native (WT) LC domain of hnRNPA2 along with 25 individual phenylalanine- and tyrosine-to-serine mutants. See also Supplemental Data, Figure S2.
Of the 25 mutants, six were found to substantially impede binding to hydrogel droplets formed from mCherry fused to the wild type LC domain of hnRNPA2. Five of the six tyrosine- or phenylalanine-to-serine mutations that substantially impede hydrogel binding occur within the region of the LC domain that is protected from NAI modification in the fibrous state (Y278S, Y283S, F291S, F309S and Y319S). The sixth mutant that significantly impeded in hydrogel binding, Y264S, occurs on the amino terminal side of the NAI protected region within a span where we failed to find acetylated side chains – a dead zone in the footprint (residues 258–282, Figure 2F). We tentatively conclude that these six residues are particularly important for polymerization of hnRNPA2, and that polymerization causes NAI protection.
The remaining 19 mutants fell into two categories with respect to hydrogel binding. Twelve mutants bound to hydrogels in a manner indistinguishable from wild type hnRNPA2. Two of these mutants, Y335S and Y341S, were located in the very C-terminal region of the LC domain, concordant with a small region that was fully accessible to NAI modification irrespective of whether the protein was in a polymeric or denatured state. Seven of these phenotype-void mutants, F95S, F197S, F207S, F215S, Y222S, F228S and Y250S were located in the amino terminal region of the LC domain that was widely accessible to NAI modification irrespective of structural state. The remaining three mutations that had no discernible effect on hydrogel binding, F244S, Y257S, Y275S, were all localized in the dead zone of the NAI footprint. Finally, seven mutants, including Y235S, Y250S, Y271S, Y288S, Y274S, Y301S and Y324S, mildly affected binding to mCherry:hnRNPA2 hydrogels. These seven mutants mapped randomly across the LC domain of hnRNPA2. We conclude that tyrosine- and phenylalanine-to-serine mutations in NAI protected regions impede hydrogel binding, whereas those in NAI accessible regions do not impede hydrogel binding. This conclusion favors functional significance of the NAI footprint.
Mutations in the LC Domain of hnRNPA2 Act Correlatively on Hydrogel Binding and Partitioning into Liquid-like Droplets
During the preparation of GFP:FUS hydrogel droplets, we have long observed that the concentrated protein solutions become cloudy prior to gelation. Reanalysis of a His6 tagged LC domain of FUS by light microscopy revealed the cloudy solution to be composed of liquid-like droplets (Figure S4). A number of investigators have recently reported that LC domains from a variety of proteins, including FUS, hnRNPA1 and DDX4, can prompt formation of liquid-like droplets (Altmeyer et al., 2015; Lin et al., 2015; Molliex et al., 2015; Nott et al., 2015; Patel et al., 2015).
It is of potential importance to know whether the physical forces leading to hydrogel formation (polymerization of LC domains) are the same or different from those leading to liquid like droplets. To this end we have followed the procedures of Parker and Rosen to create liquid-like droplets driven by the LC domain of hnRNPA2 (Lin et al., 2015). A triple fusion protein was prepared linking the LC domain of hnRNPA2 on the C-terminal side of the PTB RNA binding domain, which was in turn linked to maltose binding protein, with a TEV protease cleavage site between the MBP and PTB domains (Experimental Procedures; Figure S4B). The MBP:PTB:hnRNPA2 LC domain fusion further contained a His6 tag at its C-terminus as well as a SNAP tag for dye-labeling on the N-terminal side of the PTB domain.
Following co-expression with PPIA, purification via Ni+ and amylose resin chromatography, the protein was mixed with a synthetic RNA containing five copies of a PTB binding site and exposed to TEV protease. Within ten minutes liquid-like droplets could be observed by light microscopy (Figure 4B). We then deployed a droplet partitioning assay to assess whether GFP:hnRNPA2 LC domain fusion proteins could be incorporated into the liquid-like droplets. Recombinant GFP-alone protein was not enriched in these liquid-like droplets relative to the surrounding buffer. The GFP fusion linked to the wild type LC domain of hnRNPA2 was rapidly incorporated into liquid-like droplets. Using this assay we evaluated all 25 mutants that had been scored for hydrogel binding (Figures 4B and 4D).
Six mutants were impeded by more than 50% with respect to partitioning into liquid like droplets (Y257S, Y264S, Y278S, F291S, F309S and Y319S), another eight mutants were partially impeded (F195S, F207S, Y235S, Y250S, Y283S, Y288S Y294S and Y301S), and the remaining mutants were incorporated into liquid-like droplets in a manner indistinguishable from the wild type LC domain (Figure 4D). The correlation plot shown in Figure 4C gives evidence of a strong concordance (r = 0.83) between the effects of mutations on hydrogel binding and partitioning into liquid-like droplets. We offer that this concordance gives evidence that similar regions of the protein promote both hydrogel binding and partitioning into liquid like droplets, and that the chemical interactions that drive both processes are likely to be the same.
Liquid-like Droplets Display the NAI Footprint Found in Hydrogel Polymers and Nuclear hnRNPA2
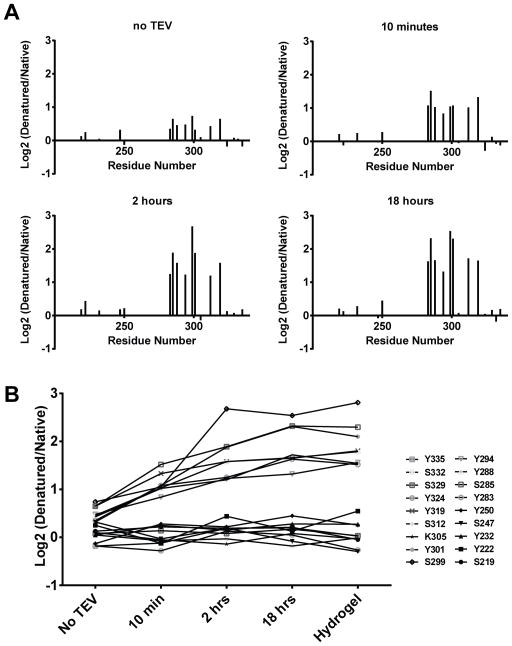
If the mutational effects driving hydrogel binding and liquid-like droplets correlate, it is possible that the LC domain of hnRNPA2 might adopt similar structures in both states. To address this question we performed NAI footprinting on the LC domain of hnRNPA2 in the context of the MBP:PTB:hnRNP LC domain fusion protein before TEV cleavage, immediately upon seeing the formation of liquid-like droplets, two hours after droplet formation, and 18 hours after droplet formation.
As shown in Figure 5, evidence of the canonical NAI footprint on the hnRNPA2 LC domain could be detected even before TEV protease cleavage. The quantitative intensity of the footprint became sequentially enhanced at each of the later time points. Specifically, the degree of difference in NAI protected residues between native and denatured samples was – across all protected residues – most pronounced in the 18 hour sample, less so in the two hour sample, further reduced in the 10 minute sample, and least pronounced in the sample assayed prior to TEV protease cleavage. For the 18 hour time point, the degree of protection from NAI-mediated acetylation of buried side chains was indistinguishable between liquid-like droplets and hydrogels.
Figure 5. Liquid-like Droplets Display the Same NAI Footprint as Found in Hydrogel Polymers and the Native hnRNPA2 Present in Nuclei Freshly Isolated from Mammalian Cells.
A. A fusion protein linking maltose binding protein (MBP) to the RNA binding domains of PTB and the LC domain of hnRNPA2 (Supplemental Data, Figure S4B) was co-expressed with the peptidyl-prolyl cis-trans isomerase enzyme (PPIA), purified, and mixed with a synthetic RNA containing five PTB binding sites. Addition of TEV protease triggered the rapid formation of liquid-like droplets (Figure 4B). Protein samples were footprinted with the NAI reagent as a function of time before and after TEV protease cleavage. Hints of the NAI footprint could be seen in the protein sample before exposure to TEV protease, and the intensity of the footprint was sequentially enhanced at the 10 minute and 2 and 18 hours post-cleavage time points.
B. The log2 ratio of NAI protection for all of the 18 acetylated amino acids is plotted on the Y-axis as a function of time post-exposure to TEV protease (X-axis). See also Supplemental Data Table S3.
Concordant with the observations of others who have studied LC domain partitioning to liquid-like droplets (Lin et al., 2015; Molliex et al., 2015; Patel et al., 2015), we conclude that as a function of time, LC polymerization is progressively enhanced within liquid-like droplets. Gorlich and colleagues have reported similar observations as a function of maturation of liquid-like droplets formed from FG repeats associated with nucleoporin proteins (Petri et al., 2012). In summary, mutational studies of the LC domain of hnRNPA2 give evidence that similar forces drive both hydrogel retention and partitioning into liquid-like droplets, and NAI footprinting studies reveal evidence that the LC domain of hnRNPA2 adopts a similar structure in both settings.
DISCUSSION
Cells display a variety of organized puncta that, unlike mitochondria, lysosomes, chloroplasts and peroxisomes, are not membrane invested. These include various nuclear structures including nucleoli, nuclear speckles and para-speckles, PML bodies, Cajal bodies and histone locus bodies (Mao et al., 2011). Cytoplasmic puncta include RNA granules, P-bodies, neuronal granules, stress granules and the polar granules of fly and worm embryos that assist in determination of the germ lineage (Anderson and Kedersha, 2009). Light microscopic studies of RNA granules have led to the idea that the granule components exist in a liquid-like state separated in phase from the cytoplasm (Brangwynne et al., 2009).
Studies that may be pertinent to the biochemical forces leading to the organization of these cellular structures have begun to appear over the past several years. A potentially common conceptualization may tie two orthogonal approaches together. Rosen and colleagues have provided evidence that multivalent, polymeric structures form when proteins containing repeated SRC homology 3 (SH3) domains are mixed with proteins containing repeated proline-rich motifs (PRMs). Upon heterotypic polymerization into dendritic assemblies, these proteins undergo phase separation into spherical, liquid-like droplets (Li et al., 2012).
Parallel and contemporary to the study of Rosen and colleagues, we have been studying the LC sequences associated with a variety of RNA binding proteins (Han et al., 2012; Kato et al., 2012). In our case, concentrated samples of these proteins have been observed to adopt a gel-like state. Reasonably clear evidence has been gathered to support the conclusion that hydrogel formation equates to polymerization of the LC sequences. Studies of hydrogels have revealed X-ray diffraction patterns consistent with cross-β structure, and electron microscopic evaluation of hydrogels has revealed homogeneous polymeric fibrils.
Of significant concern to us has been the question as to whether the polymeric structures being studied in test tube reactions are of biological relevance. Heretofore any linkage to biological utility has been limited to correlative mutagenesis. One example of this indirect approach to biological significance has been studies of the LC domains of the FET proteins, FUS, EWS and TAF15. All three of these paralogous proteins have amino terminal LC domains that can be translocated onto DNA binding domains as the causative event leading to human cancer. When fused to the DNA binding domain of GAL4, the LC domains of FET proteins function as transcriptional activation domains (Riggi et al., 2007). Unbiased mutagenesis of the LC domains of TAF15 and FUS have yielded scores of mutants that affect polymerization to varying degrees. When tested for their capacity to activate transcription in living cells, a strong correlative relationship was observed with polymerization capacity (Kwon et al., 2013). Mutants fully capable of polymerization activate gene expression potently, mutants mildly impeded in polymerization activate transcription to an intermediate degree, and mutants that are incapable of polymerization fail to activate transcription.
Here we add a more direct approach to inquire whether LC domains might function in cells via the same chemistries and structures leading to LC domain polymerization in test tubes. A footprinting method was developed using N-acetylimidazole (NAI). This chemical acetylates amino acid side chains in a manner influenced by protein structure and can be deployed as a reagent useful both for test tube biochemistry and the probing of native protein within freshly isolated nuclei (Figures 1 and S1). Using this approach we hereby demonstrate that the footprint of the LC domain of hnRNPA2 in recombinant polymers is highly related to the footprint observed in nuclei (Figure 3). These observations are consistent with the conclusion that the LC domain of at least some proportion of hnRNPA2 in nuclei adopts a similar cross-β structure as has been characterized with recombinant polymers.
In considering the virtues and properties of liquid-like droplets as compared with hydrogels, we offer two contrasting perspectives. It is possible that the physical forces leading to the two states are entirely different. Recent studies of the DDX4 protein and LC domains associated with nucleoporin proteins characterized by FG repeats favor the utility of chemical interactions deployed to intertwine otherwise unstructured, random coil LC domains (Nott et al., 2015; Petri et al., 2012). In the case of DDX4, Pi stacking between arginine and phenylalanine residues has been highlighted as a key chemical determinant for phase separation into liquid-like droplets. These interpretations are distinct from the polymerization of LC domains into cross-β structure that we consider to be the driving force for hydrogel formation.
Here we offer the alternative perspective that cross-β polymerization may be at the heart of formation of both hydrogels and liquid-like droplets. By constructing and studying 25 mutated variants of the LC domain of hnRNPA2, we have found mutants that affect hydrogel binding significantly, mildly or not at all (Figure 4A). The former category of mutants mapped almost exclusively to the region of the hnRNPA2 LC domain that was NAI-resistant in the polymeric state (Figure 2). When tested for partitioning into liquid-like droplets, a strong correlative relationship was observed with hydrogel binding (Figure 4C). Mutants strongly impeded in hydrogel retention partitioned poorly into liquid-like droplets, mutants partially impeded in hydrogel retention were mildly impeded from entering liquid-like droplets, and mutants that bound hydrogels as well as the wild type LC domain of hnRNPA2 partitioned effectively into liquid-like droplets.
We likewise deployed the NAI footprinting technique to liquid-like droplets and observed the same footprint as was found in hydrogels composed of the hnRNPA2 LC domain and nuclei freshly isolated from mammalian cells (Figure 5). Although these observations do not rule out the involvement of other chemical or physical forces in the formation of liquid-like droplets, we offer the conclusion that cross-β interactions between LC domains are an important component of the forces facilitating phase separation of LC sequences into liquid-like droplets.
If we adopt the simplistic interpretation that studies of hydrogels and liquid-like droplets are variations on essentially the same theme, one can consider the differences and utilities of the two systems. We reason that the sizes of polymers in hydrogels are much longer than those found in liquid-like droplets, and that the size distribution and dynamics of LC domains in the latter setting may be a better representation of how LC domains function in living cells. The quantitative intensity of the NAI footprint in the various settings deployed in this study may be instructive in this regard. In hydrogel samples, the degree of NAI protection in ordered regions of the protein was roughly 3X that of denatured samples. In cells, the quantitative degree of protection was roughly 1.5X. The NAI footprint observed in freshly prepared liquid-like droplets yielded a quantitative degree of protection more closely matching to that of native hnRNPA2 as probed in isolated nuclei.
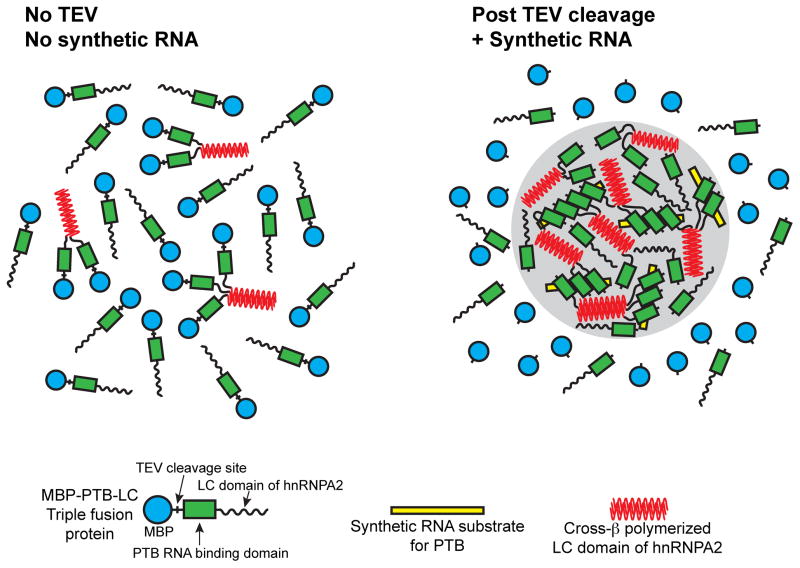
Paradoxically, evidence of the existence of a low level of cross-β structure was seen in samples of the MBP:PTB:hnRNPA2 LC domain triple fusion protein before TEV release of MBP, before exposure to the synthetic RNA containing iterative PTB binding sites, and before formation of liquid-like droplets (Figure 5). As recently articulated by Linse and colleagues, the initial nucleation of amyloid fibers is triggered within micro-seconds of protein mixing during the lag phase of polymerization (Arosio et al., 2015). Transition from lag to growth phase of amyloid polymerization reflects, of course, a profound enhancement of the proportion of molecules existing in the fibrous state. We interpret the observation of an NAI footprint in samples of the triple fusion protein before TEV cleavage and exposure to the synthetic RNA substrate, and before the formation of liquid-like droplets, to reflect the same phenomenon of fiber nucleation observed during the lag phase of polymerization of pathogenic amyloid fibers. This interpretation is displayed graphically in Figure 6.
Figure 6. Graphical Representation of Conversion of Soluble MBP:PTB:hnRNP LC Fusion Protein into Liquid-like Droplet State.
The triple fusion linking maltose binding protein (MBP = blue circle), the RNA binding domain of pyrimidine track binding protein (PTB = green rectangle), and the low complexity domain of hnRNPA2 (LC domain = wavy line) remains soluble and partially polymerized via the LC domain (red sheets) prior to TEV cleavage and exposure to synthetic RNA containing five PTB binding sites (yellow rectangle). Following TEV cleavage and exposure to RNA, MBP is left in solution and PTB:hnRNP LC domain fusion protein partitions into liquid-like droplet (grey shading) in a state of enhanced polymerization.
We have never thought or contended that LC polymers thousands of subunits in length are operative in living cells. Indeed, LC domains are a cellular sink for post-translational modification, including phosphorylation, acetylation, methylation, glycosylation and PARPylation (Choudhary et al., 2009; Lee, 2012; Zhang et al., 2013). Knowing that phosphorylation can regulate the polymerization of LC domains (Han et al., 2012), we have every reason to believe that the behavior of LC domain polymers will be far more dynamic in living cells than in the hydrogels we have been studying for the past several years.
Despite recognizing hydrogels as being aberrantly static, they have offered a number of useful advantages. They have allowed us to probe for structure – first and foremost telling us that LC domain polymerization is at the heart of hydrogel formation (Kato et al., 2012). Second, they have given us assays to probe, in an unbiased manner, for both proteins and RNAs that bind to hydrogels (Han et al., 2012). Third, they have allowed us to conduct correlative mutagenesis experiments in search of mutations that affect hydrogel binding as compared with other cellular activities (Kwon et al., 2013). Fourth, they have allowed us to study interaction with LC domains that – on their own – cannot polymerize. These include the CTD of RNA polymerase II and the SR domains of pre-mRNA splicing factors, both of which bind specifically to certain hydrogels in a manner regulated by the protein kinase enzymes known to control CTD and SR domain function (Kwon et al., 2013; Kwon et al., 2014). Finally, in this report we have employed hydrogels to develop the NAI footprinting strategy.
Since the submission of this manuscript, four new papers have been published concerning the partitioning of LC domains into liquid-like droplets. Two of the four papers conclude that there is no biologic or physiologic role for cross-β polymerization of these LC domains, and that polymerization is solely reflective of a pathologic state (Altmeyer et al., 2015; Patel et al., 2015). The two other papers conclude that cross-β polymerization of LC domains is not the driving force leading to the formation of liquid-like droplets, but that it may be of biologic utility during the maturation of liquid-like droplets and/or RNA granules (Lin et al., 2015; Molliex et al., 2015). These four papers concur with the work of the Forman-Kay and Brangwynne groups (Elbaum-Garfinkle et al., 2015; Nott et al., 2015), indicating that the primary biologic utility of LC domains is driven by forces other than cross-β polymerization, perhaps including Pi-stacking of arginine and phenylalanine residues, or other forms of weak or “fuzzy” interactions involving unfolded polypeptide domains.
Here we offer the very different perspective that cross-β polymerization commonly drives the formation of hydrogels, the retention of LC domains trapped by hydrogels, the formation of liquid-like droplets, the partitioning of LC domains into existing liquid-like droplets, and the formation and maturation of RNA granules. In other words, we submit the hypothesis that the involvement of LC domains in the formation of RNA granules, liquid-like droplets and hydrogels all rely on one in the same phenomenon – cross-β polymerization. Further experimentation, including derivation of the molecular structure of LC domains existing in the labile, polymeric state should help resolve this controversy.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures are available in Supplemental Information online.
Materials
N-acetylimidazole was purchased from Sigma-Aldrich (USA). Ring 13C6-tyrosine was purchased from Cambridge Isotope Laboratories (USA). The parental vector for expression of the triple fusion protein of MBP:PTB:hnRNPA2 LC domain was provided by Dr. Michael Rosen of University of Texas Southwestern Medical Center.
Preparation of Fusion Proteins
His6:GFP or His6:mCherry linked to the LC domain of wild-type hnRNPA2 (residues 181–341) was over-expressed alone or co-expressed with human PPIA and purified as described previously (Kato et al., 2012). Tyrosine-to-serine mutants of GFP:hnRNPA2 LC were purified in the presence of 2M guanidine hydrochloride.
Preparation of Heavy Proteins
The stable-isotope labeled (heavy) proteins (His6:hnRNPA2 LC domain and His6:GST) were prepared with ring 13C6-tyrosine (labeled with 13C on the six carbons of the phenyl ring) by following published procedures (Baxa et al., 2007).
Acetylation of Recombinant Proteins
To acetylate denatured (heavy) proteins, the proteins were denatured by 5 M GuSCN, and acetylation reactions were carried out with 30 mM NAI and 1 mg/ml proteins. The reactions were quenched by 0.8 M Tris-HCl (pH 8.8). The light proteins were acetylated in native conditions (without GuSCN). The native and denatured proteins were mixed at a 1:1 ratio, digested by chymotrypsin and analyzed by mass spectrometry.
Acetylation of Nuclei
293T cells were cultured in DMEM high glucose media containing light or 13C6-tyrosine, respectively. Intact nuclei from heavy or light cells were purified in hypotonic buffer and washed with BME free buffer. Light intact nuclei were resuspended in a nuclei buffer, and heavy nuclei were denatured in the nuclei buffer with 5 M GuSCN. Both samples were acetylated with 30 mM NAI at RT for 15 min, quenched by Tris, and mixed together at a 1:1 ratio. The mixture was digested by chymotrypsin and then analyzed by mass spectrometry.
Acetylation of Liquid-like Droplets
Liquid-like droplets of the MBP:PTB:hnRNPA2 LC triple fusion protein were prepared as described (Lin et al., 2015). For the native sample, NAI (30 mM) was added to the protein solution before TEV cleavage or after TEV cleavage at the indicated time points. After incubation for 15 min at RT, the reaction was quenched by Tris. Acetylated His6-tagged hnRNPA2 LC (heavy) protein was used for the denatured sample. The two samples were mixed, digested by chymotrypsin, and then analyzed by mass spectrometry.
Recruitment Assays with Hydrogels and Liquid-like Droplets
Hydrogel droplets of mCherry:LC domain of wild-type hnRNPA2 were prepared as described (Kato et al., 2012). GFP:hnRNPA2 LC wild-type or mutant proteins were diluted to 1 μM in 1mL of a gelation buffer, and pipetted into the hydrogel dish. After overnight incubation at 4 C, horizontal sections of the hydrogel droplets were scanned at both the mCherry and GFP excitation wavelengths by a confocal microscope. GFP signals at a boundary area of the hydrogel droplets were scanned by the program ImageJ (Schneider et al., 2012). Liquid-like droplets formed from MBP:PTB:hnRNPA2 LC and the synthetic RNA substrate were incubated with 0.1 μM of GFP:hnRNPA2 LC wild-type or mutant domains. The droplets were deposited on a cover slide and imaged by a fluorescent microscope. GFP signals inside and outside of the liquid-like droplets were measured by the program ImageJ. The partition ratio of GFP:hnRNPA2 proteins was calculated by dividing the signal inside the droplet by the signal outside.
Supplementary Material
Acknowledgments
We thank Dr. Michael Rosen for generous provision of the MBP:PTB plasmid into which the LC domain of hnRNPA2 was cloned for use in the analysis of liquid-like droplets, his suggestion that we use NAI footprinting to interrogate the structure of the LC domain of hnRNPA2 as a function of liquid-like droplet formation and maturation, and his editing of this manuscript. We also thank Drs. Deepak Nijhawan and Ting Han for valuable input, Dr. Bruce Alberts for editorial comments including recommendation for the inclusion of Figure 6, and the UTSWMC Live Cell Imaging Core for help with confocal microscopy. This work was supported by the UTSWMC Endowed Scholars Program (to Y.Y.), the Cancer Prevention and Research Institute of Texas (CPRIT grant R1103 to Y.Y.), the National Institutes of Health (NIH grant U01GM10762301 to S.L.M.) and unrestricted funding from an anonymous donor (to S.L.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grofte M, Rask M-BD, Streicher W, Jungmichel S, Nielsen ML, et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat Commun. 2015;6 doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nature reviews Molecular cell biology. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Arosio P, Knowles TP, Linse S. On the lag phase in amyloid fibril formation. Phys Chem Chem Phys. 2015;17:7606–7618. doi: 10.1039/c4cp05563b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astbury WT, Beighton E, Parker KD. The cross-beta configuration in supercontracted proteins. Biochimica et biophysica acta. 1959;35:17–25. doi: 10.1016/0006-3002(59)90330-0. [DOI] [PubMed] [Google Scholar]
- Baxa U, Wickner RB, Steven AC, Anderson DE, Marekov LN, Yau WM, Tycko R. Characterization of beta-sheet structure in Ure2p1-89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CCH, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proceedings of the National Academy of Sciences. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hope IA, Mahadevan S, Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauranzano E, Pozzi S, Pasetto L, Stucchi R, Massignan T, Paolella K, Mombrini M, Nardo G, Lunetta C, Corbo M, et al. Peptidylprolyl isomerase A governs TARDBP function and assembly in heterogeneous nuclear ribonucleoprotein complexes. Brain : a journal of neurology. 2015;138:974–991. doi: 10.1093/brain/awv005. [DOI] [PubMed] [Google Scholar]
- Lee EK. Post-translational modifications of RNA-binding proteins and their roles in RNA granules. Curr Protein Pept Sci. 2012;13:331–336. doi: 10.2174/138920312801619411. [DOI] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter David SW, Rosen Michael K, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Molecular cell. 2015 doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nature reviews Molecular cell biology. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes & development. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends in genetics : TIG. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro J, Woods KW, Park CH, Liu X, Shi Y, Johnson EF, Bouska JJ, Olson AM, Luo Y, Fry EH, et al. Synthesis and SAR of novel tricyclic quinoxalinone inhibitors of poly(ADP-ribose)polymerase-1 (PARP-1) Bioorganic & medicinal chemistry letters. 2009;19:4050–4054. doi: 10.1016/j.bmcl.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj Anderson P, Kim Hong J, Mittag T, Taylor JP. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Molecular cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Patel A, Lee Hyun O, Jawerth L, Maharana S, Jahnel M, Hein Marco Y, Stoynov S, Mahamid J, Saha S, Franzmann Titus M, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Petri M, Frey S, Menzel A, Gorlich D, Techert S. Structural characterization of nanoscale meshworks within a nucleoporin FG hydrogel. Biomacromolecules. 2012;13:1882–1889. doi: 10.1021/bm300412q. [DOI] [PubMed] [Google Scholar]
- Riggi N, Cironi L, Suva ML, Stamenkovic I. Sarcomas: genetics, signalling, and cellular origins. Part 1: The fellowship of TET. The Journal of pathology. 2007;213:4–20. doi: 10.1002/path.2209. [DOI] [PubMed] [Google Scholar]
- Riordan JF, Wacker WEC, Vallee BL. N-acetylimidazole: A reagent for determination of “free” tyrosyl residues of proteins. Biochemistry. 1965;4:1758–1765. [Google Scholar]
- Rufer AC, Thiebach L, Baer K, Klein HW, Hennig M. X-ray structure of glutathione S-transferase from Schistosoma japonicum in a new crystal form reveals flexibility of the substrate-binding site. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:263–265. doi: 10.1107/S1744309105004823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler PB. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988;333:210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- Timasheff SN, Gorbunoff MJ. Conformation of Proteins. Annual Review of Biochemistry. 1967;36:13–54. doi: 10.1146/annurev.bi.36.070167.000305. [DOI] [PubMed] [Google Scholar]
- Triezenberg SJ. Structure and function of transcriptional activation domains. Current opinion in genetics & development. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nature methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- Zydowsky LD, Etzkorn FA, Chang HY, Ferguson SB, Stolz LA, Ho SI, Walsh CT. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein science : a publication of the Protein Society. 1992;1:1092–1099. doi: 10.1002/pro.5560010903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.