ABSTRACT
Characterization and production of efficient lignocellulytic enzyme cocktails for biomass conversion is the need for biofuel industry. The present investigation reports the modeling and optimization studies of lignocellulolytic enzyme cocktail production by Cotylidia pannosa under submerged conditions. The predominant enzyme activities of cellulase, xylanase and laccase were produced in the cocktail through submerged conditions using wheat bran as a substrate. A central composite design approach was utilized to model the production process using temperature, pH, incubation time and agitation as input variables with the goal of optimizing the output variables namely cellulase, xylanase and laccase activities. The effect of individual, square and interaction terms on cellulase, xylanase and laccase activities were depicted through the non-linear regression equations with significant R2 and P-values. An optimized value of 20 U/ml, 17 U/ml and 13 U/ml of cellulase, xylanase and laccase activities, respectively, were obtained with a media pH of 5.0 in 77 h at 31C, 140 rpm using wheatbran as a substrate. Overall, the present study introduces a fungal strain, capable of producing lignocellulolytic enzyme cocktail for subsequent applications in biofuel industry.
KEYWORDS: Cotylidia pannosa, lignocellulolytic cocktail, response surface methodology, submerged fermentation, wheat bran
Introduction
The lignocellulosic biomass such as agro-industrial residues (e.g. straw, sugar cane bagasse, corn stover) and wood materials provides a low-cost feedstock for production of second generation fuels thereby offering economic, environmental and strategic advantages.1 The utilization of these lignocellulosic substrates for the production of desirable value added products such as ethanol involve three essential steps namely (i) pretreatment, which unlock the lignocellulosic interactions to make cellulose and hemicellulose, and other carbohydrate polymers more accessible for enzymatic hydrolysis; (ii) saccharification of pretreated material by hydorlytic enzymes usually cellulases and hemicellulases to release monosaccharides; and (iii) fermentation of monosaccharides, for production of desirable product viz., biofuels or other platform chemicals.2,3 The pretreatment step is very crucial as the removal of lignin from lignocellulosic substrates remains a critical issue. Even, the residual lignins after pretreatment are reported to directly affect the efficiency of saccharification by reducing the activities of cellulolytic enzymes used in saccharification.4-6 Moreover, the lignin derived products are often reported to toxic to fermenting microorganisms such as yeasts.7,8 Therefore, recalcitrance-related obstacles imposed by lignins make second generation bioethanol production a costly and energy intensive process.
In this context, the biological pretreatment for lignin removal by enzymes as biocatalysts are currently being explored as green alternatives to physico-chemical pretreatment strategies.9,10 For biological pre-treatment, laccase producing brown rot and white rot basidiomycete fungal strains have been identified as the best delignifying organisms.11 A number of white rot fungi are reported for the biological pretreatment or simultaneous saccharification and fermentation of lignocellulosic biomass.12 The application of white rot fungi is very well explored for improved enzymatic hydrolysis and its subsequent sugar yield. For this reason, these have been studied with the purpose of pretreating substrates, principally for bioethanol production. Additionally, the fungal pre-treatment do not require any extraction step for enzyme recovery, however, their main drawback is the duration of the pretreatment period (usually several weeks) as the sufficient fungal population needs to be established.12 Most of the white rot fungi studied for their potential for biomass conversion possess significant laccase activity with moderate or low cellulase and xylanase activities.13 The commercial cellulase cocktails, such as Cellic CTec3 from Novozyme (Bagsværd, Denmark) and Accellerase TRIO from DuPont Genencor (CA, USA), contain a blend of endo- and exoglucanases and β-glucosidase which acts synergistically to unlock and saccharify polysaccharides from the lignocellulose complex to fermentable sugars. The Accellerase® Trio™ is a combination of multiple enzyme activities including exoglucanase, endoglucanase, hemi-cellulases (including xylanases), and ß-glucosidase but lacks ligninolytic activity whereas Cellic CTec3 activity is limited to only cellulosic substrates.14,15 Therefore, more potent adequate enzyme preparations need to be developed for the efficient enzymatic saccharification process.
The bioconversion of lignocelluloses relies heavily on major technological innovations centered on efficient process design for production of enzymes. Response surface methodology (RSM) is a well-practiced one for creation of a process model in the form of a non-linear regression equation by considering the effect of individual, square and interaction terms of process variables on the ouput.16-18 Moreover, the response optimizer function of RSM will be helpful for predicting the process variables for getting optimum output. A white rot fungus Cotilydia pannosa reported in our recent work, possessing cellulase and xylanase activities along with the laccase activity.19,20 Therefore, the present study utilized RSM for modeling and optimization of lignnocellulosic enzymes production by Cotylidia pannosa through submerged fermentation using wheatbran as a substrate.
Results and discussion
Wheat bran due to its nutritional content and large surface area serves to be an excellent carbon source without any supplementary carbon or nitrogen source for production of lignocellulolytic enzymes. The bran is rich in nutritional contents with 27% of total carbohydrate, 14% protein, 6% lipids and around 64% digestible nitrogen.21 Additionally, no prior pretreatment is necessary for wheat bran for utilization in enzyme production.22 The list of experiments planned and executed based on the Central Composite Design (CCD) of RSM are tabulated in Table 1. Experimental outputs namely cellulase, xylanase and laccase activities from the set of experiments along with the predicted values are presented in Table 1. The accuracy of the RSM models was drawn from the close agreement of the predicted and experimental values.18
Table 1.
Composition of the various runs of the central composite design, actual and predicted values of the different compression parameters and their responses.
| Input parameters |
Response (U/ml) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Run | Temp.a(°C) | pH | ITb(h) | Agtn.c (rpm) | Cellulase | Xylanase | Laccase | |||
| Order | (A) | (B) | (C) | (D) | (Exp.) | (Predict.) | (Exp.) | (Predict.) | (Exp.) | (Predict.) |
| 1 | 25 | 4 | 120 | 150 | 16.24 | 15.81 | 14.25 | 13.05 | 2.78 | 2.87 |
| 2 | 32.5 | 5 | 72 | 100 | 16.5 | 16.51 | 14.7 | 15.6 | 12.1 | 11.77 |
| 3 | 40 | 4 | 120 | 50 | 4.86 | 4.95 | 6.59 | 6.23 | 0.4 | 0.54 |
| 4 | 25 | 4 | 24 | 150 | 12.22 | 11.85 | 6.21 | 6.95 | 0.13 | 0.28 |
| 5 | 25 | 6 | 24 | 150 | 11.33 | 11.02 | 2.16 | 1.44 | 1.38 | 0.90 |
| 6 | 32.5 | 5 | 120 | 100 | 13.45 | 13.35 | 12.89 | 13.55 | 6.9 | 6.74 |
| 7 | 25 | 6 | 120 | 150 | 11.12 | 11.80 | 3.40 | 4.25 | 2.86 | 3.06 |
| 8 | 40 | 4 | 24 | 50 | 0.18 | −0.70 | 2.14 | 2.05 | 0 | −0.21 |
| 9 | 32.5 | 4 | 72 | 100 | 14.9 | 15.77 | 14.56 | 15.07 | 11.9 | 12.1 |
| 10 | 32.5 | 6 | 72 | 100 | 15.7 | 16.47 | 11.48 | 12.26 | 10.9 | 12.08 |
| 11 | 40 | 4 | 120 | 150 | 8.1 | 8.51 | 5.4 | 6.3 | 1.62 | 2.11 |
| 12 | 25 | 5 | 72 | 100 | 11.8 | 12.9 | 8.6 | 9.16 | 4.19 | 4.90 |
| 13 | 32.5 | 5 | 72 | 100 | 16.5 | 16.51 | 14.7 | 15.57 | 12 | 11.77 |
| 14 | 25 | 6 | 24 | 50 | 9.45 | 8.85 | 1.71 | 1.58 | 0 | −0.50 |
| 15 | 25 | 4 | 24 | 50 | 4.7 | 4.97 | 5.1 | 4.94 | 0 | 0.05 |
| 16 | 32.5 | 5 | 72 | 100 | 17.5 | 16.5 | 16.7 | 15.57 | 12 | 11.77 |
| 17 | 40 | 6 | 120 | 50 | 7 | 7.18 | 6.1 | 6.12 | 0.05 | −0.11 |
| 18 | 40 | 6 | 24 | 150 | 6.2 | 6.06 | 2.91 | 3.27 | 0.55 | 0.94 |
| 19 | 32.5 | 5 | 72 | 100 | 17.5 | 16.51 | 16.7 | 15.57 | 13 | 11.77 |
| 20 | 40 | 4 | 24 | 150 | 5.3 | 5.38 | 3 | 2.25 | 0.54 | 0 |
| 21 | 32.5 | 5 | 72 | 100 | 17.5 | 16.51 | 16.7 | 15.57 | 13 | 11.77 |
| 22 | 32.5 | 5 | 72 | 100 | 17.5 | 16.51 | 16.7 | 15.57 | 13 | 11.77 |
| 23 | 32.5 | 5 | 72 | 50 | 17.26 | 18.33 | 17.56 | 18.1 | 11.3 | 12.58 |
| 24 | 40 | 6 | 24 | 50 | 4.49 | 4.70 | 5.1 | 5.21 | 0 | −0.43 |
| 25 | 40 | 6 | 120 | 150 | 6.51 | 6.02 | 4.97 | 4.04 | 3.02 | 2.63 |
| 26 | 32.5 | 5 | 72 | 100 | 17.5 | 16.51 | 16.7 | 15.57 | 11.6 | 11.77 |
| 27 | 32.5 | 5 | 72 | 150 | 20.63 | 21.19 | 17.3 | 18.05 | 14 | 14.07 |
| 28 | 40 | 5 | 72 | 100 | 6.62 | 7.17 | 6.87 | 7.60 | 3.85 | 4.55 |
| 29 | 25 | 6 | 120 | 50 | 12.46 | 12.16 | 4.87 | 4.53 | 0.1 | 0.30 |
| 30 | 32.5 | 5 | 24 | 100 | 8.39 | 10.13 | 9.43 | 10.06 | 3.71 | 5.29 |
| 31 | 25 | 4 | 120 | 50 | 11.5 | 11.45 | 10.77 | 11.17 | 1.68 | 1.27 |
Temperature;
Incubation Time;
Agitation Rate
Considering the individual, square and interaction terms of SmF variables on outputs, the following non-linear regression equations (uncoded form) were developed for cellulase, xylanase and laccase activities as follows:
| (1) |
| (2) |
| (3) |
The predictive ability of developed non-linear regression models was further confirmed through significance test and ANOVA tests. Significance test results for cellulase, xylanase and laccase activities are tabulated in Table 2. In case of cellulase, individual, square and interaction effects of SmF variables seemed to have significant effect (P < 0 .05) except the individual, square and interaction effects of pH. In case of xylanase, except the individual and interaction effects of agitation, the individual, square and interaction effects of other SmF variables (Incubation time, temp and pH) determined significant impact (P < 0 .05). Whereas the important factors for determining the laccase activity included only the individual and square effects of incubation time and agitation. From the results of ANOVA test (Table 3), significant contributors toward cellulase activity were observed to be the linear, squared, and interaction terms of temp, pH and agitation. In case of xylanase activity, the linear, square and interaction effects of temp, incubation time, pH were also observed to play significant role. The most significant contributions for laccase activity included the linear, square and interaction effects of incubation time and agitation. The fitness and adequacy of the developed non-regression model was further confirmed through the R2 and adjusted R2 values (Cellulase: R2 - 98.20 %, Adj. R2 - 96.63%; Xylanase: R2 - 98.14 %, Adj. R2 - 96.503%; Laccase: R2-98.45%, Adj. R2 - 97.10%).18,23
Table 2.
Results of significance test on the non-linear model-coefficients, standard errors, T statistics, and P values for for cellulase (U/ml), xylanase (U/ml) and laccase (U/ml) (coded form).
| Coef |
SE Coef |
T - Value |
P - Value |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Term | Cellu.a | Xylan.b | Lacca.c | Cellu.a | Xylan.b | Lacca.c | Cellu.a | Xylan.b | Lacca.c | Cellu.a | xylan.b | Lacca.c |
| Constant | 16.51, | 15.57, | 11.77 | 0.29, | 0.31, | 0.27 | 57.55, | 50.38, | 43.30 | 0.00, | 0.00, | 0.00 |
| Temp | −2.86, | −0.78, | −0.17 | 0.23, | 0.25, | 0.22 | −12.57, | −3.17, | −0.80 | 0.00, | 0.01, | 0.44 |
| pH | 0.39, | −1.41, | −0.01 | 0.23, | 0.25, | 0.22 | 1.53, | −5.73, | −0.04 | 0.15, | 0.00, | 0.97 |
| IT | 1.61, | 1.75, | 0.73 | 0.23, | 0.25, | 0.22 | 7.06, | 7.12, | 3.37 | 0.00, | 0.00, | 0.00 |
| Agtn | 1.43, | −0.02, | 0.74 | 0.23, | 0.25, | 0.22 | 6.28, | −0.08, | 3.44 | 0.00, | 0.94, | 0.00 |
| Temp*Temp | −6.48, | −7.19, | −7.04 | 0.60, | 0.65, | 0.57 | −10.79, | −11.12, | −12.38 | 0.00, | 0.00, | 0.00 |
| pH*pH | −0.39, | −1.91, | 0.32 | 0.60, | 0.65, | 0.57 | −0.65, | −2.95, | 0.56 | 0.53, | 0.01, | 0.58 |
| IT*IT | −4.77, | −3.77, | −5.75 | 0.60, | 0.65, | 0.57 | −7.94, | −5.82, | −10.12 | 0.00, | 0.00, | 0.00 |
| Agtn*Agtn | 3.25, | 2.50, | 1.56 | 0.60, | 0.65, | 0.57 | 5.42, | 3.87, | 2.75 | 0.00, | 0.00, | 0.01 |
| Temp *Ph | 0.38, | 1.63, | 0.08 | 0.24, | 0.26, | 0.23 | 1.57, | 6.27, | 0.36 | 0.14, | 0.00, | 0.72 |
| Temp*IT | −0.21, | −0.51, | −0.12 | 0.24, | 0.26, | 0.23 | −0.86, | −1.97, | −0.52 | 0.40, | 0.07, | 0.61 |
| Temp.*Agtn | −0.20, | −0.45, | −0.01 | 0.24, | 0.26, | 0.23 | −0.83, | −1.73, | −0.02 | 0.42, | 0.10, | 0.98 |
| pH*IT | −0.79, | −0.82, | −0.11 | 0.24, | 0.26, | 0.23 | −3.28, | −3.14, | −0.47 | 0.01, | 0.01, | 0.65 |
| pH*Agtn | −1.18, | −0.54, | 0.29 | 0.24, | 0.26, | 0.23 | −4.87, | −2.06, | 1.27 | 0.00, | 0.06, | 0.22 |
| IT*Agtn | −0.63, | −0.03, | 0.34 | 0.24, | 0.26, | 0.23 | −2.61, | −0.13, | 1.49 | 0.02, | 0.90, | 0.16 |
| S = 0.97 | R-Sq = 98.20% | R-Sq(adj) = 96.63% (For cellulase) | ||||||||||
| S = 1.04 | R-Sq = 98.14% | R-Sq(adj) = 96.50% (For xylanase) | ||||||||||
| S = 0.91 | R-Sq = 98.45% | R-Sq(adj) = 97.10% (For laccase) | ||||||||||
cellulase;
xylanase;
laccase
Table 3.
ANOVA for quadratic model for cellulase (U/ml), xylanase (U/ml) and laccase (U/ml).
| Source | DF |
Adj SS |
Adj MS |
F - Value |
P - Value |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C.a | X.b | L.c | C.a | X.b | L.c | C.a | X.b | L.c | C.a | X.b | L.c | C.a | X.b | L.c | |
| Model | 14, | 14, | 14 | 816.61, | 914.46, | 854.46 | 58.33, | 65.32, | 61.03 | 62.36, | 60.15, | 72.74 | 0.00, | 0.00, | 0.00 |
| Linear | 4, | 4, | 4 | 233.38, | 101.55, | 20.00 | 58.34, | 25.39, | 5.00 | 62.38, | 23.38, | 5.96 | 0.00, | 0.00, | 0.00 |
| Temp. | 1, | 1, | 1 | 147.70, | 10.88, | 0.53 | 147.67, | 10.89, | 0.53 | 157.90, | 10.02, | 0.63 | 0.00, | 0.01, | 0.44 |
| pH | 1, | 1, | 1 | 2.178, | 35.60, | 0.00 | 2.18, | 35.60, | 0.00 | 2.33, | 32.79, | 0.00 | 0.15, | 0.00, | 0.97 |
| IT | 1, | 1, | 1 | 46.66, | 55.06, | 9.53 | 46.66, | 55.06, | 9.53 | 49.89, | 50.71, | 11.36 | 0.00, | 0.00, | 0.00 |
| Agtn. | 1, | 1, | 1 | 36.84, | 0.01, | 9.94 | 36.84, | 0.01, | 9.94 | 39.39, | 0.01, | 11.84 | 0.00, | 0.94, | 0.00 |
| Square | 4, | 4, | 4 | 540.98, | 747.40, | 830.73 | 135.24, | 186.85, | 207.68 | 144.59, | 172.08, | 247.50 | 0.00, | 0.00, | 0.00 |
| Temp.*Temp. | 1, | 1, | 1 | 108.95, | 134.24, | 128.57 | 108.95, | 134.24, | 128.57 | 116.48, | 123.63, | 153.22 | 0.00, | 0.00, | 0.00 |
| pH*pH | 1, | 1, | 1 | 0.39, | 9.44, | 0.26 | 0.39, | 9.44, | 0.26 | 0.42, | 8.69, | 0.32 | 0.53, | 0.01, | 0.58 |
| IT*IT) | 1, | 1, | 1 | 59.03, | 36.83, | 85.87 | 59.03, | 36.83, | 85.87 | 63.11, | 33.92, | 102.33 | 0.00, | 0.00, | 0.00 |
| Agtn.*Agtn. | 1. | 1, | 1 | 27.48, | 16.28, | 6.33 | 27.48, | 16.28, | 6.33 | 29.38, | 14.99, | 7.54 | 0.00, | 0.00, | 0.01 |
| 2-Way Inter | 6, | 6, | 6 | 2.26, | 65.51, | 3.73 | 7.04, | 10.92, | 0.62 | 7.53, | 10.06, | 0.74 | 0.00, | 0.00, | 0.62 |
| Temp.*pH | 1, | 1, | 1 | 2.29, | 42.69, | 0.11 | 2.30, | 42.69, | 0.11 | 2.45, | 39.31, | 0.13 | 0.14, | 0.00, | 0.73 |
| Temp.*IT | 1, | 1, | 1 | 0.69, | 4.21, | 0.23 | 0.69, | 4.21, | 0.23 | 0.74, | 3.87, | 0.27 | 0.40, | 0.07, | 0.61 |
| Temp.*Agtn | 1, | 1, | 1 | 0.65, | 3.26, | 0.00 | 0.65, | 3.26, | 0.00 | 0.69, | 3.01, | 0.00 | 0.42, | 0.10, | 0.98 |
| pH*IT | 1, | 1, | 1 | 10.05, | 10.72, | 0.18 | 10.05, | 10.72, | 0.18 | 10.74, | 9.87, | 0.22 | 0.01, | 0.01, | 0.65 |
| pH*Agtn. | 1, | 1, | 1 | 22.23, | 4.62, | 1.36 | 22.23, | 4.62, | 1.36 | 23.76, | 4.25 | 1.62 | 0.00, | 0.06, | 0.22 |
| IT*Agtn. | 1, | 1, | 1 | 6.35, | 0.02, | 1.86 | 6.35, | 0.02, | 1.87 | 6.79, | 0.02, | 2.21 | 0.02, | 0.90, | 0.16 |
| Error | 16, | 16, | 16 | 4.97, | 17.37, | 13.43 | 0.93, | 1.09, | 0.84 | ||||||
| Lack-of-Fit | 10, | 10, | 10 | 13.54, | 11.66, | 11.20 | 1.35, | 1.17, | 1.12 | 5.69, | 1.22, | 3.02 | 0.02, | 0.42, | 0.09 |
| Pure Error | 6, | 6, | 6 | 1.43, | 5.71, | 2.22 | 0.24, | 0.95, | 0.37 | ||||||
| Total | 30, | 30, | 30 | 831.58, | 931.83, | 867.88 | |||||||||
cellulase;
xylanase;
laccase
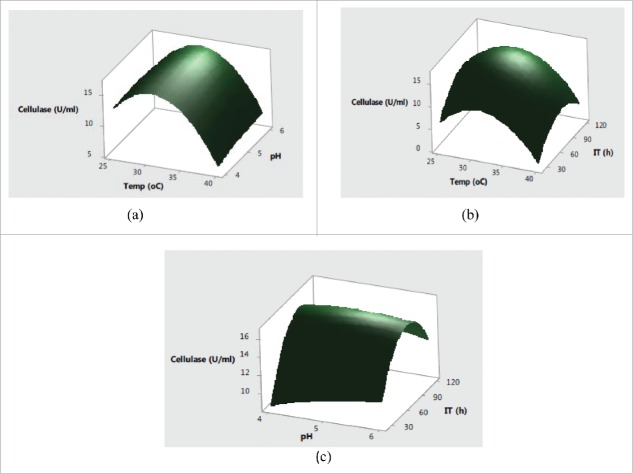
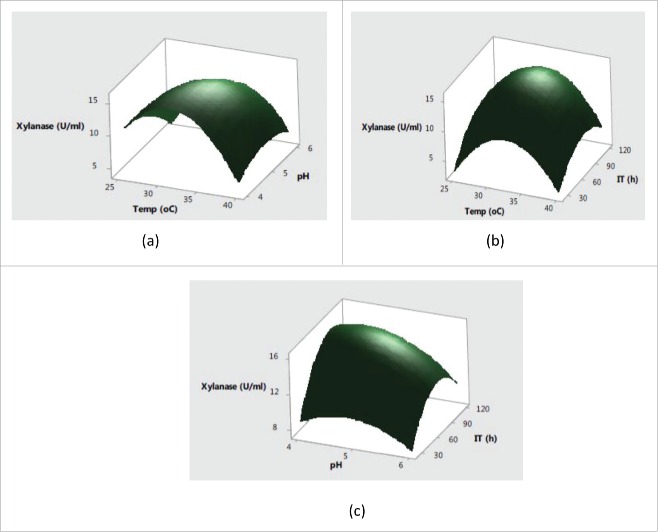
Further, the interaction effects of variables selected on production of enzymes were studied by plotting three dimensional surface curves to determine the optimum level of each variable for maximum enzyme activity. The interaction effects of SmF variables on cellulase activity are depicted in response surface plots from Fig. 1(a)–Fig. 1(c). Fig. 1(a) indicated that the higher cellulase activities were obsreved at intermediate values of temperature and pH. Later on, irrespective of increase or decrease in pH and temperature conditions, no significant effect on cellulase production was observed. The pH of the substrate plays a critical role in production of lignocellulolytic enzymes by affecting either the microbial growth or denaturating the enzymes.24 The change in pH is also reported to affect the transport of various components across the cell membrane and denaturation of the enzyme activities.25 Fig. 2(b) showed that the increase in temperature with incubation time resulted higher cellulase production with a maximum cellulase production at 32°C in 72 h. Usually, the optimum temperatures and incubation time for lignocellulolyitc enzymes production vary with the use of different strains. The cumulative effect of pH and incubation time on cellulase production revealed that the higher cellulase production was mainly recorded in the mid values of pH and incubation time with decreased trend after pH of 5.5 and prolonged incubation time of 90 h. This type of behavior is mainly attributed to the in effecient transport across membranes at higher pH conditions and denaturation of cellulase activity at longer incubation times. Fig. 2(a) showed that the maximum xylanase production was obtained around 32°C and pH 5.0 after mid values of incubation time. Further increase in temperature and incubation time resulted in significant decrease in xylanase activities (Fig. 2(b). From cumulative graph Fig. 2(c) maximum xylanase production was observed at pH 5.0 after 92 h of incubation. The cumulative effect of submerged fermentation variables on laccase production are presented in Fig. 3(a-c). The results in Fig. 3(a) indicated that the higher laccase activity was obtained at mid values of temperature with no significant effect of change in pH. The combined effect of temperature and incubation time on laccase production from Fig. 3(b) demonstrated that the maximum laccase production could be acheived at middle values of temperature and incubation time. Moreover, the significant effect of incubation time and non-significant behavior of change of pH on laccase production was also observed in Fig. 3(c).
Figure 1.
Response surface plots showing the effect of (a) temperature and pH (b) temperature and incubation time (c) pH and incubation time on cellulase production.
Figure 2.
Interaction effects of (a) temperature and pH (b) temperature and incubation time (c) pH and incubation time on xylanase production.
Figure 3.
Effect of (a) temperature and pH (b) temperature and incubation time (c) pH and incubation time on laccase production.
The maximum lignocellulolytic activities of fungi have been reported at pH 5.0 in case of Agaricus bisporus, Trametes versicolor, Trichoderma harzianum and Penicillium janczewskii.26,27 The optimum temperature for lignocellulolytic enzyme production was similar to those of other mesophilic fungi such as Aspergillus japonicas C03, Aspergillus glaucas XC8, A. niger MS82, Trichoderma reesei Rut C30, Trichoderma viride strain EU2-77, Penicillum echinulatum and Fusarium oxysporum. The maximum xylanase production has already been reported at a temperature range of 25–30°C by Trichoderma viride28 and P. glabrum.29 Usually, the maximal reported temperature for these filamentous fungus is 35°C, with an indication of the absence of growth 37°C.30,31 The activity of lignocellulolytic enzymes also increased on increasing the agitation up to 100 rpm. As for the submerged fermentation, mechanical agitation is known to be a crucial factor because of its effectiveness in mixing the contents of the medium, uniform air distribution and prevention of cell clumping. The lower activity levels at an agitation of above 100 rpm could be attributed to the possible damage to the filamentous structure thereby lowering the enzyme production. The shear stress sensitivity of mycelium has also been reported in the case of xylanase production by Thermactinomyces thalophile subgroup C.32 Furthermore, the shearing of mycelium at high agitation rates also release intracellular proteins in broth, which increase foam generation during the fermentation process and thus reduce xylanase yield by affecting the oxygen transfer ratios.21
Response optimizer function of MINITAB 16 was used to predict the submerged fermentation variables for optimal cellulase, xylanase and laccase activities. The predicted variables seems to be equivalent for the SmF performance using a substrate pH of 5.0 at 31°C, 140 rpm for 77 h. The experimental results obtained through the triplicate runs by utilizing the predicted variables i.e 20.6 U/ml of cellulase activity, 17.3 U/ml of xylanase activity and 14 U/ml of laccase activity, were in close agreement with the the predicted outputs i.e cellulase: 20.1 U/ml; xylanase: 17.2 U/ml; laccase: 13.1 U/ml. The close agreement between the predicted and experimental responses indicated the adequacy and accuracy of the model for further scale-up studies. Moreover, the lignocellulolytic enzyme activities attianed through this study determined significant improvement over the one variable at time (OVAT) experiments i.e cellulase: 10 U/ml; xylanase: 7 U/ml; laccase: 5 U/ml. The higher activities obtained through RSM studies in case of individual lignocellulolytic enzyme production through SmF have been documented very well in the literature. Overall, the present study introduces a white rot fungal strain, capable of producing three essential lignocellulolytic enzymes namely cellulase, xylanase and laccase for lignocellulosic bioethanol sector.
Materials and methods
Microbial culture and maintenance
The culture of C.pannosa (Gene Bank Accession No. KT008117.1)19,20 was obtained from Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya (CSKHPKV), Palampur, India. The culture was maintained on yeast extract peptone dextrose (YEPD) agar at 4°C. All the chemicals and reagents used were of analytical grade. The Birchwood xylan, ABTS [2,2-azinobis (3-ethylbenzathiazoline-6-sulfonic acid) and carboxymethyl cellulose were purchased from Sigma (St. Louis, USA).
Growth conditions and enzyme production set up
For enzyme production, 2% wheat bran added to the YEP culture media consisting of 1% yeast extract and 2% peptone followed by sterilization at 121°C for 15 min at 15 psi. A spore inoculum of C.pannosa (2.9×108 fungal spores/mL) was inoculated aseptically into 100 mL of sterilized growth medium. The enzyme production was carried out at different pH (4–6) when incubated at a different temp. (25–40°C) for 24–120 h at 50 −150 rpm based on the Central Composite Design (CCD) of RSM (Table 4 and Table 1). An aliquot of fermentation broth was withdrawn from the flask after a regular interval and centrifuged at 4°C at 7000 rpm for 15 min. The supernatant after centrifugation was collected and used for measuring cellulase, xylanase and laccase enzyme activities, respectively.
Table 4.
Experimental range of variables for the central composite design in terms of actual and coded factors.
| Range of variables |
||||
|---|---|---|---|---|
| Variables | Symbol coded | Low (−1) | Mid(0) | High(+1) |
| Temperature (°C) | A | 25.0 | 32.5 | 40.0 |
| pH | B | 4.0 | 5.0 | 6.0 |
| Incubation Time (h) | C | 24 | 72 | 120 |
| Agitation (rpm) | D | 50 | 100 | 150 |
Lignocellulolytic enzyme assays
The carboxymethyl cellulase and xylanase activities of crude enzyme were estimated by measuring the reducing sugars through DNS method33,34 The laccase activity was determined spectrophotometrically with ABTS [2,2-azinobis (3-ethylbenzathiazoline-6-sulfonic acid) as a substrate.35
One unit of CMCase activity was defined as an amount of enzyme that releases one micromole of glucose per minute under standard reaction conditions. One unit of xylanase activity was defined as an amount of enzyme that releases one micromole of xylose per minute under standard reaction conditions. One unit of laccase activity was defined as the quantity of enzyme required to oxidize one μmol of ABTS per minute. A control with inactivated enzyme was measured simultaneously for all the enzyme assays.
Modeling and optimization studies
The values of SmF variables for the CCD design of RSM were selected based on literature review and initial screening data (data not shown) through MINITAB 16 software. In the present study, the outputs included the cellulase, xylanase and laccase activities. The set of variables and its ranges and levels utilized in the present study are tabulated under Table 4. The set of experiments based on the CCD (shown in Table 1) was executed in triplicate runs. The nonlinear regression analysis was carried out based on the experimental results of cellulase, xylanase and laccase activities using MINITAB 16 resulting in a second-order polynomial equation. The individual, square and interaction effects of SmF variables on cellulase, xylanase and laccase activities were studied through significance and analysis of variance (ANOVA) tests. The adequacy of the developed model was further checked through R2 and adjusted R2 values. For prediction of a set of SmF variables, the response optimizer function of MINITAB 16 software was utilized to predict maximal cellulase, xylanase and laccase activities.36
Conclusions
The CCD of response surface methodology was successfully utilized to determine the effect of individual, square and interaction of SmF parameters on lignocellulolytic enzyme cocktail production by Cotylidia pannosa. A maximum enzyme activity of 20 U/ml for cellulase, 17 U/ml for xylanase and 13 U/ml for laccase, was obtained when the fungus has been cultivated under submerged conditions of pH 5.0, temperature of 31°C, and a shaking speed of 140 rpm in 77 h using 2% wheat bran as a substrate. The individual square terms of incubation temperatutre and incubation time and their interaction with each other and with pH were the main factors that affected enzyme cocktail production, whereas the interaction effects of agitation with other variables did not promote any change in the range studied. The developed non-linear regression models with good accuracy could be further used for scaleup studies of the enzyme cocktail. Moreover, the introduced white-rot fungi Cotylidia pannosa, could be a potential strain to work on by bioethanol researchers for enhanced yields through integrated industrial- systems biology approach.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors gratefully acknowledge JUIT, India, for providing research facilities to Deepika Sharma.
References
- [1].Sukumaran RK, Surender VJ, Sindhu R, Binod P, Janu KU, Sajna KV, Rajasree KP, Pandey A. Lignocellulosic ethanol in India: Prospects, challenges and feedstock availability. Bioresour Technol 2010; 101:4826-33; PMID:20018505; http://dx.doi.org/ 10.1016/j.biortech.2009.11.049 [DOI] [PubMed] [Google Scholar]
- [2].Khare SK, Pandey A, Larroche C. Current perspectives in enzymatic saccharification of lignocellulosic biomass. Biochem Eng J 2015; 102:38-44; http://dx.doi.org/ 10.1016/j.bej.2015.02.033 [DOI] [Google Scholar]
- [3].Swana J, Yang Y, Behnam M, Thompson R. An analysis of net energy production and feedstock availability for biobutanol and bioethanol. Bioresour Technol 2011; 102:2112-17; PMID:20843683; http://dx.doi.org/ 10.1016/j.biortech.2010.08.051 [DOI] [PubMed] [Google Scholar]
- [4].Mansfield SD, Mooney C, Saddler JN. Substrate and enzyme characteristics that limit cellulose hydrolysis. Biotechnol Prog 1999; 15:804-16; PMID:10514250; http://dx.doi.org/ 10.1021/bp9900864 [DOI] [PubMed] [Google Scholar]
- [5].Eriksson T, Borjesson J, Tjerneld F. Mechanism of surfactant effect in enzymatic hydrolysis of lignocellulose. Enzyme Microb Technol 2002; 31:353-64; http://dx.doi.org/ 10.1016/S0141-0229(02)00134-5 [DOI] [Google Scholar]
- [6].Moilanen U, Kellock M, Galkin S, Viikari L. The laccase-catalyzed modification of lignin for enzymatic hydrolysis. Enzyme Microb Technol 2011; 49:492-98; PMID:22142723; http://dx.doi.org/ 10.1016/j.enzmictec.2011.09.012 [DOI] [PubMed] [Google Scholar]
- [7].Palmqvist H, Hahn-Hagerdal B. Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresour Technol 2000; 74:25-33; http://dx.doi.org/ 10.1016/S0960-8524(99)00161-3 [DOI] [Google Scholar]
- [8].Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Biomass recalcitrance: Engineering plants and enzymes for biofuels production Science 2007; 315:804-7; PMID:17289988; http://dx.doi.org/ 10.1126/science.1137016 [DOI] [PubMed] [Google Scholar]
- [9].Heap L, Green A, Brown D, van Dongen B, Turner N. Role of laccase as an enzymatic pretreatment method to improve lignocellulosic saccharification. Catal Sci Technol 2014; 4:2251-59; http://dx.doi.org/ 10.1039/c4cy00046c [DOI] [Google Scholar]
- [10].Ulla Moilanen U, Kellock M, Várnai A, Andberg M, Viikari L. Mechanisms of laccase-mediator treatments improving the enzymatic hydrolysis of pre-treated spruce. Biotechnol Biofuels 2014; 7:177; PMID:25648942; http://dx.doi.org/ 10.1186/s13068-014-0177-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Manavalan T, Manavalan A, Heese K. Characterization of lignocellulolytic enzymes from white-rot fungi. Curr Microbiol 2015; 70:485-98; PMID:25487116; http://dx.doi.org/ 10.1007/s00284-014-0743-0 [DOI] [PubMed] [Google Scholar]
- [12].Chandel AK, Caroline BMG, Strap JL, Silva SS. Biodelignification of lignocellulosic substrates: an intrinsic and sustainable pretreatment strategy for clean energy production. Crit Rev Biotechnol 2015; 35:281-93; PMID:24156399; http://dx.doi.org/ 10.3109/07388551.2013.841638 [DOI] [PubMed] [Google Scholar]
- [13].Chaturvedi V, Verma P. An overview of key pretreatment processes employed for bioconversion of lignocellulosic biomass into biofuels and value added products. Three Biotech. 2013; 3:415-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sun FF, Hong J, Hu J, Saddler JN, Fang X, Zhang Z, Shen S. Accessory enzymes influence cellulase hydrolysis of the model substrate and the realistic lignocellulosic biomass. Enzyme Microb Technol 2015; 79-80:42-8; PMID:26320713; http://dx.doi.org/ 10.1016/j.enzmictec.2015.06.020 [DOI] [PubMed] [Google Scholar]
- [15].Rosales-Calderon O, Trajano HL, Duff SJB. Stability of commercial glucanase and β-glucosidase preparations under hydrolysis conditions. Palomo J, ed. Peer J 2014; 2:e402; PMID:24949230; http://dx.doi.org/ 10.7717/peerj.402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Myers RH, Montgomery DC, Anderson-Cook CM. Response Surface Methodology: process and product optimization using designed experiments. John Wiley & Sons: New York, 2009 [Google Scholar]
- [17].Chauhan M, Garlapati VK. Modeling embedded optimization strategy for formulation of bacterial lipase based bio-detergent. Indus Eng Chem Res 2014; 53:514-20; http://dx.doi.org/ 10.1021/ie401357h [DOI] [Google Scholar]
- [18].Mahapatra P, Kumari A, Garlapati VK, Banerjee R, Nag A. Enzymatic synthesis of fruit flavor esters by immobilized lipase from Rhizopus oligosporus optimized with response surface methodology. J Mol Catal B: Enzym 2009; 60:57-63; http://dx.doi.org/ 10.1016/j.molcatb.2009.03.010 [DOI] [Google Scholar]
- [19].Sharma D, Goel G, Sud A, Chauhan RS. A novel laccase from newly isolated Cotylidia pannosa and its application in decolorization of synthetic dyes. Biocatal Agric Biotechnol 2015b; 4:661-66 [Google Scholar]
- [20].Sharma D, Goel G, Bansal S, Mahajan R, Sharma BM, Chauhan RS. Characterization of cellulolytic activities of newly isolated Thelephora sowerbyi from North-Western Himalayas on different lignocellulosic substrates. J Basic Microbiol 2015a; DOI: 10.1002/jobm.201500107. [DOI] [PubMed] [Google Scholar]
- [21].Khurana S, Kapoor M, Gupta S, Kuhad RC. Statistical optimization of alkaline xylanase production from Streptomyces violaceoruber under submerged fermentation using response surface methodology. Ind J Microbiol 2007; 47:144-52; http://dx.doi.org/ 10.1007/s12088-007-0028-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Thygesen A, Thomsen AB, Schmidt AS, Jørgensen H, Ahring BK, Olsson L. Production of cellulose and hemicellulose-degrading enzymes by filamentous fungi cultivated on wet-oxidised wheat straw. Enzyme Microb Tech 2003; 32:606-15; http://dx.doi.org/ 10.1016/S0141-0229(03)00018-8 [DOI] [Google Scholar]
- [23].Kishore D, Kayastha AM. Optimisation of immobilisation conditions for chick pea β-galactosidase (CpGAL) to alkylamine glass using response surface methodology and its applications in lactose hydrolysis. Food Chem 2012; 134:1650-7; PMID:25005995; http://dx.doi.org/ 10.1016/j.foodchem.2012.03.055 [DOI] [PubMed] [Google Scholar]
- [24].Ramesh MV, Lonsane BK. Regulation of α-amylase production in Bacillus licheniformis M 27 by enzyme end-products in submerged fermentation and its overcoming in solid state fermentation system. Biotechnol Lett 1991; 13:355-60; http://dx.doi.org/ 10.1007/BF01027682 [DOI] [Google Scholar]
- [25].Moon SH, Parulekar SJ. A parametric study of protease production in batch and fed-batch cultures of Bacillus firmus. Biotechnol Bioeng 1991; 37:467-83; PMID:18597393; http://dx.doi.org/ 10.1002/bit.260370509 [DOI] [PubMed] [Google Scholar]
- [26].Isil S, Nilufer A. Investigation of factors affecting xylanase activity from Trichoderma harzianum1073 D3. Brazilian Arch Biol Technol 2005; 48:187-93; http://dx.doi.org/ 10.1590/S1516-89132005000200004 [DOI] [Google Scholar]
- [27].Terrasan CRF, Temer B, Durate MCT, Carmona EC. Production of xylanolytic enzymes by Penicillium janczewskii. Bioresour Technol 2010; 101:4139-43; PMID:20122825; http://dx.doi.org/ 10.1016/j.biortech.2010.01.011 [DOI] [PubMed] [Google Scholar]
- [28].Goyal M, Kalra KL, Sareen VK, Soni G. Xylanase production with xylan rich lignocellulosic wastes by a local soil isolate of Trichoderma viride. Braz J Microbiol 2008; 39:535-41; PMID:24031262; http://dx.doi.org/ 10.1590/S1517-83822008000300025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Knob A, Beitel SM, Fortkamp D, Terrasan CR, de Almeida AF. Production, purification, and characterization of a major Penicillium glabrum xylanase using brewer's spent grain as substrate. BioMed Res Intern 2013; 2013:728-35; http://dx.doi.org/ 10.1155/2013/728735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sinigaglia M, Corbo MR, Ciccarone C. Influence of temperature, pH and water activity on “in vitro” inhibition of Penicillium glabrum (Wehmer) westling by yeasts. Microbiol Res 1998; 153:137-43; PMID:9760746; http://dx.doi.org/ 10.1016/S0944-5013(98)80031-1 [DOI] [PubMed] [Google Scholar]
- [31].Nevarez L, Vasseur V, Le Madec A, Le Bras MA, Coroller L, Leguérinel I, Barbier G. Physiological traits of Penicillium glabrum strain LCP 08.5568, a filamentous fungus isolated from bottled aromatised mineral water. Int J Food Microbiol 2009; 130:166-71; PMID:19233496; http://dx.doi.org/ 10.1016/j.ijfoodmicro.2009.01.013 [DOI] [PubMed] [Google Scholar]
- [32].Kohli U, Nigam P, Singh D, Chaudhary K. Thermostable, alkalophilic and cellulase free xylanase production by Thermonoactinomyces thalophilus subgroup C. Enzym Microb Technol 2001; 28:606-10; http://dx.doi.org/ 10.1016/S0141-0229(01)00320-9 [DOI] [PubMed] [Google Scholar]
- [33].Nguyen LT, Neo KRS, Yang KL. Continuous hydrolysis of carboxymethyl cellulose with cellulase aggregates trapped inside membranes. Enzyme Microb Technol 2015; 78:34-39; PMID:26215342; http://dx.doi.org/ 10.1016/j.enzmictec.2015.06.005 [DOI] [PubMed] [Google Scholar]
- [34].Mendis M, Simsek S. Production of structurally diverse wheat arabinoxylan hydrolyzates using combinations of xylanase and arabinofuranosidase. Carbohyd Polym 2015; 132:452-59; http://dx.doi.org/ 10.1016/j.carbpol.2015.05.083 [DOI] [PubMed] [Google Scholar]
- [35].Bhattacharya SS, Garlapati VK, Banerjee R. Optimization of laccase production using response surface methodology coupled with differential evolution. New Biotechnol 2011; 28:31-39; http://dx.doi.org/ 10.1016/j.nbt.2010.06.001 [DOI] [PubMed] [Google Scholar]
- [36].Basş D, Boyacá IH. Modeling and optimization I: Usability of response surface methodology. J Food Eng 2007; 78:836-45; http://dx.doi.org/ 10.1016/j.jfoodeng.2005.11.024 [DOI] [Google Scholar]