ABSTRACT
Heme biosynthesis is a highly conserved pathway which is present in all kingdoms, from Archaea to higher organisms such as plants and mammals. The heme molecule acts as a prosthetic group for different proteins and enzymes involved in energy metabolism and reactions involved in electron transfer. Based on our recent findings and other recent reports, we here illustrate that heme is more than a co-factor. We also discuss the necessity to gain more insight into the heme biosynthesis pathway regulation, as this interacts closely with overall stress control. Understanding heme biosynthesis and its regulation could impact our ability to develop more efficient yeast cell factories for heterologous protein production.
KEYWORDS: cell stress, Hap1, heme biosynthesis, protein production, yeast cell factories
Introduction
Metabolic engineering is a well-established discipline that is in the center of the growing interest and effort in the development of so-called microbial cell factories for the production of bulk and fine chemicals, second generation biofuels, as well as proteins or enzymes of industrial interest or for therapeutic applications. The variety of potential microbial cell factories is wide, but yeasts have proved to be very versatile hosts1,2 and therefore are increasingly becoming the preferred choice for cell factory development. Yeasts are particularly attractive for production of recombinant proteins, since they have the eukaryal processing machinery and hence are able to produce more complex proteins that can be secreted.
We have been working for the past few years on the establishment of the yeast Saccharomyces cerevisiae as a robust platform system for production of therapeutic proteins, such as insulin precursor and, more recently, human hemoglobin (HbA). The latter can be used for the development of artificial blood substitutes3,4 and we can currently produce up to 7% HbA out of the total cell protein in its active form. However, we are aware that there is plenty of room for improvement.
The importance of heme for microbial physiology
Heme is an ubiquitous molecule, present in all kingdoms, and very well known to be an essential co-factor involved in many reactions related to respiration and electron transfer.5 However heme is more than a co-factor: its role in microbial cells has important implications for their metabolism, since internal heme levels can determine cell physiology under certain conditions. However in most cases, very little is known about how endogenous heme levels are actually influenced by the cell environment. One of the best known examples that shows the variety of heme-regulation diversity is the response to environmental oxygen: high levels of extracellular oxygen induce heme biosynthesis in yeast, while it has the opposite effect in bacteria.6 Heme levels may determine the pathogenicity of many bacterial strains according to environmental oxygen and/or iron levels, thus heme indirectly also acts as a pathogenicity factor.7 In yeasts, on the other hand, heme has a wide variety of functions, from energy metabolism to cell protection against several biological stresses including oxidative stress response activation.8 Intracellular heme levels in yeast are responsible for the activation of either the genes involved in respiration and/or fermentation. All these functions are also closely related to the environmental oxygen levels. However, the intracellular heme levels need to be tightly regulated, since free heme is toxic as it catalyzes the generation of reactive oxygen species (ROS).9 The existence of proteins responsible for controlling internal heme homeostasis has therefore been hypothesized in bacteria, although the function of many of the putative candidates identified still remains unclear.5 More information is available for yeasts, in particular S. cerevisiae, although still some important points need to be solved. For example, it is very well described how intracellular heme levels can control the cellular fate via activation of Hap1p, a transcription factor responsible for activating aerobic respiration, oxidative stress response and, furthermore, heme homeostasis. It triggers a negative feedback response for the endogenous heme biosynthetic pathway, thus keeping intracellular heme levels within the non-toxic range (Fig. 1). How oxygen activates heme biosynthesis is, however, still unknown.
Figure 1.
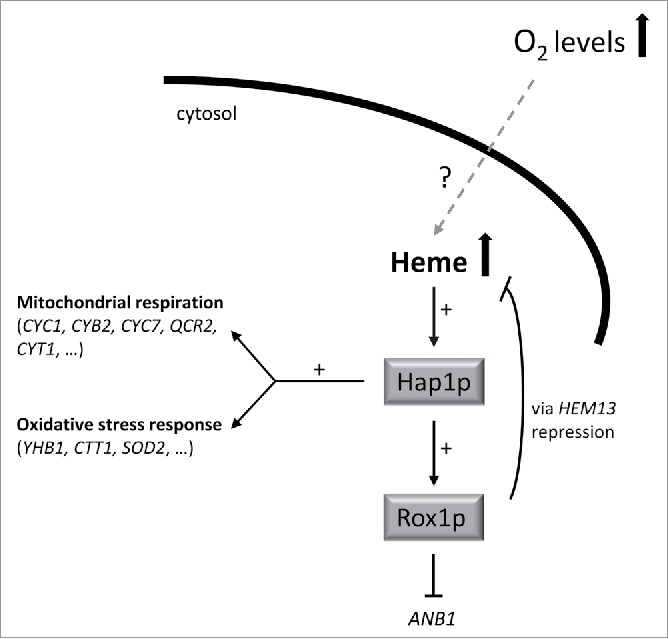
Schematic representation of the mechanism of action of the transcripton factor Hap1p in response to high heme levels, as a consequence of an increase in the environmental oxygen concentration. High intracellular heme activates Hap1p which subsequently induces the transcription of genes involved in both mitochondrial respiration and the oxidative stress response. It also induces the activation of Rox1p, a transcriptional repressor that blocks ANB1 transcription (Anb1p is a transcription factor which initiates anaerobic metabolism) and is also responsible for heme homeostasis by repressing HEM13.
Heme metabolism and its applications in heterologous protein production
Interestingly, the relation between heme biosynthesis and its intracellular levels and heterologous protein production is not only restricted to the overexpression of heme-containing proteins. Previous studies reported that integration of heme biosynthesis pathway from the bacterium Vitreoscella stercoraria, also in combination with recombinant bacterial hemoglobin, improved respiration and heterologous protein synthesis in the yeast Pichia pastoris.10,11 How the Vitreoscella hemoglobin accounts for improved respiration as well as how it enhances protein production is not understood. Furthermore, other studies have demonstrated that deletion of HAP1, a gene encoding the main transcription factor responsive for intracellular heme levels regulation, has a positive impact on recombinant protein production in the yeast Kluyveromyces lactis.12 It is not known how HAP1 deletion results in a better recombinant protein yield.
In our recent study on recombinant human hemoglobin production in S. cerevisae 4 we aimed to increase the pool of heme while avoiding negative feedback of the biosynthesis pathway, so we knocked-out the HAP1 gene, which resulted in increased capacity for protein production. We additionally performed genome-wide transcription analysis to study how the transcriptional regulation was affected. The analysis showed that the deletion of HAP1 inactivated respiration and enhanced the endogenous heme biosynthesis. We speculate that by allowing down-regulation of respiration and up regulation of heme-synthesis the cell found it beneficial to produce high levels of recombinant HbA by draining the toxic excess of intracellular heme.
Even more surprisingly, the deletion of HAP1 also resulted in up-regulation of all the cellular pathways related to protein synthesis such as the ribosomal subunits assembly, the cytosolic translation process and aminoacyl-tRNA biosynthesis among others, meanwhile the protein processing related reactions in the endoplasmic reticulum were downregulated.
We reported previously a direct relation between recombinant protein production and high levels of oxidative stress in yeast cells, due to the burden of additional protein folding processes generated in the ER.13 We showed that using anaerobic fermentation conditions mitigated this negative effect, decreasing oxidative stress by reducing the intracellular ROS content.14 However, further analysis of the transcriptional data (using a transcription factor enrichment algorithm) revealed that not only oxidative stress related processes were downregulated after HAP1 deletion, but also several other transcription factors involved in other stresses in the yeast cells (MSN2, MSN4, POS5 among others).4 At this stage we do not know how is the relation between heme metabolism and cell stress regulation mediated.
Concluding remarks
Internal heme levels and its main transcriptional regulator of biosynthesis Hap1p seem to be 2 very important elements when studying and developing more efficient yeast cell factories for protein production. For production of heme-proteins, the heme level is typically increased by adding external supplements such as the heme precursor 5-aminolevulinic acid (ALA) or hemin, prior to or throughout the fermentation. This is, however, a very expensive solution and is therefore a limiting factor for process scale up to industrial production levels. However, our previous work demonstrates that a hap1- strain can be a very good starting point for further engineering and thus overcome problems with heme depletion and hereby enhance productivity in a cost efficient manner.
In terms of other recombinant proteins, our work also confirms that there is a positive correlation between HAP1 deletion and cell performance, most likely due to reduced cellular stress, although more experimental work at this stage is required.
It would be also of interest to investigate the entire Hap1p regulatory network and learn more about pathways, genes and proteins which could lead us to other interesting engineering targets with a similar result. This kind of investigation would require a systems biology approach, combined with detailed cell physiology characterization and follow-up with molecular and cell biology studies, but it would lead toward new knowledge about the host cell, and hence the design of more robust production platforms.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The authors would like to thank funding from the Novo Nordisk Foundation, the Knut and Alice Wallenberg Foundation, FORMAS and the Sweblood consortium funded by the Swedish Strategic Research Foundation.
References
- 1.Nielsen J, Larsson C, van Maris A, Pronk J. Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol 2013; 24:398-404; PMID:23611565; http://dx.doi.org/ 10.1016/j.copbio.2013.03.023 [DOI] [PubMed] [Google Scholar]
- 2.Martinez JL, Liu L, Petranovic D, Nielsen J. Pharmaceutical protein production by yeast: towards production of human blood proteins by microbial fermentation. Curr Opin Biotechnol 2012; 23:965-71; PMID:22503236; http://dx.doi.org/ 10.1016/j.copbio.2012.03.011 [DOI] [PubMed] [Google Scholar]
- 3.Liu L, Martinez JL, Liu Z, Petranovic D, Nielsen J. Balanced globin protein expression and heme biosynthesis improve production of human hemoglobin in Saccharomyces cerevisiae. Metab Eng 2014; 21:9-16; PMID:24188961; http://dx.doi.org/ 10.1016/j.ymben.2013.10.010 [DOI] [PubMed] [Google Scholar]
- 4.Martinez JL, Liu L, Petranovic D, Nielsen J. Engineering the oxygen sensing regulation results in an enhanced recombinant human hemoglobin production by Saccharomyces cerevisiae. Biotechnol Bioeng 2015; 112:181-8; PMID:25082441; http://dx.doi.org/ 10.1002/bit.25347 [DOI] [PubMed] [Google Scholar]
- 5.Tiburzi F, Imperi F, Visca P. Is the host heme incorporated in microbial heme-proteins? IUBMB Life 2009; 61:80-3; PMID:18702108; http://dx.doi.org/ 10.1002/iub.123 [DOI] [PubMed] [Google Scholar]
- 6.Hardison R. Hemoglobins from bacteria to man: evolution of different patterns of gene expression. J Exp Biol 1998; 201:1099-117; PMID:9510523 [DOI] [PubMed] [Google Scholar]
- 7.Frankenberg N, Moser J, Jahn D. Bacterial heme biosynthesis and its biotechnological application. Appl Microbiol Biotechnol 2003; 63:115-27; PMID:13680202; http://dx.doi.org/ 10.1007/s00253-003-1432-2 [DOI] [PubMed] [Google Scholar]
- 8.Zitomer RS, Lowry CV. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev 1992; 56:1-11; PMID:1579104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anzaldi LL, Skaar EP. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect Immun 2010; 78:4977-89; PMID:20679437; http://dx.doi.org/ 10.1128/IAI.00613-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu JM, Fu WC. Intracellular co-expression of Vitreoscilla hemoglobin enhances cell performance and beta-galactosidase production in Pichia pastoris. J Biosci Bioeng 2012; 113:332-7; PMID:22099376; http://dx.doi.org/ 10.1016/j.jbiosc.2011.10.014 [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Sun Y, Shen X, Ke F, Zhao H, Liu Y, et al.. Intracellular expression of Vitreoscilla hemoglobin improves production of Yarrowia lipolytica lipase LIP2 in a recombinant Pichia pastoris. Enzyme Microb Technol 2012; 50:22-8; PMID:22133436; http://dx.doi.org/ 10.1016/j.enzmictec.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 12.Yu J, Jiang J, Fang Z, Li Y, Lv H, Liu J. Enhanced expression of heterologous inulinase in Kluyveromyces lactis by disruption of hap1 gene. Biotechnol Lett 2010; 32:507-12; PMID:20013302; http://dx.doi.org/ 10.1007/s10529-009-0182-3 [DOI] [PubMed] [Google Scholar]
- 13.Tyo KE, Liu Z, Petranovic D, Nielsen J. Imbalance of heterologous protein folding and disulfide bond formation rates yields runaway oxidative stress. BMC Biol 2012; 10:16; PMID:22380681; http://dx.doi.org/ 10.1186/1741-7007-10-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Osterlund T, Hou J, Petranovic D, Nielsen J. Anaerobic alpha-amylase production and secretion with fumarate as the final electron acceptor in Saccharomyces cerevisiae. Appl Environ Microbiol 2013; 79:2962-7; PMID:23435897; http://dx.doi.org/ 10.1128/AEM.03207-12 [DOI] [PMC free article] [PubMed] [Google Scholar]


