Abstract
The mammalian transcriptome has recently been revealed to encompass a large number of noncoding RNAs (ncRNAs) that play a variety of important regulatory roles in gene expression and other biological processes. MicroRNAs (miRNAs), the best studied of the short noncoding RNAs (sncRNAs), have been extensively characterized with regard to their biogenesis, function and importance in tumorigenesis. Another class of sncRNAs called piwi-interacting RNAs (piRNAs) has also gained attention recently in cancer research owing to their critical role in stem cell regulation. Long noncoding RNAs (lncRNAs) of >200 nucleotides in length have recently emerged as key regulators of developmental processes, including mammary gland development. lncRNA dysregulation has also been implicated in the development of various cancers, including breast cancer. In this review, we describe and discuss the roles of sncRNAs (including miRNAs and piRNAs) and lncRNAs in the initiation and progression of breast tumorigenesis, with a focus on outlining the molecular mechanisms of oncogenic and tumor-suppressor ncRNAs. Moreover, the current and potential future applications of ncRNAs to clinical breast cancer research are also discussed, with an emphasis on ncRNA-based diagnosis, prognosis and future therapeutics.
Keywords: noncoding RNAs, microRNAs, piwi-interacting RNAs, long noncoding RNAs, breast cancer
Introduction
Breast cancer is the most commonly diagnosed cancer and the second-leading cause of cancer death for women worldwide [1]. Although recent advances in early detection and cancer therapeutics have led to a decrease in breast cancer mortality rates, breast cancer is a heterogeneous, complicated disease that remains a major public health concern. The currently accepted model of breast tumorigenesis involves the stepwise, pathological progression from normal breast → breast hyperplasia → in situ carcinoma → invasive carcinoma → metastatic cancer [2]. This paradigm is strongly supported by clinical and epidemiological evidence as well as molecular clonality studies [3–5]. Malignant breast tumor progression is caused by multiple genetic and epigenetic alterations, which activate the various hallmarks of cancer [6]. The accumulation of these aberrations facilitates malignant transformation and confers cancerous phenotypes. Noncoding RNAs (ncRNAs) have recently gained wide spread attention as one cause of genetic and epigenetic dysregulation [7, 8].
Recent advances in DNA and RNA-sequencing techniques have revealed that only 2% of the human genome is composed of protein-coding genes [9]. However, >70–90% of the genome is actively transcribed into ncRNA molecules [10–13]. A growing body of evidence demonstrates that ncRNA molecules are critical regulators of gene expression, acting at both transcriptional and posttranscriptional levels with crucial roles in a variety of biological processes [7, 8]. ncRNAs can be divided into two major classes based on transcript size: small ncRNAs (sncRNAs) and long noncoding RNAs (lncRNAs) [14]. sncRNAs are <200 nucleotides in length and encompass microRNAs (miRNAs), endogenous small interfering RNAs, piwi-interacting RNAs (piRNAs) and the recently discovered transcription initiation RNAs [7, 14, 15]. miRNAs have been extensively investigated in cancer studies, and the deregulation of oncogenic and tumor-suppressor miRNAs in cancers has been shown to play a critical role in carcinogenesis [16]. The mammalian genome also transcribes a large number of ncRNAs that are longer than 200 nucleotides, called lncRNAs [17]. LncRNAs are a heterogeneous group of RNA molecules that have recently been shown to exploit multiple modes of action to regulate gene expression, and are involved in a wide spectrum of cellular processes [17]. Accumulating evidence has shown that lncRNAs have roles in both oncogenic and tumor-suppressor pathways [18].
This review focuses on the roles of miRNAs and lncRNAs in the development and progression of breast cancer, and introduces the emerging roles of piRNAs in breast cancer. This review also covers the translational applications of ncRNAs in the diagnosis, prognosis and therapy of breast cancer.
Biogenesis and function of ncRNAs
In this section, we summarize the recent advances in understanding the biogenesis and function of sncRNAs (with a special emphasis on miRNAs and piRNAs) and lncRNAs. In addition, we discuss how alterations in miRNA biogenic pathway components contribute to breast cancer development.
MicroRNAs
It is indisputable that miRNAs remain the best-characterized class of sncRNAs. The pathways for miRNA biogenesis and miRNA function have been widely reviewed by others [7, 19, 20] and will be only briefly described here. Sequence analysis has revealed that the majority of miRNAs are transcribed from the intergenic regions of the human genome [21, 22]. However, some miRNAs are transcribed from exonic or intronic regions as well [22, 23]. miRNA biogenesis is a multistep process, starting with the transcription of primary miRNAs (pri-miRNAs) by RNA polymerase II [7, 19, 20]. pri-miRNAs are processed into precursor miRNAs (pre-miRNAs, ∼70 nucleotides in length) by the RNase III Drosha-DGCR8-DDX5 microprocessor complex [7, 19, 20, 24], and are then exported to the cytoplasm by Exportin (a Ran-GFP-dependent transporter) [7, 19, 20]. In the cytoplasm, pre-miRNAs are cleaved by the RNase Dicer-TAR RNA-binding protein (TRBP) complex, producing mature, single-strand miRNAs with a length of 19–23 nucleotides [7, 19, 20]. Not all miRNAs go through the canonical miRNA biogenesis pathway. Special miRNAs known as mirtrons are produced from spliced introns with structural features similar to pre-miRNAs, and undergo a miRNA-processing pathway that bypasses the Drosha-mediated cleavage step [25].
It is estimated that miRNAs can regulate the expression of >60% of human genes via guiding a diverse set of multi-protein RNA-induced silencing complexes (RISC) to specifically target mRNAs [26]. The miRNA-associated RISC complexes consist of the argonaute (Ago) and glycine-tryptophan (GW) repeat-containing protein of 182 kDa (GW182) families of proteins, as well as other accessory proteins [27, 28]. The mode of miRNA-mediated gene expression silencing (mRNA decay or translational repression) is determined by the combinatory nature of the RISC complex components and the degree of the complementarity between the 8 nt miRNA seed sequence and the miRNA-targeting site in the 3′-untranslated region (3′-UTR) of mRNA [27, 28]. In addition to these miRNA-based silencing mechanisms targeting mRNAs, some miRNAs are complementary to gene promoters and mediate transcriptional activation and silencing through targeting of Ago/GW182-containing complexes to promoter regions [29, 30]. Therefore, miRNAs can modulate gene expression via multiple distinct mechanisms.
Since their discovery, extensive studies of miRNA function have been conducted in almost every cancer, and deregulated miRNAs have been recognized to play key roles in the carcinogenic process [16]. Dysregulation of oncogenic and tumor-suppressor miRNA expression in cancer can result from multiple pathological mechanisms occurring at the transcriptional or posttranscriptional level. For instance, DNA hypermethylation of the miRNA promoter has been identified in various cancers, including breast cancer, leading to silencing of miRNA expression at the transcriptional level [31]. Alterations affecting the functionality of protein regulators involved in pri-miRNA and pre-miRNA processing and miRNA maturation can also result in dysregulation of miRNA expression in cancer. Investigations using immunohistochemistry have shown that Dicer expression is progressively lost during breast cancer development, and is associated with features of aggressive behavior including higher histological grade, loss of hormone receptors and breast cancer 1 (BRCA1) protein expression, and with shorter disease-free survival (DFS) [32]. These findings suggest that global mature miRNA biogenesis is downregulated during breast tumorigenesis. This hypothesis is supported by a genome-wide miRNA profiling study of a large cohort of breast cancers that found a global decrease of miRNA expression in breast tumors compared with adjacent normal tissue, and a gradual decline in expression with progressive tumor grade [33]. In addition, several oncogenic and tumor-suppressor factors that are known to be altered in breast cancer have been identified to regulate expression or functionality of miRNA biogenic pathway components, which in turn alter miRNA expression. For example, oncogenic protein Cyclin D1 functions as a positive regulator that induces Dicer expression and is required for pre-miRNA processing [34]. Cyclin D1 and Dicer are coexpressed in luminal-A and basal-like subtypes of breast cancer, and Dicer function was demonstrated to be critical for the oncogenic functions mediated by Cyclin D1 (e.g. proliferation and migration) [34]. In contrast, epidermal growth factor receptor (EGFR) inhibits the biogenesis of tumor-suppressor miRNAs in response to hypoxia by phosphorylating Argonaute RISC catalytic component 2 (AGO2) at Tyr 393 and inhibiting its binding to Dicer [35]. Tumor suppressor proteins have also been shown to affect miRNA biogenesis. The tumor-suppressor BRCA1 directly interacts with Drosha and DDX5 of the Drosha microprocessor complex, accelerating the processing of pri-miRNA transcripts [36]. Alterations of these factors in certain breast cancers such as Cyclin D1-overexpressing, BRCA1-deficient and erb-b2 receptor tyrosine kinase 2 (HER2)-amplified breast cancers can result in dysregulation of miRNA biogenesis, leading to aberrations in miRNA-dependent gene expression networks and triggering tumorigenic progression.
Piwi-interacting RNAs
piRNAs are a recently discovered class of small ncRNAs that were first identified in Drosophila germ line cells, and range from 24 to 32 nucleotides in length [37]. In Drosophila, mature piRNAs are generated from the processing of single-stranded RNAs transcribed from genomic ‘piRNA clusters', the genomic intra- and inter-genic regions composed of the remnants of transposable elements (TEs), by PIWI family proteins and other factors [37]. In addition, piRNAs can be generated from another mechanism in the cytoplasm termed the ‘ping-pong' cycle, involving the primary antisense piRNA-directed cleavage of transposon transcripts by Aubergine and PIWI proteins [37].
In Drosophila, piRNAs are essential for sustaining genomic stability via the suppression of TEs at the transcriptional and posttranscriptional level [37]. piRNAs, in association with PIWI family factors, silence expression of TEs via triggering DNA methylation of TE loci [37]. However, only ∼20% of known mammalian piRNAs are mapped to transposons and other repeat genomic regions, suggesting that piRNAs may have other functional roles in addition to suppressing TEs [38, 39]. Compelling new evidence has revealed that in the soma, piRNAs can modulate histone modifications and DNA methylation in a sequence-specific manner, enabling piRNAs to modulate the chromosomal conformation and regulate gene expression [39]. Dysregulation of piRNAs and proteins (e.g. PIWI family proteins) involved in piRNA biogenesis has been found in a variety of cancers, including breast cancer [39, 40], highlighting the emerging roles of piRNA-mediated epigenetic events in tumorigenic processes of human somatic cancers.
Long noncoding RNAs
LncRNAs represent the most numerous and functionally diverse class of ncRNAs [8, 14, 18]. LncRNAs are transcribed from introns or intergenic regions in either the sense or antisense orientation relative to protein-coding genes. The majority of lncRNAs are polyadenylated and transcribed by RNA polymerase II [41–43], whereas lncRNAs lacking poly(A) tails are generally transcribed by RNA Polymerase III [44, 45]. LncRNAs are regulated in both spatial and temporal manners as the expression of protein-coding genes [13, 46]. The primary nucleotide sequences as well as complex secondary structures of lncRNAs and their cell-type/developmental stage-specific expression allow them to target genomic loci or interact with other RNA molecules (mRNA or ncRNAs) and proteins to regulate a variety of biological processes in the cell in a temporal and spatial manner [14, 47, 48].
LncRNAs can regulate chromatin remodeling, protein functionality and gene expression through multiple mechanisms [47, 49, 50]. LncRNAs can serve either as guiding scaffolds to assist nuclear transcriptional regulators to bind specific DNA elements or as scaffolds for the formation of cellular substructures (e.g. paraspeckles) or protein complexes [8, 18, 51, 52]. In addition, some nuclear lncRNAs have been shown to interact with splicing factors or directly with pre-mRNAs to affect RNA splicing [51]. By hybridizing with RNA transcripts or with regulatory sncRNAs (e.g. miRNAs), lncRNAs can regulate the translational activity and decay of mRNAs. Some lncRNAs have been found to contain miRNA targeting sites and can serve as endogenous miRNA sponges to suppress the inhibitory effects of miRNAs on mRNA translation and stability [8, 18, 53]. This lncRNA-mediated sponge function has been shown to be crucial for a number of cellular processes, including cell differentiation and pluripotency [8, 18, 49, 53]. LncRNAs serve as important regulatory molecules of gene expression and protein functionality at multiple levels, and their deregulation plays a key role in tumorigenesis [8, 18, 53].
miRNAs in breast tumorigenesis
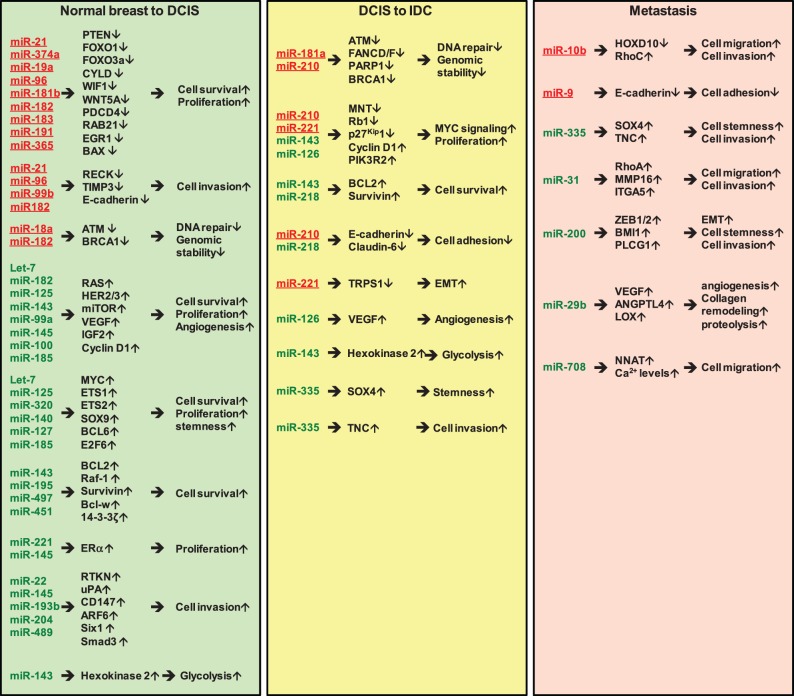
The recent progress in RNA deep sequencing techniques has significantly advanced miRNA research and the understanding of their roles in breast cancer initiation and progression. Volinia et al. recently used custom-designed algorithms to analyze small-RNA deep-sequencing data generated by Farazi et al. and identified key miRNAs involved in the transition from normal breast-to-ductal carcinoma in situ (DCIS) and DCIS-to-Invasive Ductal Carcinoma (IDC) [54, 55]. In all, 6 normal breast, 8 DCIS and 80 IDC cases were analyzed in their study [55]. We summarized these key miRNAs, their gene targets and functional roles in these pathological transitions in Table 1. Seven miRNAs that are implicated in the metastatic process of breast cancer are also summarized in Table 1.
Table 1.
Summary of the miRNAs involved in breast cancer progression
| Cancer progression stage | miRNA | Expression | Target genes | Function | References | ||
|---|---|---|---|---|---|---|---|
| Normal breast to DCIS | miR-18a | Up | ATM | miR-18a impairs DNA damage response via downregulation of ATM. | [90] | ||
| miR-19a | Up | PTEN | miR-19 enhances chemoresistance via the activation of oncogenic PI3K-AKT signaling. | [92] | |||
| miR-21 | Up | PDCD4, PTEN, TIMP3, RECK | miR-21 suppresses apoptosis and activates PI3K signaling to promote cell survival and tumorigenesis. | [91, 99, 101] | |||
| miR-26b | Up | CHD1, GREB1, KPNA2 | miR-26b is the downstream target of the ERα-Myc signaling axis and is implicated in regulating ERα downstream target gene expression. | [89] | |||
| miR-93 | Up | JAK1, STAT3, AKT3, SOX4, EZH1, and HMGA2 | miR-93 suppresses TGFβ signaling to promote mesenchymal-to-epithelial transition (MET) and luminal tumorigenesis. | [110] | |||
| miR-96 | Up | FOXO3a, FOXO1, RECK | miR-96 promotes cell proliferation and survival. | [93, 94, 102] | |||
| miR-99b | Up | E-cadherin, ZO-1 | miR-99b is the downstream target of TGFβ signaling and promote EMT. | [113] | |||
| miR-106b | Up | EP300, Smad7 | miR-106b is involved in modulating TGFβ signaling to promote EMT for increasing the motility and invasiveness of breast cancer cells | [114] | |||
| miR-181b | Up | CYLD | miR-181b is the STAT3 downstream target to mediate NFkB activation for enhancing tumorigenic cell transformation. | [95] | |||
| miR-182 | Up | FOXO1, BRCA1, RECK | miR-182 suppresses BRCA1-dependent DNA repair to enhance genomic instability and promotes the invasiveness of breast cancer cells. | [93, 96, 103] | |||
| miR-183 | Up | RAB21 | miR-183 increases cell proliferation and migration. | [104] | |||
| miR-191 | Up | EGR1 | miR-191 is positively regulated by ERα signaling and mediates survival of ER+ breast cancer cells. | [97] | |||
| miR-200b, miR-200c | Up | ZEB1, ZEB2, BMI1, PLCG1 | miR-200c suppresses TGFβ signaling and EMT. | [109] | |||
| miR-365 | Up | SHC1, BAX | miR-365 targets pro-apoptotic regulators to promote cell survival. | [100] | |||
| miR-374a | Up | WIF1, PTEN, WNT5A | miR-374a activates WNT/β-catenin signaling to promote tumorigenesis. | [98] | |||
| miR-429 | Up | ZEB1 | miR-429 inhibits cell migration and invasion of breast cancer cells. | [109] | |||
| miR-449a | Up | FOS, Met | miR-449a suppresses EMT by targeting FOS and Met. | [111] | |||
| let-7b, 7c, 7d | Down | MYC, HMGA2, H-RAS | Let-7 miRNAs act as tumor suppressors to inhibit oncogenic pathways. | [62, 66] | |||
| miR-22 | Down | TET1-3, CD147 | miR-22-mediated suppression of ten eleven translocation (TET) family proteins (TET1-3) induce the silencing of miR-200 expression. miR-22 overexpression suppresses breast cancer cell invasion, metastasis, and proliferation by targeting CD147. | [78, 81] | |||
| miR-99a | Down | mTOR | miR-99a decreases cell viability and induces apoptosis via targeting mTOR signaling. | [60] | |||
| miR-100 | Down | IGF2 | miR-100 suppresses oncogenic insulin-like growth factor 2 (IGF2)-mTOR signaling to inhibit breast tumorigenesis. | [61] | |||
| miR-125a, miR-125b | Down | HER2, HER3, ETS1 | miR-125 targets the oncogenic tyrosine kinase receptors and transcription factor to suppress tumorigenesis. | [58, 68] | |||
| miR-127-3p | Down | BCL6 | miR-127-3p inhibits carcinogenesis. | [67] | |||
| miR-140 | Down | SOX9, ALDH1 | miR-140 targets stem-cell factors to inhibit the self-renewal of breast cancer cells. | [56] | |||
| miR-143 | Down | HER3, Cyclin D1, BCL-2, Survivin, hexokinase 2 | miR-143 targets oncogenic and anti-apoptotic molecules to suppress proliferation and induce apoptosis of breast cancer cells. It also inhibits glycolysis of cancer cells. | [59, 72, 74, 85] | |||
| Normal breast to DCIS | miR-145 | Down | RTKN, ERα, N-RAS, VEGFA, HER3, ARF6 | miR-145 targets multiple signal transduction pathways (Rho, ERa, Ras, HER3, ARF6 and VEGF) to inhibit breast tumor cell growth and angiogenesis. | [59, 63, 79, 80, 88] | ||
| miR-185 | Down | VEGFA, E2F6, DNMT1 | miR-185 acts as a tumor suppressor to inhibit the proliferation and invasion of breast cancer cells via alleviating the expression of oncogenic molecules E2F6, DNMT1 and VEGFA. | [70, 77] | |||
| miR-193b | Down | uPA | miR-193b targets the urokinase-type plasminogen activator to suppress tumor progression and invasion in breast cancer. | [84] | |||
| miR-195 | Down | Raf-1, BCL-2 | miR-195 targets survival factors and signaling pathways to sensitize breast cancer cells to chemotherapy. | [64, 65] | |||
| miR-204 | Down | Six1 | miR-204 suppresses EMT via downregulating Six1 expression. | [82] | |||
| miR-221/222 | Down | ERα | miR-221/222 suppresses ER-dependent tumorigenesis. | [87] | |||
| miR-320 | Down | ETS2 | miR-320 targets the oncogenic transcription factor ETS2 to suppress the self-renewal of breast CSCs. | [69] | |||
| miR-451 | Down | 14-3-3ζ | miR-451 decreases breast cancer cell survival via downregulating 14-3-3ζ expression. | [76] | |||
| miR-489 | Down | Smad3 | miR-489 is an EMT suppressor via targeting TGFβ1-Smad3 signaling. | [83] | |||
| miR-497 | Down | Bcl-w | miR-497 is a pro-apoptotic regulator to induce cell death. | [75] | |||
| DCIS to IDC | miR-181a | Up | ATM | miR-181a promotes EMT and cell survival to enhance the self-renewal of breast cancer stem cells. | [121] | ||
| miR-210 | Up | FANCD/F, PPP2CA, Rb1, NLK, PARP1, BRCA1, CDH1, MNT | miR-210 targets the MYC inhibitor MNT to activate oncogenic MYC signaling. It may also directly or indirectly target multiple factors involved in tumor suppression, DNA repair and cell adhesion. | [55, 127] | |||
| miR-221/222 | Up | p27Kip1, TRPS1 | miR-221/222 promotes cell proliferation by targeting the CDK inhibitor p27Kip1 and facilitates EMT by targeting the ZEB2 inhibitor TRPS1. | [124, 125] | |||
| miR-126 | Down | VEGFA, PIK3R2 | miR-126 acts as a tumor suppressor to inhibit breast cancer cell proliferation. | [118] | |||
| miR-143 | Down | HER3, Cyclin D1, BCL-2, Survivin, hexokinase 2 | miR-143 inhibits tumor cell survival, proliferation and glycolysis. | [59, 72, 74, 85] | |||
| miR-218 | Down | HOXB3, Survivin | miR-218 activates tumor-suppressive molecules (RASSF1A and claudin-6) to inhibit tumorigenesis. | [117, 119] | |||
| miR-335-5p | Down | SOX4, TNC | miR-335-5p suppresses the invasive transition of breast cancer cells. | [120] | |||
| Metastasis | miR-10b | Up | HOXD10, TIAM1 | miR-10b is the first identified microRNA upregulated in metastatic breast cancer cells by the EMT transcription factor Twist and is required for tumor cell invasion and metastasis. | [116, 130] | ||
| miR-9 | Up | E-cadherin | miR-9 expression is induced by Myc in breast cancer cells to promote EMT and angiogenesis in breast tumors. | [131] | |||
| miR-335 | Down | SOX4, PTPRN2, TNC, MERTK | miR-335 acts as a metastatic suppressor by inhibiting the stemness and metastatic transformation of breast cancer cells. | [120] | |||
| miR-31 | Down | ITGA5, RhoA, RDX, Fzd3, MMP16 | miR-31 inhibits pro-metastatic signaling pathways. | [132] | |||
| miR-200 | Down | ZEB1, ZEB2, BMI1, PLCG1. moesin | miR-200 inhibits EMT to suppress breast cancer metastasis. | [109, 133] | |||
| miR-29b | Down | VEGFA, ANGPTL4, PDGF, LOX, MMP9 | miR-29b suppresses breast cancer cell metastasis by inhibiting signaling networks associated with angiogenesis, collagen remodeling and proteolysis. | [135] | |||
| miR-708 | Down | NNAT | miR-708 inhibits breast cancer migration and metastasis by regulating intracellular Ca2+ levels. | [136] | |||
The overlapped miRNA genes between Volinia's and Hannafon's miRNA profiling datasets (see Figure 1) are underlined.
From their analysis, 66 miRNAs were identified to be differentially expressed in DCIS compared to normal breast [55]. This differentially expressed miRNA profile was largely maintained in the DCIS to IDC transition [55], consistent with previous studies showing that the genetic profiles of DCIS tumors are remarkably similar to IDC tumors when histological grade and hormone receptor status are matched [3–5]. Therefore, miRNA profiling analysis of DCIS and IDC by Volinia et al. [55] supports the well-accepted paradigm that DCIS is a nonobligate precursor of IDC. Among the miRNAs that are differentially expressed in DCIS relative to normal breast, 19 were upregulated ≥3-fold and 25 downregulated ≥3-fold in DCIS. Among these 44 differentially expressed miRNAs, 12 upregulated and 17 downregulated miRNAs with reported roles in breast cancer are listed in Table 1. miR-140, recently identified by us to be progressively downregulated in DCIS, is also included in the list [86].
Hannafon et al. [121] performed expression profiling of 365 miRNA by real-time quantitative polymerase chain reaction assays to generate a miRNA expression signature comparing normal breast epithelium from reduction mammoplasty (n = 9) to paired samples of histologically normal epithelium and DCIS (n = 16). We compared the 30 relevant miRNAs from Volinia's studies with 31 miRNAs identified by Hannafon et al. [121], which were differentially expressed in DCIS tumors relative to normal tissue controls and found that 11 differentially expressed miRNAs were consistently detected in these two independent analyses (Figure 1 and Table 1). These six upregulated (miR-21, miR-93, miR-182, miR-183, miR-200 b, miR-200 c) and five downregulated (let-7 c, miR-99 a, miR-125 b, miR-127-3 p, miR-145) may represent the key miRNA signature for the pathological progression from normal breast to DCIS. Six upregulated (miR-18 a, miR-99 b, miR-181 b, miR-191, miR-365 and miR-449 a) and three downregulated (miR-195, miR-204 and miR-489) miRNAs found only in Hannafon's studies have reported roles in breast tumorigenesis and are also included in Table 1. This comparison shows that only one-third of their respective breast-cancer-relevant miRNAs can be commonly identified by these two data sets. This may be owing to a variation in tissue samples used in these two independent studies. It has also recently been shown that dysregulation of miRNA expression can be breast cancer subtype specific [33].
Figure 1.
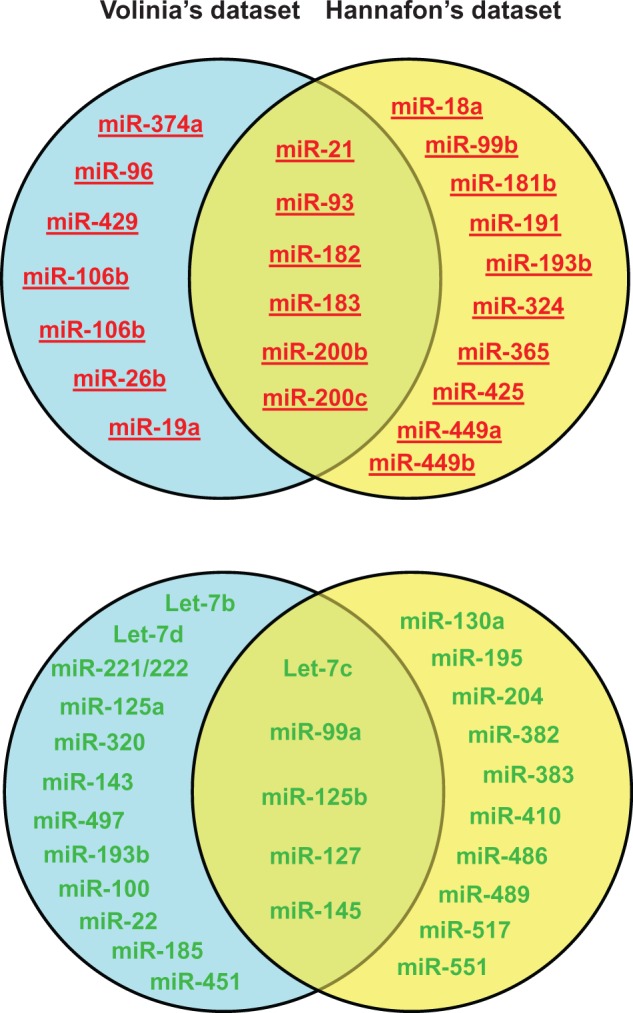
The diagram to show miRNAs differentially expressed in DCIS relative to normal breast tissue from two different miRNA profiling data sets. Upregulated and downregulated miRNAs are indicated by red/underlining and green colors, respectively. The upregulated and downregulated miRNAs in the overlapped areas are differentially expressed miRNAs commonly identified in both Volinia’s and Hannafon’s data sets. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
miRNAs downregulated in DCIS
All 21 miRNAs downregulated in DCIS tumors relative to normal counterparts are classified as tumor-suppressive miRNAs, and are involved in regulating multiple biological processes and signal transduction pathways to suppress breast tumorigenesis. For instance, oncogenic signaling pathways that are involved in promoting cell proliferation, survival and invasiveness including HER2/HER3 (miR-125 a, miR-125 b, miR-143 and miR-145) [83, 87], mechanistic target of rapamycin (mTOR, miR-99 a) [81], insulin-like growth factor 2 (IGF2; miR-100) [82], RAS (let-7 b, 7 c, 7 d and miR-145) [77, 91] and Raf-1 (miR-195) [98, 99], are activated in DCIS via the downregulation of their respective inhibitory miRNAs (Table 1). Moreover, some tumor-suppressor miRNAs inhibit oncogenic transcription factors, including Myc (let-7 b, 7 c and 7 d) [78], B-cell CLL/lymphoma 6 (BCL6, mir-127-3p) [85], v-ets avian erythroblastosis virus E26 oncogene homolog 1 (ETS1, mir-125 a, miR-125 b) [84], v-ets avian erythroblastosis virus E26 oncogene homolog 2 (ETS2, mir-320) [102], SRY box 9, (SOX9, miR-140) [86] and E2F transcription factor 6 (E2F6, miR-185) [95], all of which were found to be downregulated in DCIS (Table 1).
During the tumorigenic transformation process, premalignant breast epithelial cells lose cell polarity and gain deregulated proliferation, leading to rapid growth and formation of the tumor mass. This pathological progression involves enhanced survival signaling and inhibition of anoikis, the apoptotic response that is normally activated when cell polarity is lost [122]. Some of the miRNAs downregulated in DCIS are implicated in the downregulation of anti-apoptotic factors, including BCL2 (miR-143, miR195) [88, 99, 123], Survivin (miR-143) [89], Bcl-w (miR-497) [105] and 14-3-3ζ (miR-451) [103] (Table 1). In addition, these identified tumor-suppressive miRNAs also functionally target other biological processes including cell-cycle progression (Cyclin D1, targeted by miR-143) [89], angiogenesis (vascular endothelial growth factor A [VEGFA], targeted by miR-145 and miR-185) [91, 96], epigenetic regulation (TET1-3, targeted by miR-22; DNA (cytosine-5)-methyltransferase 1 [DNMT1], targeted by miR-185) [79, 95], Rho-dependent signal transduction (RTKN, targeted by miR-145) [92], invasion (ADP ribosylation factor 6 [ARF6], targeted by miR-145; CD147, targeted by miR-22; Six1, targeted by miR-204; Smad3, targeted by miR-489) [80, 93, 100, 104] and degradation of the extracellular matrix (ECM) (urokinase plasminogen activator, uPA, targeted by miR-193 b) [97] (Table 1).
In addition to targeting HER3, Cyclin D1, B-cell CLL/lymphoma 2 [BCL2] and Survivin, miR-143 has been shown to target hexokinase 2, leading to suppression of glycolysis (Table 1) [90]. This metabolic regulation mechanism is preferentially exploited by cancer cells to metabolize glucose to generate energy [124]. Downregulation of miR-143 in DCIS releases inhibition of hexokinase 2 and increases glycolysis in breast cancer cells. This leads to the ‘Warburg effect', the observation that most cancer cells undergo high rates of glycolysis followed by lactic acid fermentation, instead of the low rates of glycolysis followed by mitochondrial oxidation of pyruvate via the citric acid cycle that occurs in most normal cells [124].
Importantly, two miRNAs (miR-221/222 and miR-145) known to target estrogen receptor α (ERα) [94, 101] were identified to be downregulated in DCIS (Table 1). Given that the majority of DCIS tumors are ER-positive, downregulation of miR-221/222 and miR-145 results in upregulation of ERα and increased ERα signaling, promoting DCIS tumor cell growth. Consistent with this, miR-26 b, a downstream target of the ERα-Myc signaling axis [61], is upregulated in DCIS (Table 1). Collectively, the decreased expression of these 21 tumor-suppressive miRNAs contributes to increased cell survival, proliferation and glycolysis of DCIS tumor cells, as well as enhanced angiogenesis and ECM degradation in the tissue microenvironment (Figure 2).
Figure 2.
The flow diagram for the roles of miRNAs in breast cancer progression. Upregulated and downregulated miRNAs are indicated by red/underlining and green colors, respectively. The gene targets and outcomes for the dysregulated miRNAs are presented in the flow diagram. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
miRNAs upregulated in DCIS
From the upregulated miRNA signature, 10 oncogenic miRNAs, including miR-18 a, miR-19 a, miR-21, miR-96, miR-181 b, miR-182, miR-183, miR-191, miR-365 and miR-374 a, were identified in DCIS (Table 1). Their gene targets encompass eight tumor suppressors (ATM, PTEN, FOXO1, FOXO3a, CYLD, BRCA1, EGR1 and WIF1) [56–58, 63, 64, 69, 72, 75], three pro-apoptotic factors (programmed cell death 4, PDCD4, SHC1 and BAX) [58, 59, 74], two inhibitors of matrix metalloproteinases (TIMP3 and RECK) implicated in regulating ECM degradation [60, 65, 70] and a GTPase (RAB21) involved in the control of cellular membrane traffic [71] (Table 1). The protein ataxia telangiectasia mutated (ATM), targeted by miR-18 a, is a kinase that plays a pivotal role in the maintenance of genomic integrity through the activation of cell cycle checkpoints, enhancing repair of DNA double-strand breaks [56]. A low level of ATM in breast cancer has been shown to correlate with poor outcome [125]. In addition to ATM, BRCA1, targeted by miR-182, is another critical factor involved in maintaining genomic stability through DNA double-stranded break repair [69]. BRCA1 is also implicated in transcriptional regulation to suppress the tumorigenic progression of breast and ovarian cancers [126]. Phosphatase and tensin homolog (PTEN), targeted by miR-19 a, miR-21 and miR-374 a [57, 58, 75], is a well-known tumor suppressor that functions as a phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase to negatively regulate PI3K-AKT signaling, a critical oncogenic signaling pathway for breast tumorigenesis [127]. WNT inhibitory factor 1 (WIF1), targeted by miR-374 a, is known to function as an inhibitor of WNT-β-catenin signaling by competing with WNT ligands to bind their cognate receptors [75]. The tumor suppressor cylindromatosis (CYLD), targeted by miR-181 b, is a deubiquitinating enzyme that inhibits the NFκB and mitogen-activated protein kinase (MAPK) activation pathways by deubiquitinating upstream regulatory factors [68, 128]. Early growth response gene 1 (EGR1) is a tumor-suppressive transcription factor involved in mediating hormone starvation-induced apoptosis in ER-positive breast cancer cells [72]. Through targeting EGR1, miR-191 protects ER-positive breast cancer cells from apoptosis induced by hormone deprivation [72]. FOXO1 and FOXO3a, targeted by miR-96 and miR-182, are two well-known forkhead box-containing transcription factors with tumor-suppressive roles in regulating cell cycle progression, DNA repair and apoptosis [63, 64]. Rab21, targeted by miR-183, has been shown to attenuate EGF-mediated MAPK signaling by promoting the internalization and degradation of EGFR [71]. Therefore, upregulation of these oncogenic miRNAs in DCIS is expected to facilitate cell survival, proliferation, genomic instability and degradation of the ECM (Figure 2), potentiating the tumor-promoting effects from the downregulation of tumor-suppressive miRNAs.
Several miRNAs involved in negative regulation of the TGFβ-mediated epithelial-to-mesenchymal transition (EMT), including miR-200 family members (miR-200 b, miR-200 c, miR-429), miR-93 and miR-449 a [62, 73, 76], were found to be upregulated in DCIS (Table 1). Upregulation of miR-200 is in part owing to downregulation of miR-22, which inhibits expression of the demethylating enzymes ten eleven translocation (TET 1-3). TET proteins catalyze DNA demethylation of the miR-200 gene promoter, inducing miR-200 expression [129]. Intriguingly, miR-99 b and miR-106 b, which are both activators of TGFβ-mediated EMT [66, 67], were upregulated in DCIS (Table 1). Moreover, as shown in Table 1, non-histone chromatin-binding protein HMGA2, a mediator of TGFβ signaling, is a target of let-7 [77]. Downregulation of let-7 leads to increased HMGA2 expression and TGFβ-mediated EMT [130]. Therefore, in the transition from normal breast to DCIS, TGFβ-mediated EMT may be regulated either by tumor-subtype-specific miRNAs (this topic is discussed in the following section) or by a complicated network of both positive and negative miRNA regulators. Further studies are needed to address the role of this regulation in the progression to DCIS.
miRNAs dysregulated in the DCIS-to-IDC transition
By comparing IDC biopsies with DCIS tumors, Volinia et al. [55] identified nine miRNAs that are differentially expressed in IDC compared with DCIS. They defined these nine miRNAs as the ‘invasiveness microsignature' that can be used to differentiate IDC from DCIS: let-7 d, miR-181 a, -210 and -221 were activated, whereas miR-10 b, -126, -218, -335-5 p and -143 were repressed [55]. While upregulation of let-7 d and downregulation of miR-10 b in IDC relative to DCIS are not consistent with their reported functional roles in breast cancer [77, 78, 114], the rest of the invasiveness microsignature miRNAs exhibit consistency between their functional roles in breast cancer and their expression statuses in the DCIS-to-IDC transition. We list these seven key miRNAs, their gene targets and functions in Table 1.
The four miRNAs (miR-126, -218, -335-5 p and -143) downregulated in the DCIS-to-IDC transition are all tumor suppressors, and are involved in promoting cell adhesion (by activating claudin-6) [111] and inhibiting VEGF-dependent angiogenesis [110], oncogenic PI3K-AKT signaling (PIK3R2) [110], G1-S transition (Cyclin D1) [89], cell survival (BCL2, Survivin) [88, 89, 112], stemness pluripotency (SOX4) [113] and invasiveness (tenascin C, TNC) [113] (Table 1).
Three miRNAs (miR-181 a, miR-210 and miR-221/222) which were activated during the DCIS-to-IDC transition [55], are implicated in stimulating cell proliferation and EMT, and in inhibiting cellular DNA repair ability and cell adhesion (Table 1). miR-181a is a downstream target of TGFβ signaling that enhances EMT, migration and invasiveness [106, 131]. Loss of ATM, a target of miR-181 a in breast cancer [106], has been shown to accelerate pancreatic cancer formation and EMT [132], suggesting miR-181 a-mediated repression of ATM may contribute to aggressiveness in breast cancer [106]. miR-221/222 is downstream of the RAS pathway and its expression is also transactivated by the basal-like transcription factor FOSL1 [108]. miR-221/222 promotes EMT by targeting trichorhinophalangeal 1 (TRPS1), a member of the GATA family that transcriptionally represses the expression of Zinc finger E-box-binding homeobox 2 (ZEB2) to suppress ZEB2-induced EMT [108]. miR-221/222 can also enhance cell proliferation via targeting the cylin-dependent kinase (CDK) inhibitor p27Kip1 [109]. miR-210 has been reported to be inducible by hypoxia and to regulate genes implicated in tumor initiation [133]. It has also been shown that miR-210 can target the Myc antagonist MNT (MAX network transcriptional repressor) to activate Myc-dependent oncogenic signaling [107]. Analysis by Volinia et al. [55] also indicates that a number of breast cancer-related genes were expressed in an antagonistic fashion to miR-210 along the DCIS-IDC progression axis, suggesting they represent potential targets of miR-210 or downstream effectors. These genes are functionally involved in the apoptotic caspase cascade, HER2 receptor recycling, tumor necrosis factor receptor 1 (TNFR1) signaling, Fas cell surface death receptor (FAS) signaling and BRCA1-related cancer susceptibility [55]. Overall, multiple studies show that signal transduction pathways and molecules implicated in promoting cell survival, proliferation, EMT, invasion and angiogenesis elicit the progression of DCIS to IDC by deregulating their corresponding miRNA regulators (Figure 2).
The roles of miRNAs in breast cancer metastasis
Metastasis, the leading cause of breast and other cancer deaths, is a multistep process involving extensive vascularization at the primary tumor site, tumor cell mobilization and invasion, tumor cell dissemination via blood circulation and colonization at distant tissue sites. Genetic and epigenetic dysregulation of molecular mechanisms controlling these processes is believed to be the main cause of metastasis. The acquisition of EMT and angiogenic ability is a key requirement for tumor cells to become metastatic [134]. Seven representative miRNAs that have been identified to target crucial factors implicated in regulating these metastatic processes are listed in Table 1.
miR-10 b is a metastatic promoter in breast cancer that is induced by the EMT transcription factor Twist [114]. Through targeting HOXD10, miR-10 b activates the expression of RhoC, a pro-metastatic factor promoting cell migration and invasion [114]. However, three lines of contradictory evidence indicate that miR-10 b may serve as a metastatic suppressor: (1) miR-10 b is not associated with metastasis and prognosis of breast cancer, but is inversely correlated with tumor size and grade [135]; (2) it inhibits tumor cell migration and invasion via targeting TIAM1 (T lymphoma invasion and metastasis 1) [115]; (3) its expression is repressed in the DCIS-to-IDC transition [55]. This discrepancy may be attributable to the distinct contexts existing in their experimental systems and more evidence is needed to clarify the exact role of miR-10 b in breast cancer metastasis. miR-9 has also been reported to be a metastatic promoter. miR-9 is a downstream target of Myc and represses E-cadherin expression, activating β-catenin signaling and stimulating VEGF-mediated angiogenesis and metastasis [116]. Tavazoie et al. [113] identified miR-335 as a metastatic suppressor that impedes migration and metastasis through targeting progenitor cell transcription factor SOX4 and ECM component tenascin C. miR-335 is frequently lost in primary breast tumors from patients with relapse, and its loss is associated with poor distal metastasis-free survival [113]. In addition to miR-335, miR-31 was reported by Valastyan et al. [117] as another metastatic suppressor that represses a cohort of metastasis-promoting genes, including RhoA and ITGA5, to inhibit several steps of metastasis, including local invasion, extravasation, initial survival at a distant site and metastatic colonization.
The miR-200 family members are well-known inhibitors of E-cadherin transcriptional repressors ZEB1 and ZEB2, and are also implicated in the negative regulation of breast cancer metastasis through targeting of either ZEB1/2 or moesin [73, 118]. miR-200 is negatively regulated by PELP1 (proline, glutamic acid and leucine rich protein 1), a metastatic activator, releasing miR-200-mediated suppression of EMT [136]. Chou et al. [119] recently identified miR-29 b as the transcriptional target of GATA3, a transcription factor that specifies and maintains mammary luminal epithelial cell fate. The GATA3-miR29b signaling axis was found to be inactivated in invasive breast cancer and to be responsible for suppressing metastatic progression via miR-29 b-mediated targeting of a network of pro-metastatic regulators involved in angiogenesis, collagen remodeling and proteolysis [119]. miR-29 b is enriched in luminal breast cancers, and its loss increases metastasis and promotes a mesenchymal phenotype [119]. Intriguingly, miR-708 was recently identified by Ryu et al. [120] as a metastatic inhibitor via targeting of the endoplasmic reticulum protein neuronatin (NNAT) to decrease intracellular calcium levels, giving rise to a reduction in activation of ERK and FAK, decreased cell migration and impaired metastases.
The roles of miRNAs in breast cancer molecular subtypes
Breast cancer is a heterogeneous disease that manifests variable responses to anti-cancer therapy in a tumor-specific manner. For clinicians to predict patients’ outcomes and determine the most effective therapeutic strategy, it is necessary to completely characterize and classify the differences between tumor types. Therefore, the development of detailed classification systems is needed to stratify breast cancer into specific subtypes. Gene expression profiling has emerged as an effective system for breast cancer classification [137–139], and has been used to define five intrinsic molecular subtypes of invasive breast carcinoma: luminal A, luminal B, HER2-enriched, normal- and basal-like [137, 138]. Recently Dvinge et al. [33] conducted miRNA profiling analysis of a large cohort of breast IDC tumors (n = 1302) using microarrays. In their studies, the expression status of 103 miRNAs was characterized across the five breast cancer molecular subtypes. Through interrogation of their analysis results, we found that 21 of the 53 miRNAs involved in breast cancer progression (listed in Table 1) display molecular subtype-specific expression patterns (Table 2). This analysis indicates that variation in the molecular-subtype composition of analyzed breast tumor samples can significantly affect miRNA profiling results. This may explain why only one-third of differentially expressed miRNAs overlaps between Volinia's and Hannafon's data sets (Figure 1).
Table 2.
Molecular subtype-specific expression of miRNAs involved in breast cancer (BC) progression
| miRNA | The role in BC progression | Expression status in BC | Expression status in molecular subtypes of IDC |
|---|---|---|---|
| miR-18 a | Transition to DCIS | Upregulation | Upregulation in basal-like IDC; downregulation in luminal-A IDC |
| miR-19 a | Transition to DCIS | Upregulation | Upregulation in basal-like IDC |
| miR-26 b | Transition to DCIS | Upregulation | Downregulation in basal-like IDC; upregulation in luminal-A IDC |
| miR-93 | Transition to DCIS | Upregulation | Upregulation in basal-like IDC |
| miR-96 | Transition to DCIS | Upregulation | Upregulation in luminal-A IDC |
| miR-106 b | Transition to DCIS | Upregulation | Upregulation in basal-like IDC |
| miR-183 | Transition to DCIS | Upregulation | Upregulation in HER2-enriched IDC |
| miR-365 | Transition to DCIS | Upregulation | Downregulation in basal-like IDC |
| miR-449 a/449 b | Transition to DCIS | Upregulation | Downregulation in basal-like IDC; upregulation in luminal-A/B IDC |
| miR-99 a | Transition to DCIS | Downregulation | Downregulation in luminal-B IDC |
| miR-100 | Transition to DCIS | Downregulation | Downregulation in luminal-B IDC |
| miR-125 b | Transition to DCIS | Downregulation | Downregulation in luminal-B IDC |
| miR-193 b | Transition to DCIS | Downregulation | Downregulation in basal-like IDC; upregulation in luminal-B IDC |
| miR-195 | Transition to DCIS | Downregulation | Downregulation in basal-like and HER2-enriched IDC; Upregulation in luminal-A IDC |
| miR-497 | Transition to DCIS | Downregulation | Downregulation in basal-like and HER2-enriched IDC |
| miR-210 | DCIS-to-IDC transition | Upregulation | Upregulation in basal-like and HER2-enriched IDC |
| miR-126 | DCIS-to-IDC transition | Downregulation | Downregulation in basal-like IDC |
| miR-143 | Transition to DCIS; DCIS-to-IDC transition | Downregulation | Downregulation in basal-like IDC |
| miR-9 | Metastasis | Upregulation | Upregulation in basal-like IDC |
| miR-10 b | Metastasis | Upregulation | Downregulation in basal-like IDC; upregulation in luminal-A IDC |
The analyzed results shown in Table 2 provide useful information for delineating the roles of the 21 miRNAs in the pathological development of molecular subtypes, and in their physiological as well as molecular phenotypes. Expression of six miRNAs (miR-9, miR-18 a, miR-19 a, miR-93, miR-106 b, miR-210) is upregulated in basal-like IDC, whereas expression of 10 miRNAs (miR-10 b, miR-26 b, miR-126, miR-143, miR-193 b, miR-195, miR-326, miR-449 a, miR-449 b, miR497) is downregulated in the same molecular subtype (Table 2). Dysregulation of these 16 miRNAs contributes to the tumorigenic development of basal-like breast tumors, an aggressive breast cancer subtype characterized by high tumor grade, proliferation rate, frequency of recurrence and the presence of p53 mutations. Patients with basal-like breast cancers frequently have poor prognosis, and are difficult to treat owing to the frequent lack of hormone receptors (a triple-negative phenotype with loss of ER and PR, and no HER2 amplification) that can be therapeutically targeted [138–140]. Six of these 16 basal-related miRNAs have been reported to be associated with basal-like/triple-negative breast cancer, including miR-9, miR-18 a, miR-19 a, miR-93, miR-106 b and miR-126 [141–146]. Noticeably, three miRNAs (miR-126, miR-143, miR-210) implicated in the DCIS-to-IDC transition are specifically dysregulated in basal-like IDC, suggesting that their deregulation plays a critical role in driving the progression of DCIS lesions to malignant basal-like IDC tumors (Table 2). Moreover, expression of seven basal-related miRNAs (miR-10 b, miR-18 a, miR-26 b, miR-193 b, miR-195, miR-449 a, miR-449 b) is dysregulated in luminal-type IDC in an opposite manner from basal-like IDC, suggesting that their alterations are involved in determining molecular subtypes of breast cancer. It is worthy to note that miR-26 b is a downstream mediator of the ERα-Myc signaling axis [61]. ERα signaling is known to positively and negatively regulate luminal and basal phenotypes, respectively [147]. Therefore, upregulation of miR-26 b potentiates the luminal phenotype via the activation of ERα signaling, whereas its downregulation promotes the basal phenotype owing to decreased ERα signaling activity. Similarly, upregulation of the EMT inhibitors miR-449 a/449 b [76] facilitates the development of luminal IDC, and their downregulation enhances basal-like IDC development.
Among luminal subtypes, luminal-B is a more aggressive subtype than luminal-A owing to low/negative expression of PR and/or HER2 overexpression, and high proliferation indicated by high Ki-67 [148]. Three miRNAs (miR-99 a, miR-100 and miR-125 b) that are involved in the transition to DCIS are specifically downregulated in luminal-B IDC. This indicates that deregulation of their protein targets contributes to luminal-B IDC development, including aberrant activation of IGF2/PI3K/AKT/mTOR and HER2/3 signaling pathways. This association between the IGF2/PI3K/AKT/mTOR pathway and luminal-B IDC is further supported by a recent study showing that activation of the mTOR/S6K signaling axis correlated with breast tumors manifesting the luminal-B molecular profile of high Ki-67, ER(+) and HER2(−) [149]. Targeting the IGF2/PI3K/AKT/mTOR signaling axis is a potential strategy to prevent DCIS from developing into malignant luminal-B tumors or to treat this subtype of breast cancer.
The roles of miRNAs in breast cancer stem cells and drug resistance
A large body of evidence has suggested that cancer stem cells (CSCs) found in most types of cancer (including breast cancer) contribute to tumor development and progression. Since Al-Hajj et al. [150] identified CD44+/CD24− Lineage cells in breast tumors as enriched breast cancer stem cells (BCSCs), the roles of miRNAs in CSCs of breast cancer have been actively investigated. Yu et al. [77] reported that let-7 targeted and repressed expression of H-RAS and HMGA2 to restrict the self-renewal of normal mammary stem cells and was significantly downregulated in BCSCs. By miRNA profiling analysis, Shimono et al. [151] identified 37 differentially expressed miRNAs between human BCSCs and nontumorigenic cancer cells. Among them, three clusters, miR-200 c-141, miR-200 b-200 a-429 and miR-183-96-182, were found to be downregulated in human BCSCs, normal human and murine mammary stem/progenitor cells and embryonic carcinoma cells [151]. They further showed that ectopic miR-200 c expression substantially inhibited the ability of normal mammary stem cells to generate mammary ducts. Overexpression of miR-200 c in human BCSCs decreased in vivo xenograft tumor formation, suggesting the biological similarity between BCSCs and normal stem cells [151]. We also recently identified miR-140 as a negative regulator in BCSCs, where its expression is aberrantly downregulated compared with normal breast stem cells [86]. miR-140 is able to target the stem cell factors SOX9 and ALDH to restrict the self-renewal of BCSCs [86].
It is noteworthy that in addition to its critical role in breast tumor cell invasion and metastasis, EMT has been functionally associated with BCSC phenotypes [152]. Therefore, miRNAs involved in regulating EMT are also critical regulators of BCSC phenotypes. For instance, the aforementioned miR-22 negatively regulates expression of miR-200 via targeting of TET1-3 [79]. This miR-22-mediated effect promotes metastasis in an animal breast tumor model through alleviating the inhibitory effect of miR-200 on EMT [79]. The oncogenic role of miR-22 is consistent with the positive association of miR-22 overexpression with poor clinical outcomes, and with silencing of the TET-miR-200 axis in patients [79]. In addition, Martello et al. [153] revealed that miR-103/107 potentiates EMT by targeting Dicer, resulting in attenuation of overall miRNA biogenesis in tumor cells, and miR-103/107 expression is positively associated with metastasis and poor breast cancer outcomes.
Drug resistance in cancer patients is a serious clinical problem resulting in treatment failure. The main molecular pathological mechanisms responsible for drug resistance are the dysregulated expression of the drug transporters responsible for anti-cancer drug efflux, and the aberrant activation of anti-apoptotic, cell-survival and oncogenic pathways. As mentioned above, during breast tumorigenesis, many miRNAs are aberrantly regulated to promote tumor cell survival (Table 1). Additionally, several miRNAs have been identified to target drug transporter genes in breast cancer cells. For instance, miR-451 and miR-326 are required for the chemosensitivity of breast cancer cells to doxorubicin via direct targeting of ABC family transporter genes ABCB1 and ABCC1 (MRP1), respectively [154, 155]. Similarly, miR-487 a has been found to target ABCG2 to modulate the chemoresponsiveness of breast cancer cells to mitoxantrone [156]. Furthermore, it has been reported that miR-221/222 is responsible for tamoxifen resistance in luminal-type breast cancers by targeting p27Kip1, a cell-cycle inhibitor and tumor suppressor [109]. Rao et al. [157] also found that aberrant overexpression of miR-221/222 makes breast cancer cells resistant to fulvestrant through deregulating multiple signaling pathways associated with drug resistance, including activation of β-catenin signaling and suppression of TGFβ signaling.
Exosomal miRNAs in breast cancer development
Exosomes are 50–100 nm membrane microvesicles that can be secreted from various types of cells in both physiological and pathological conditions [158]. Exosomes carry regulatory molecules (e.g. miRNAs, mRNAs and proteins) between cells, and are emerging as an important mechanism for intercellular communication [158–161]. Accumulating evidence has revealed that exosomal communications between cancer cells themselves and between cancer cells and other types of normal cells (e.g. immune cells, adipocytes, endothelial cells, stromal fibroblasts) significantly contribute to tumor development and malignancy [162–164].
Exosomal miRNAs (extracellular miRNAs) have recently attracted a high level of attention owing to their involvement in various aspects of breast cancer development and their potential roles in the diagnosis and prognostic prediction of breast cancer. For example, bone marrow stroma has been shown to inhibit breast cancer cell proliferation via secreting exosomal miRNAs (miR-127, -197, -221/222 and -223) that directly target the CXCL12 chemokine gene [165]. On the other hand, breast cancer invasion and metastasis are facilitated by exosomal miR-223 derived from tumor-associated macrophages, which targets myocyte enhancer factor 2 C to activate β-catenin signaling [166]. Moreover, exosomal miRNAs secreted from breast cancer cells have been demonstrated to promote the metastatic process. Zhou et al. [162] reported that exosomal miR-105 secreted from breast cancer cells mediated the destruction of tight junctions in endothelial cells, promoting the metastatic spreading of cancer cells through targeting the tight junction protein ZO-1. The oncogenic role of exosomal miR-105 was in line with the positive correlation between the secreted levels of miR-105 and the metastatic states of breast cancer [162].
Two lines of evidence have functionally linked exosomal miRNAs to drug resistance and CSC regulation. Chen et al. [167] recently found that exosomal miRNAs derived from drug-resistant breast cancer cells targeted PTEN in drug-sensitive breast cancer cells to enhance their resistance to adriamycin and docetaxel. We recently showed that the composition of miRNAs carried by exosomes secreted from CSCs of DCIS tumor cells was altered compared with exosomes from normal stem-like breast cells [168]. Remarkably, CSC-secreted exosomes carried less miR-140, a tumor-suppressor miRNA mentioned above, than those secreted from normal stem-like cells [168], indicating that the tumorigenic process alters exosomal contents to dysregulate the function of exosomes.
piRNAs in breast cancer
While piRNAs account for the largest class of the sncRNA superfamily, the role of piRNAs in human carcinogenesis is virtually unknown. In addition to germ line cells, Piwi-related orthologous proteins and piRNAs have been detected in other cell types in various species, suggesting that they may have somatic functions [39]. Hiwi, the human ortholog of Drosophila Piwi, is overexpressed in a variety of human cancers, demonstrating Hiwi-piRNA dysregulation in cancer development [39]. The expression profile and function of piRNAs in human tumorigenesis is just beginning to be investigated. Nevertheless, some compelling lines of evidence indicate that the Hiwi-piRNA regulatory axis may contribute to human tumorigenesis by transcriptionally silencing tumor-suppressing genes through epigenetic mechanisms [39, 169]. This supports a new paradigm, which has been proposed, that cancer cells may hijack the Hiwi-piRNA pathway to aberrantly modulate the chromatin conformation and silence tumor suppressor gene expression for inducing and maintaining a ‘stem-like' state [39].
Three lines of studies have recently defined the role of piRNAs in breast cancer. Zhang et al. reported that piwi-like RNA-mediated gene silencing 2 (Piwil2), an Ago family protein, was overexpressed in CD44+CD24− BCSCs. Aberrant expression of Piwil2 is associated with clinicopathological characteristics including age, tumor size, histological type, tumor stage and lymph node metastasis [169]. piR-932, a Piwil2-associated piRNA, is induced in breast cancer cells with EMT [169], suggesting that piRNA-mediated epigenetic mechanisms are involved in modulating the stemness of breast cancer cells. Through deep-sequencing analysis of the breast cancer transcriptome, Hashim et al. identified numerous piRNAs that are differentially expressed in breast cancer cell lines and tumor biopsies when compared with normal counterparts. The potential mRNA targets for cancer-specific piRNAs were also analyzed [170]. Through genotypic screening of a panel of single-nucleotide polymorphism (SNP)-containing piRNAs, Fu et al.[171] identified the breast cancer-specific variant of piR-021285 with SNP rs1326306 . Intriguingly, variant piR-021285 mimic-transfected MCF7 cells exhibited a distinct methylome and enhanced invasiveness in comparison with wild-type-mimic-transfected cells [171]. Owing to these exciting findings, it is anticipated that more piRNA-related studies in breast cancer will be reported in the future.
lncRNAs in breast tumorigenesis
As mentioned above, while miRNAs primarily function posttranscriptionally through binding to mRNA 3′-UTRs, lncRNAs regulate gene functions through diverse mechanisms, and some of them target genes or proteins with higher specificity than miRNAs by their more specific regulatory roles. These advantageous features may mean that lncRNAs are more suitable to be used as therapeutic targets in cancer treatment, and studies of lncRNAs in cancer have blossomed into an important emerging field of research. Here we provide a concise review of the roles of representative lncRNAs in breast tumorigenesis. Comprehensive reviews of lncRNAs in breast cancer are available elsewhere [172–175].
Epigenetic regulators
The lncRNA HOTAIR (HOX transcript antisense RNA) has been recently recognized as a key oncogenic player in breast cancer metastasis [176]. HOTAIR is an antisense RNA transcribed from the HOXC locus and acts in trans to repress the expression of the HOXD locus [174]. HOTAIR is an RNA scaffold, and the 5′ and 3′ domains of HOTAIR are involved in recruiting two critical epigenetic protein complexes, Polycomb Repressive Complex 2 (PRC2) and the LSD1-CoREST-REST H3K4 demethylase complex, to regulate gene expression [177]. In addition, Wang et al. [178] recently found that BRCA1 can compete with HOTAIR to bind EZH2, a component of PRC2, to attenuate PRC2 activity. Therefore, loss of BRCA1 or upregulation of HOTAIR results in the reprogramming of the genome-wide PRC2-binding pattern in breast epithelial cells, leading to the altered methylation signature found in aggressive breast cancer cells [178].
Recently HOTAIR has been shown to be upregulated in tamoxifen-resistant breast cancer tissues compared with their primary counterparts [179]. This facilitates the ligand-independent activation of ER, which contributes to tamoxifen resistance [179]. Moreover, upregulation of HOTAIR was also found in triple-negative breast cancer (TNBC) cell lines in addition to primary breast tumors [180]. Suppression of HOTAIR expression by co-treatment with clinically validated inhibitors of c-ABL (imatinib) and EGFR (lapatinib) resulted in synergistic growth inhibition in TNBC cells [180]. This dual treatment effect was mediated by blocking nuclear expression of β-catenin and preventing its recruitment to the HOTAIR promoter [180]. A recent report has also shown that HOTAIR is involved in triggering EMT in breast cancer cells and maintaining BCSCs [181]. These lines of evidence together indicate that HOTAIR is a multifunctional regulator that is implicated in a variety of oncogenic events and promotes breast cancer development and metastasis.
Protein synthesis regulators
Three oncogenic lncRNAs, BC200, TreRNA (translational regulatory lncRNA) and UCA1 (urothelial carcinoma-associated 1) that have regulatory roles in protein synthesis have been identified to be deregulated in breast cancer. BC200, a neuronal and germ-cell-specific 200 nt lncRNA, was found to be aberrantly expressed in invasive breast cancer, but not in benign tumors and normal breast tissue [182]. Its expression in DCIS is associated with high nuclear grade, suggesting BC200 may serve as a prognostic indicator of breast cancer progression [182]. BC200 binds polyA binding protein via its oligo (A) rich region, interfering with the protein translation process [183]. However, it is uncertain if this function contributes to the oncogenic role of BC200 in breast cancer. TreRNA expression is required for the invasive and metastatic ability of breast cancer cells [184]. By modulating the formation of a ribonucleoprotein complex consisting of RNA-binding proteins (hnRNP K, FXR1 and FXR2), PUF60 and SF3B3, TreRNA acts as a translational repressor of E-cadherin mRNA, promoting EMT and tumor cell invasiveness [184]. UCA1 expression is also upregulated in breast cancer, and competes with p27Kip1 mRNA to bind heterogeneous nuclear ribonucleoprotein 1 (hnRNP1), resulting in repression of p27Kip1 mRNA translation and promoting cell cycle progression [185].
Hormone signaling regulators
GAS5 (Growth Arrest-Specific 5) and SRA (Steroid Receptor RNA Activator) are two well-known lncRNAs involved in modulating hormone signaling. GAS5 is a tumor suppressor whose expression is found to be downregulated in breast cancer [186], and functions as an RNA transcript decoy to prevent the glucocorticoid receptor (GR) from binding to its target genes [187]. This is mediated by a stem-loop structure formed by the 3′ end of GAS5, which resembles the structure of the GR DNA-binding element. Through the repression of anti-apoptotic GR target gene expression, GAS5 promotes cell apoptosis [187]. SRA lncRNA is transcribed from the SRA gene locus, which also encodes protein-coding RNA isoforms without the intron 1 sequence [188, 189]. The stem-loop structure formed by SRA enables it to interact with multiple proteins, including steroid receptors (estrogen, progesterone and androgen receptors) [188, 189]. SRA expression is associated with the ER+/PR− and ER−/PR+ status of breast tumors and its variant isoform correlates with tumor grade [190]. Lanz et al. [191] showed that transgenic SRA mice manifested increased epithelial proliferation and apoptosis in mammary glands, indicating SRA overexpression alone is not sufficient for inducing mammary tumors and the collaboration with other oncogenic signals is likely required for driving breast carcinogenesis.
Nuclear architectural RNAs
Recent studies have shown that lncRNAs act as the scaffolding backbone for certain nuclear bodies including nuclear speckles and paraspeckles [192]. MALAT1 and NEAT1 are two of the best characterized scaffolding lncRNAs. MALAT1 (Metastasis associated lung adenocarcinoma transcript 1), also known as NEAT2, was first discovered as a prognostic indicator of later metastasis in lung cancer [193, 194]. MALAT1 is a core component of nuclear speckles, which are implicated in RNA processing, splicing and export [192, 195]. MALAT1 can regulate RNA splicing and gene expression via its interaction with multiple factors involved in RNA splicing, transcription and chromatin remodeling [192, 195]. MALAT1 is required for the G1-S transition and mitotic progression via the regulation of cell cycle gene expression [196]. Although high MALAT1 expression is associated with poorer outcomes in a number of cancer types [197], it is downregulated in mammary tumors in c-myc transgenic mice [198]. However, the exact role of MALAT1 in breast cancer is still uncertain at this stage, as another line of investigation found that MALAT1 is upregulated in primary breast cancer [199].
NEAT1 (Nuclear Enriched Abundant Transcript 1) is a core component of nuclear paraspeckles. Paraspeckles are nuclear bodies that are composed of functionally diverse proteins, and regulate the sequestration of hyperedited RNA transcripts to modulate RNA splicing and transcription [200]. In prostate cancer, NEAT1 modulates the epigenomic pattern to co-activate ER target genes and promote tumor cell malignancy [201]. Recently it has been shown that genetic knockout of Neat1 in a mouse model causes aberrant morphogenesis of mammary glands, leading to defects in lactation, indicating the critical role of Neat1 in murine mammary gland development [202]. This lactation defect results from the decreased proliferation of Neat1-mutant mammary epithelial cells during lobular-alveolar development [202]. This study is the first to show an important biological function of Neat1 in mammary gland development, and also links NEAT1 to the paraspeckle nuclear bodies present in in vivo mammary luminal epithelial cells. Although its role in breast cancer is unknown, it was recently shown that NEAT1 is a hypoxia-inducible gene, and its presence is required for alleviating hypoxia-induced apoptosis in breast cancer cells [203].
miRNA regulators
As mentioned above, a class of lncRNAs (also known as competitive endogenous RNA, ceRNAs) serves as miRNA sponges that compete with mRNA targets, resulting in suppressing miRNA regulatory function. While not a lncRNA, Yang et al. [204] recently reported that the noncoding 3′-UTR of FOXO1 functions as a ceRNA in repressing breast cancer metastasis via alleviating the miR-9-mediated inhibition of E-cadherin expression. Moreover, we also recently showed that the lincRNA-RoR serves as a ceRNA to suppress the inhibitory effect of miR-145 on triple-negative breast cancer invasiveness through targeting of ARF6 [93]. Using computational analysis, Paci et al. [205] showed that the sponge interaction between ceRNAs and mRNAs in normal breast epithelium is altered in breast cancer, suggesting that dysregulation of ceRNAs may play a significant role in breast tumorigenesis.
Translational implications of ncRNAs in breast cancer
NcRNAs as diagnostic and prognostic biomarkers
miRNAs are stable in body fluids such as blood, urine and nipple discharge and are therefore ideal biomarkers for noninvasive early detection of breast cancer. Multiple studies (Table 3) have demonstrated that miRNA biomarkers provide excellent sensitivity and specificity for detecting early- and even advanced-stage breast cancer, strongly suggesting that they are potential diagnostic tools for identifying early and advanced breast cancers [206–224]. The most commonly reported miRNAs in body fluids and best candidates for biomarkers are miR-21, miR-155, miR-10 b, miR-92 a, miR-133 a, miR-145, miR-148 b and miR-451 [207, 211–217, 219–223]. It is worthy to note that among these identified diagnostic miRNA biomarkers, three miRNAs, including miR-195, miR-145 and miR-451, were reported to be breast cancer specific and can be potentially exploited to distinguish breast cancer from other types of cancer [206, 219]. Moreover, miRNAs have been shown to be a promising diagnostic tool to detect early-stage breast cancer. For instance, a combination of miR-145 and miR-451 has been shown to sensitively detect DCIS [219]. A five-miRNA signature (including miR-127-3 p, miR-148 b, miR-409-3 p, miR-652 and miR-801) is another diagnostic signature for early detection of stage I or II of breast cancer [215]. Multiple lines of study have demonstrated that miRNAs can be potentially used to diagnose metastasis of breast cancer to lymph nodes and internal organs. Upregulation of miR-10 b, miR-21, miR-214 and miR-373 and downregulation of miR-92 a have been shown to correlate with the lymph node status [208, 213, 216]. In addition, elevated levels of miR-10 b and miR-21 positively correlated with internal-organ metastasis of breast cancer [207, 211].
Table 3.
MicroRNAs in breast cancer diagnosis and prognosis
| Application | Tested biopsies | miRNA biomarkers | Clinical relevance | References |
|---|---|---|---|---|
| Diagnosis | blood | miR-195 | Elevated circulating miR-195 was found to be breast cancer specific and could differentiate breast cancer from other cancers and from controls with a sensitivity of 88% at a specificity of 91%. | [206] |
| serum | miR-21 | High circulating miR-21 concentrations correlated significantly (P < 0.001) with visceral metastasis of breast cancer. | [207] | |
| blood | miR-214 | Serum miR-214 distinguished malignant from benign tumors and healthy controls (P = 0.0001). | [208] | |
| Elevated miR-214 levels correlated with a positive lymph node status (P = 0.039). | ||||
| tissue, serum | miR-222 | miR-222 was significantly increased in the serum of BC patients by further validation (P < 0.05), which may be a useful biomarker for differentiating BC patients from controls with receiver operating characteristic (ROC) curve area 0.67 of (95% CI = 0.5649–0.7775). | [209] | |
| serum | miR-181 a | Median miR-181 a levels were significantly lower in patients with BC compared with healthy controls (P = 0.001). | [210] | |
| ROC analysis demonstrated the sensitivity and specificity of miR-181 a for BC diagnosis at 70.7 and 59.9%, respectively. | ||||
| serum | miR-10 b | Serum miR-10 b concentrations were significantly higher in patients with bone metastases than in patients without bone metastases or control subjects. | [211] | |
| serum | miR-145, miR-155, miR-382 | ROC curve analyses revealed that three serum miRNAs could be valuable biomarkers for distinguishing BC from normal controls. Additionally, a combination of ROC curve analyses of miR-145, miR-155 and miR-382 showed better sensitivity and specificity of our assay. | [212] | |
| tissue, serum | miR-21, miR-92 a | The level of miR-92 a was significantly lower, while miR-21 was higher, in tissue and serum samples of BC than that of healthy controls (P < 0.001). | [213] | |
| Decreased levels of miR-92 a and increased levels of miR-21 were associated with tumor size and a positive lymph node status (P < 0.001). | ||||
| tissue, serum | miR-1, miR-92 a, miR-133 a, miR-133 b | They were identified as the most relevant diagnostic markers for detecting breast cancer in the study. | [214] | |
| plasma | miR-127-3 p, miR-148 b, miR-409-3 p, miR-652, miR-801 | miR-127-3 p, miR-148 b, miR-409-3 p, miR-652 and miR-801 can detect stage I or stage II breast cancer thus making them attractive candidates for early detection. | [215] | |
| plasma | miR-10 b, miR-373 | The plasma levels of circulating miR-10 b and miR-373 were significantly higher in 10 breast cancer patients with lymph node metastasis compared with 10 N (0) patients and 10 normal donors (P < 0.01). | [216] | |
| A combination of the two circulating miRNAs further enhanced the sensitivity to 72% and the specificity to 94.3 %. | ||||
| Circulating miRNA-10 b and miRNA-373 are potential biomarkers for detecting the lymph node status of breast cancer. | ||||
| plasma | miR-21, miR-146 a | The circulating level of miR-21 and miR-146 a were significantly higher in plasma samples of breast cancer patients, when compared with those of healthy controls (P < 0.0004 and P < 0.005, respectively). | [217] | |
| plasma | miR-30 a | ROC analysis showed the sensitivity and specificity of miRNA-30 a for breast cancer diagnosis at 74.0 and 65.6%, respectively. | [218] | |
| Diagnosis | tissue, plasma | miR-145, miR-451 | A combination of miR-145 and miR-451 was the best biomarker (P < 0.0001) in discriminating breast cancer from healthy controls and all other types of cancers with 88% of the positive predictive value and 92% of the negative predictive value. | [219] |
| The positive predictive value for ductal carcinoma in situ (DCIS) cases was 96%. | ||||
| serum | miR-21 | The level of serum miR-21 was significantly higher in BC patients than controls (P < 0.001). | [220] | |
| The sensitivity and specificity of miR-21 were 87.6% and 87.3%. | ||||
| plasma | miR-148 b, miR-133 a | Plasma levels of miR-148 b and miR-133 a were significantly higher in breast cancer patients. | [221] | |
| urine | miR-21, miR-155, miR-125 b, miR-451 | The ROC including all four miRNAs as well as the group of the four significant deregulated miRNAs separated BC patients from healthy controls with a high accuracy (AUC = 0.887). | [222] | |
| serum | miR-29 b-2, miR-155, miR-197, miR-205 | The expression level of miR-29 b-2, -155, -197 and -205 was significantly increased in the serum of breast cancer patients. | [223] | |
| Nipple Discharge | miR-4484, miR-K12-5-5 p, miR-3646, miR-4732-5 p | Expression levels of these four miRNAs were elevated in breast cancer samples compared with those of normal controls. | [224] | |
| Prognosis | tissue | miR-210 | Expression levels of miR-210 showed an inverse correlation with disease-free and overall survival of breast cancer patients, significant in both univariate and multivariate analyses. | [225] |
| FFPE tissue | Let-7 b, miR-205 | let-7 b expression was shown to be associated with luminal tumors and to have an independent significant positive prognostic value in this group. | [226] | |
| miR-205 is associated with tumors of ductal morphology and is of significant positive prognostic value within these tumors. | ||||
| tissue | miR-30 a | Reduced tumor expression of miR-30 a in breast cancer patients was associated with an unfavorable outcome, including late tumor stage, lymph node metastasis and worse progression. | [227] | |
| plasma | miR-141, miR-200 b, miR-200 c, miR-210, miR-768-3 p | The study identified miR-200 b alone or combinations of miRNAs as surrogate markers for differentiating MBC cases from controls. | [228] | |
| tissue | miR-187 | High miR-187 was significantly associated with reduced breast cancer-specific survival in the entire cohort (P = 0.021) and in lymph node-positive patients (P = 0.012). | [229] | |
| tissue | miR-210 | Overexpression of miR-210 was significantly associated with the higher risk of recurrence, metastasis and overall decreased survival rates for breast cancer patients. | [230] | |
| IDC tissue | miR-1307, miR-103, miR-328, miR-93, miR-874, miR-484, miR-148 b | Thirty mRNAs and seven miRNAs were associated with overall survival across different clinical and molecular subclasses of a 466-patient IDC cohort from TCGA. | [231] | |
| FFPE tissue | miR-21, miR-205 | Univariate analysis revealed that both miR-21 and miR-205 were significantly associated with disease-free interval and only miR-205 with overall survival. | [232] | |
| tissue | Let-7 b | let-7 b expression in breast cancer patients was inversely associated with tumor lymph node metastasis (P = 0.001), patient overall survival (P = 0.027), relapse-free survival (P = 0.016). | [233] | |
| tissue | miR-196 a | The study indicates that the miR-196 a rs11614913T>C polymorphisms are possible prognostic biomarker for patients with hormone receptor-expressing early breast cancer. | [234] | |
| Prognosis | tissue | miR-22 | miR-22 was significantly correlated with the TNM stage, local relapse, distant metastasis and survival of breast cancer patients. | [235] |
| tissue | miR-34 a | Loss of miR34a is associated with poor outcome, independent of age, node status, receptor status and tumour size. | [236] | |
| tissue | miR-127 | Patients with low miR-127 showed poorer overall survival than those with high miR-127. Multivariate analyses indicated that the status of miR-127 was an independent prognostic factor. | [237] | |
| tissue, plasma | miR-106 b | Patients with high miR-106 b expression levels tended to have shorter DFS times and overall survival times (P < 0.001). In a Cox regression model, high-level tissue and plasma miR-106 b expression were unfavorable prognostic factors. | [238] | |
| tissue | miR-200 b | The low expression of miR-200 b was correlated with late TNM stage, negative oestrogen receptor and positive HER-2 status. Multivariate analysis showed that miR-200 b expression was an independent prognostic predictor for BC patients. | [239] | |
| tissue | miR-30 e* | miR-30 e* was identified as independent protective prognostic factor in lymph node-negative untreated patients with ESR1+/ERBB2- tumors | [240] |
Prognostic biomarkers are in high demand, as early diagnosis of aggressive cancer helps clinicians provide the appropriate therapy and clinical care. Expression levels of a number of miRNAs, which are either positively or inversely associated with metastatic status, disease-free time and overall survival of breast cancer patients have been identified [225–240] and are detailed in Table 3. The most relevant miRNA with prognostic value in breast cancer is miR-210, the aforementioned oncogenic miRNA implicated in the DCIS-to-IDC transition. Elevated expression levels of miR-210, also known to be a hypoxia-inducible miRNA, are positively correlated with higher risk of recurrence, metastasis and overall decreased survival rates for breast cancer patients [225, 228, 230], and miR-210 may serve as an independent prognostic predictor. Through locked nucleic acid probe in situ hybridization analysis of a large cohort of 2919 breast tumors using tissue microarrays, let-7 b and miR-205 were identified to be frequently lost in a range of malignant tumors [226]. let-7 b was demonstrated to be an independent surrogate marker with a significant positive prognostic value in luminal-type breast tumors [226]. miR-205 downregulation was associated ductal tumors and was shown to be of significant positive prognostic value within this tumor type [226]. From survival analysis of miRNAs and mRNAs in a 466-patient IDC cohort from TCGA, several miRNAs such as miR-328, miR-484 and miR-874 were discovered to be associated with overall survival across different clinical and molecular subclasses of breast cancer, and had the highest prognostic value in early stage I and II tumors when miRNA and mRNA predictors were combined together as an integrated signature [231]. Furthermore, a long follow-up study demonstrated that miR-21 and miR-205 were independent biomarkers predicting early disease relapse, whereas only miR-205 overexpression was associated with overall survival [232]. Recently, from a study of three different cohorts composed of over 1500 breast cancer patients, the tumor-suppressor miRNA miR-34 a has been shown to be a promising prognostic biomarker as its loss can predict the poor outcome in the lymph-node-negative, not lymph-node-positive, patient population [236].
The findings highlighted above were derived from studies of tumor tissue, which are not as practical as circulating prognostic biomarkers identified from blood specimens. To seek potential circulating miRNAs present in serum/plasma with prognostic values, Madhavan et al. [228] profiled circulating levels of miRNAs in a cohort of metastatic breast cancer (MBC) patients, which encompassed three sub-cohorts including circulating tumor cells (CTC)-positive, CTC-negative and CTC-low groups based on the levels of CTC. Their results showed that compared with CTC-negative MBC and controls, CTC-positive displayed significantly higher levels of miR-141, miR-200 a, miR-200 b, miR-200 c, miR-203, miR-210, miR-375 and miR-801. Combinations of these miRNAs and miR-200 b alone were demonstrated to be promising circulating prognostic markers for progression-free and overall survival [228]. In addition to sncRNAs, lncRNAs have gradually gained attention for their potential as prognostic biomarkers. The lncRNA HOTAIR is the best example that has been shown to be an independent biomarker for the prediction of the risk of metastasis in ER-positive breast cancer patients [241]. It is anticipated that more surrogate lncRNA biomarkers with prognostic values for breast cancer will be unveiled in the future.
ncRNAs as therapeutic targets
Owing to their crucial roles in regulating multiple cancer-related pathways, miRNAs have been actively investigated for clinical application and therapeutic intervention [242]. The current approach for developing miRNA-based therapeutics is through the administration of a synthetic anti-miRNA or a miRNA-mimic oligonucleotide [242]. To improve the therapeutic efficacy, Lennox et al. [243] conjugated a novel compound, N,N-diethyl-4 -(4-nitronaphthalen-1-ylazo)-phenylamine, to these miRNA-based oligonucleotides to increase their in vivo stability and binding affinity to endogenous RNA targets.
Given that naked nucleic acid molecules are not generally suitable for in vivo applications owing to their intrinsic negative charge, packaging of the gene silencing agents (e.g. siRNAs and miRNAs) into carrier vehicles is a promising strategy for delivery to target tissues [244]. Multiple nanotechnology-based vehicle platforms have been developed to deliver gene silencing agents for cancer therapy. The most exploited strategy is to package the double-stranded RNA into lipid- or non-lipid-based nanoparticles with a size <200 nm in diameter [244]. Because the disorganization of the tumor blood vessel endothelium leads to cell-junction gaps ranging from 100 to 500 nm [245], this enhanced permeability of the tumor vasculature enables RNA-containing nanoparticles to easily cross the fenestration to reach tumor interstitium. By conjugating active targeting moieties on the surface, nanoparticles can be targeted to specific tissues. This nanotechnology-based approach has been successfully applied to miRNA-based therapeutic agents for in vivo cancer therapy. Systemic delivery of antisense peptide nucleic acids encapsulated in unique polymer nanoparticles inhibited miR-155 function in a mouse lymphoma model and resulted in regression of lymphadenopathy [246]. It is expected that the nanoparticle-based approach will be developed in the future for miRNA-based therapy of breast cancer. Moreover, the most promising therapeutic strategy is the administration of a combination of miRNA therapeutic agents or miRNA-based agents with other anticancer therapies such as chemotherapy and radiotherapy [242]. Advances in nanotechnology are crucial for implementing these therapeutic ideas. In addition to the therapeutic application, miRNA-based treatment is a potential strategy for chemoprevention of breast cancer. Recently, ingestion of plants bioengineered to produce human cancer-related miRNAs was reported to be a compelling approach for cancer chemoprevention [247]. Although lncRNAs are critical in breast cancer progression and metastasis, therapeutic application is still challenging owing to technical hurdles [174], and more investigation of lncRNAs is necessary.
Concluding remarks
Research on the functional and clinical implication of miRNAs in human diseases has exploded since they were discovered a decade ago. They have been demonstrated to be valuable in diagnosis, prognosis and therapy of cancer. However, more large-scale studies are warranted to demonstrate whether these findings can eventually benefit cancer patients. As miRNAs are able to target multiple genes and signaling pathways, it will be necessary to comprehensively characterize their mechanistic actions in cells to avoid any side effects. Functional investigations of lncRNAs and piRNAs in breast cancer are at an early stage, and further research is necessary to provide new avenues for the development of cancer biomarkers and therapeutics.
Key Points
MicroRNAs (miRNAs) are crucial small noncoding RNA regulators that modulate the functionality of multiple genes and signaling pathways in cells at the posttranscriptional level.
Long noncoding RNAs have recently emerged as important cellular RNAs that regulate a variety of biological processes via multiple mechanistic actions.
Genome-wide analysis of miRNA profiles in breast cancer biopsies provides new insights into the roles of miRNAs in the development and progression of breast cancer.
A number of lncRNAs are found to be critical for the invasive and metastatic progression of breast cancer.
MiRNAs are potential targets for translational applications in breast cancer diagnosis, prognosis and therapy.
Funding
This work was supported by National Institutes of Health (NIH) R01 grants 5R01CA157779-03 and 5R01CA163820-04, and American Cancer Society (ACS) grant RSG-12-006-01-CME to Q.Z.
Biographies
Pang-Kuo Lo is a Research Associate Faculty in the Department of Biochemistry and Molecular Biology, University of Maryland Baltimore, USA. He has been working in the fields of cancer epigenetics and cancer stem cell biology for over 9 years.
Benjamin Wolfson is a PhD student focusing on noncoding RNAs in Adipogenesis/Breast Cancer in the Molecular Medicine Program and the Department of Biochemistry and Molecular Biology at the University of Maryland Baltimore, USA.
Xipeng Zhou is a medical student in Clinical Medicine at Jiangsu University School of Medicine, Zhenjiang, Jiangsu, China 202013.
Nadire Duru received her PhD in radiation oncology at Purdue University, USA. She currently works as a postdoctoral researcher on epigenetic regulation and cancer stem cells in breast cancer at the University of Maryland Baltimore, USA.
Ramkishore Gernapudi received his PhD in Environmental Toxicology at the Southern University and A&M College, Baton Rouge, LA. He currently works as postdoctoral researcher on noncoding RNAs in Adipogenesis/Breast Cancer at the University of Maryland Baltimore, USA.
Qun Zhou is an Associate Professor in Biochemistry and Molecular Biology at the University of Maryland Baltimore, USA. He has been working in the fields of cancer epigenetics and breast cancer biology for over 12 years.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5–29. [DOI] [PubMed] [Google Scholar]
- 2.Bombonati A, Sgroi DC. The molecular pathology of breast cancer progression. J Pathol 2011;223:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buerger H, Otterbach F, Simon R, et al. Comparative genomic hybridization of ductal carcinoma in situ of the breast-evidence of multiple genetic pathways. J Pathol 1999;187:396–402. [DOI] [PubMed] [Google Scholar]
- 4.Buerger H, Otterbach F, Simon R, et al. Different genetic pathways in the evolution of invasive breast cancer are associated with distinct morphological subtypes. J Pathol 1999;189:521–6. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Garcia MA, Geyer FC, Lacroix-Triki M, et al. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology 2010;57:171–92. [DOI] [PubMed] [Google Scholar]
- 6.Byler S, Goldgar S, Heerboth S, et al. Genetic and epigenetic aspects of breast cancer progression and therapy. Anticancer Res 2014;34:1071–7. [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861–74. [DOI] [PubMed] [Google Scholar]
- 8.Spizzo R, Almeida MI, Colombatti A, et al. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 2012;31:4577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexander RP, Fang G, Rozowsky J, et al. Annotating non-coding regions of the genome. Nat Rev Genet 2010;11:559–71. [DOI] [PubMed] [Google Scholar]
- 10.Bertone P, Stolc V, Royce TE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science 2004;306:2242–6. [DOI] [PubMed] [Google Scholar]
- 11.Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science 2005;309:1559–63. [DOI] [PubMed] [Google Scholar]
- 12.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol 2009;21:416–25. [DOI] [PubMed] [Google Scholar]
- 15.Taft RJ, Glazov EA, Cloonan N, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet 2009;41:572–8. [DOI] [PubMed] [Google Scholar]
- 16.Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene 2006;25:6170–5. [DOI] [PubMed] [Google Scholar]
- 17.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011;1:391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 20.Chua JH, Armugam A, Jeyaseelan K. MicroRNAs: biogenesis, function and applications. Curr Opin Mol Ther 2009;11:189–99. [PubMed] [Google Scholar]
- 21.Rodriguez A, Griffiths-Jones S, Ashurst JL, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res 2004;14:1902–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J 2007;26:775–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature 2007;448:83–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003;425:415–9. [DOI] [PubMed] [Google Scholar]
- 25.Berezikov E, Chung WJ, Willis J, et al. Mammalian mirtron genes. Mol Cell 2007;28:328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schraivogel D, Meister G. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem Sci 2014;39:420–31. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004;305:1437–41. [DOI] [PubMed] [Google Scholar]
- 28.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 2007;131:1097–108. [DOI] [PubMed] [Google Scholar]
- 29.Matsui M, Chu Y, Zhang H, et al. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res 2013;41:10086–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DH, Saetrom P, Snove O, Jr, et al. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA 2008;105:16230–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita S, Takahashi RU, Yamashita R, et al. Genome-wide analysis of DNA methylation and expression of microRNAs in breast cancer cells. Int J Mol Sci 2012;13:8259–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khoshnaw SM, Rakha EA, Abdel-Fatah TM, et al. Loss of Dicer expression is associated with breast cancer progression and recurrence. Breast Cancer Res Treat 2012;135:403–13. [DOI] [PubMed] [Google Scholar]
- 33.Dvinge H, Git A, Graf S, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 2013;497:378–82. [DOI] [PubMed] [Google Scholar]
- 34.Yu Z, Wang L, Wang C, et al. Cyclin D1 induction of Dicer governs microRNA processing and expression in breast cancer. Nat Commun 2013;4:2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen J, Xia W, Khotskaya YB, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 2013;497:383–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol 2012;197:201–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev 2012;26:2361–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim VN. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev 2006;20:1993–7. [DOI] [PubMed] [Google Scholar]
- 39.Siddiqi S, Matushansky I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem 2012;113:373–80. [DOI] [PubMed] [Google Scholar]
- 40.Cheng J, Deng H, Xiao B, et al. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett 2011;315:12–7. [DOI] [PubMed] [Google Scholar]
- 41.Kapranov P, Drenkow J, Cheng J, et al. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res 2005;15:987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013;20:300–7. [DOI] [PubMed] [Google Scholar]
- 44.Dieci G, Fiorino G, Castelnuovo M, et al. The expanding RNA polymerase III transcriptome. Trends Genet 2007;23:614–22. [DOI] [PubMed] [Google Scholar]
- 45.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007;316:1484–8. [DOI] [PubMed] [Google Scholar]
- 46.Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Heesch S, van Iterson M, Jacobi J, et al. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol 2014;15:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011;477:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009;33:717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013;341:78992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 2012;9:703–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farazi TA, Horlings HM, Ten Hoeve JJ, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res 2011;71:4443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volinia S, Galasso M, Sana ME, et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci USA 2012;109:3024–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song L, Lin C, Wu Z, et al. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS One 2011;6:e25454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang Z, Li Y, Huang K, et al. Regulation of miR-19 to breast cancer chemoresistance through targeting PTEN. Pharm Res 2011;28:3091–100. [DOI] [PubMed] [Google Scholar]
- 58.Qi L, Bart J, Tan LP, et al. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 2009;9:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frankel LB, Christoffersen NR, Jacobsen A, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 2008;283:1026–33. [DOI] [PubMed] [Google Scholar]
- 60.Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 2008;28:536980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan S, Ding K, Li R, et al. Identification of miR-26 as a key mediator of estrogen stimulated cell proliferation by targeting CHD1, GREB1 and KPNA2. Breast Cancer Res 2014;16:R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu S, Patel SH, Ginestier C, et al. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet 2012;8:e1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem 2009;284:23204–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin H, Dai T, Xiong H, et al. Unregulated miR-96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS One 2010;5:e15797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Kong X, Li J, et al. miR-96 promotes tumor proliferation and invasion by targeting RECK in breast cancer. Oncol Rep 2014;31:1357–63. [DOI] [PubMed] [Google Scholar]
- 66.Turcatel G, Rubin N, El-Hashash A, et al. MIR-99a and MIR-99b modulate TGF-beta induced epithelial to mesenchymal plasticity in normal murine mammary gland cells. PLoS One 2012;7:e31032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith AL, Iwanaga R, Drasin DJ, et al. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene 2012;31:5162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iliopoulos D, Jaeger SA, Hirsch HA, et al. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 2010;39:493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moskwa P, Buffa FM, Pan Y, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell 2011;41:210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chiang CH, Hou MF, Hung WC. Up-regulation of miR-182 by beta-catenin in breast cancer increases tumorigenicity and invasiveness by targeting the matrix metalloproteinase inhibitor RECK. Biochim Biophys Acta 2013;1830:3067–76. [DOI] [PubMed] [Google Scholar]
- 71.Yang X, Zhang Y, Li S, et al. Rab21 attenuates EGF-mediated MAPK signaling through enhancing EGFR internalization and degradation. Biochem Biophys Res Commun 2012;421:651–7. [DOI] [PubMed] [Google Scholar]
- 72.Di Leva G, Piovan C, Gasparini P, et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet 2013;9:e1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park SM, Gaur AB, Lengyel E, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008;22:894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamada S, Masamune A, Miura S, et al. MiR-365 induces gemcitabine resistance in pancreatic cancer cells by targeting the adaptor protein SHC1 and pro-apoptotic regulator BAX. Cell Signal 2014;26:179–85. [DOI] [PubMed] [Google Scholar]
- 75.Cai J, Guan H, Fang L, et al. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J Clin Invest 2013;123:566–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen SP, Liu BX, Xu J, et al. MiR-449a suppresses the epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by multiple targets. BMC Cancer 2015;15:706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007;131:1109–23. [DOI] [PubMed] [Google Scholar]
- 78.Kim HH, Kuwano Y, Srikantan S, et al. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 2009;23:1743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song SJ, Poliseno L, Song MS, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 2013;154:311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kong LM, Liao CG, Zhang Y, et al. A regulatory loop involving miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer invasion and metastasis. Cancer Res 2014;74:3764–78. [DOI] [PubMed] [Google Scholar]
- 81.Hu Y, Zhu Q, Tang L. MiR-99a antitumor activity in human breast cancer cells through targeting of mTOR expression. PLoS One 2014;9:e92099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gebeshuber CA, Martinez J. miR-100 suppresses IGF2 and inhibits breast tumorigenesis by interfering with proliferation and survival signaling. Oncogene 2013;32:3306–10. [DOI] [PubMed] [Google Scholar]
- 83.Scott GK, Goga A, Bhaumik D, et al. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem 2007;282:1479–86. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y, Yan LX, Wu QN, et al. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res 2011;71:3552–62. [DOI] [PubMed] [Google Scholar]
- 85.Chen J, Wang M, Guo M, et al. miR-127 regulates cell proliferation and senescence by targeting BCL6. PLoS One 2013;8:e80266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li Q, Yao Y, Eades G, et al. Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene 2014;33:2589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan X, Chen X, Liang H, et al. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer 2014;13:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang H, Cai X, Wang Y, et al. microRNA-143, down-regulated in osteosarcoma, promotes apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol Rep 2010;24:1363–9. [DOI] [PubMed] [Google Scholar]
- 89.Yu X, Zhang X, Dhakal IB, et al. Induction of cell proliferation and survival genes by estradiol-repressed microRNAs in breast cancer cells. BMC Cancer 2012;12:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jiang S, Zhang LF, Zhang HW, et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. Embo J 2012;31:1985–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zou C, Xu Q, Mao F, et al. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle 2012;11:2137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang S, Bian C, Yang Z, et al. miR-145 inhibits breast cancer cell growth through RTKN. Int J Oncol 2009;34:1461–6. [PubMed] [Google Scholar]
- 93.Eades G, Wolfson B, Zhang Y, et al. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res 2014;13:330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spizzo R, Nicoloso MS, Lupini L, et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ 2010;17:246–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tang H, Liu P, Yang L, et al. miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol Cancer Ther 2014;13:3185–97. [DOI] [PubMed] [Google Scholar]
- 96.Wang R, Tian S, Wang HB, et al. MiR-185 is involved in human breast carcinogenesis by targeting Vegfa. FEBS Lett 2014;588:4438–47. [DOI] [PubMed] [Google Scholar]
- 97.Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene 2009;28:3937–48. [DOI] [PubMed] [Google Scholar]
- 98.Li D, Zhao Y, Liu C, et al. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin Cancer Res 2011;17:1722–30. [DOI] [PubMed] [Google Scholar]
- 99.Yang G, Wu D, Zhu J, et al. Upregulation of miR-195 increases the sensitivity of breast cancer cells to Adriamycin treatment through inhibition of Raf-1. Oncol Rep 2013;30:877–89. [DOI] [PubMed] [Google Scholar]
- 100.Zeng J, Wei M, Shi R, et al. MiR-204-5p/Six1 feedback loop promotes epithelial-mesenchymal transition in breast cancer. Tumour Biol 2015;1–7. [DOI] [PubMed] [Google Scholar]
- 101.Zhao JJ, Lin J, Yang H, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem 2008;283:31079–86. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 102.Bronisz A, Godlewski J, Wallace JA, et al. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol 2011;14:159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bergamaschi A, Katzenellenbogen BS. Tamoxifen downregulation of miR-451 increases 14-3-3zeta and promotes breast cancer cell survival and endocrine resistance. Oncogene 2012;31:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang L, He D, Yang D, et al. MiR-489 regulates chemoresistance in breast cancer via epithelial mesenchymal transition pathway. FEBS Lett 2014;588:2009–15. [DOI] [PubMed] [Google Scholar]
- 105.Shen L, Li J, Xu L, et al. miR-497 induces apoptosis of breast cancer cells by targeting Bcl-w. Exp Ther Med 2012;3:475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y, Yu Y, Tsuyada A, et al. Transforming growth factor-beta regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene 2011;30:1470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang Z, Sun H, Dai H, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle 2009;8:2756–68. [DOI] [PubMed] [Google Scholar]
- 108.Stinson S, Lackner MR, Adai AT, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal 2011;4:ra41. [DOI] [PubMed] [Google Scholar]
- 109.Miller TE, Ghoshal K, Ramaswamy B, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem 2008;283:29897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhu N, Zhang D, Xie H, et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem 2011;351:157–64. [DOI] [PubMed] [Google Scholar]
- 111.Li Q, Zhu F, Chen P. miR-7 and miR-218 epigenetically control tumor suppressor genes RASSF1A and Claudin-6 by targeting HoxB3 in breast cancer. Biochem Biophys Res Commun 2012;424:28–33. [DOI] [PubMed] [Google Scholar]
- 112.Hu Y, Xu K, Yague E. miR-218 targets survivin and regulates resistance to chemotherapeutics in breast cancer. Breast Cancer Res Treat 2015;151:269–80. [DOI] [PubMed] [Google Scholar]
- 113.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008;451:147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007;449:682–8. [DOI] [PubMed] [Google Scholar]
- 115.Moriarty CH, Pursell B, Mercurio AM. miR-10b targets Tiam1: implications for Rac activation and carcinoma migration. J Biol Chem 2010;285:20541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ma L, Young J, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 2010;12:247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 2009;137:1032–46. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 118.Li X, Roslan S, Johnstone CN, et al. MiR-200 can repress breast cancer metastasis through ZEB1-independent but moesin-dependent pathways. Oncogene 2014;33:4077–88. [DOI] [PubMed] [Google Scholar]
- 119.Chou J, Lin JH, Brenot A, et al. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol 2013;15: 201–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ryu S, McDonnell K, Choi H, et al. Suppression of miRNA-708 by polycomb group promotes metastases by calcium-induced cell migration. Cancer Cell 2013;23:63–76. [DOI] [PubMed] [Google Scholar]
- 121.Hannafon BN, Sebastiani P, de las Morenas A, et al. Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast Cancer Res 2011;13:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhan L, Rosenberg A, Bergami KC, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 2008;135:865–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhu J, Ye Q, Chang L, et al. Upregulation of miR-195 enhances the radiosensitivity of breast cancer cells through the inhibition of BCL-2. Int J Clin Exp Med 2015;8:9142–8. [PMC free article] [PubMed] [Google Scholar]
- 124.Warburg O. On the origin of cancer cells. Science 1956;123:309–14. [DOI] [PubMed] [Google Scholar]
- 125.Rondeau S, Vacher S, De Koning L, et al. ATM has a major role in the double-strand break repair pathway dysregulation in sporadic breast carcinomas and is an independent prognostic marker at both mRNA and protein levels. Br J Cancer 2015;112:1059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene 2006;25:5854–63. [DOI] [PubMed] [Google Scholar]
- 127.Davis NM, Sokolosky M, Stadelman K, et al. Deregulation of the EGFR/PI3K/PTEN/Akt/mTORC1 pathway in breast cancer: possibilities for therapeutic intervention. Oncotarget 2014;5:4603–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nikolaou K, Tsagaratou A, Eftychi C, et al. Inactivation of the deubiquitinase CYLD in hepatocytes causes apoptosis, inflammation, fibrosis, and cancer. Cancer Cell 2012;21:738–50. [DOI] [PubMed] [Google Scholar]
- 129.BSong SJ, Poliseno L, Song MS, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 2013;154:311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Morishita A, Zaidi MR, Mitoro A, et al. HMGA2 is a driver of tumor metastasis. Cancer Res 2013;73:4289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Taylor MA, Sossey-Alaoui K, Thompson CL, et al. TGF-beta upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest 2013;123:150–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Russell R, Perkhofer L, Liebau S, et al. Loss of ATM accelerates pancreatic cancer formation and epithelial-mesenchymal transition. Nat Commun 2015;6:7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang X, Ding L, Bennewith KL, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell 2009;35:856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Geiger TR, Peeper DS. Metastasis mechanisms. Biochim Biophys Acta 2009;1796:293–308. [DOI] [PubMed] [Google Scholar]
- 135.Gee HE, Camps C, Buffa FM, et al. MicroRNA-10b and breast cancer metastasis. Nature 2008;455:E8–9. [DOI] [PubMed] [Google Scholar]
- 136.Roy SS, Gonugunta VK, Bandyopadhyay A, et al. Significance of PELP1/HDAC2/miR-200 regulatory network in EMT and metastasis of breast cancer. Oncogene 2014;33:3707–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–52. [DOI] [PubMed] [Google Scholar]
- 138.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001;98:10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 2003;100:8418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Badve S, Dabbs DJ, Schnitt SJ, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol 2011;24:157–67. [DOI] [PubMed] [Google Scholar]
- 141.Gwak JM, Kim HJ, Kim EJ, et al. MicroRNA-9 is associated with epithelial-mesenchymal transition, breast cancer stem cell phenotype, and tumor progression in breast cancer. Breast Cancer Res Treat 2014;147:39–49. [DOI] [PubMed] [Google Scholar]
- 142.Jonsdottir K, Janssen SR, Da Rosa FC, et al. Validation of expression patterns for nine miRNAs in 204 lymph-node negative breast cancers. PLoS One 2012;7:e48692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mouw JK, Yui Y, Damiano L, et al. Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat Med 2014;20:360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Farazi TA, Ten Hoeve JJ, Brown M, et al. Identification of distinct miRNA target regulation between breast cancer molecular subtypes using AGO2-PAR-CLIP and patient datasets. Genome Biol 2014;15:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de Rinaldis E, Gazinska P, Mera A, et al. Integrated genomic analysis of triple-negative breast cancers reveals novel microRNAs associated with clinical and molecular phenotypes and sheds light on the pathways they control. BMC Genomics 2013;14:643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu Y, Cai Q, Bao PP, et al. Tumor tissue microRNA expression in association with triple-negative breast cancer outcomes. Breast Cancer Res Treat 2015;152:183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guttilla IK, Adams BD, White BA. ERalpha, microRNAs, and the epithelial-mesenchymal transition in breast cancer. Trends Endocrinol Metab 2012;23:73–82. [DOI] [PubMed] [Google Scholar]
- 148.Inic Z, Zegarac M, Inic M, et al. Difference between luminal A and luminal B subtypes according to Ki-67, tumor size, and progesterone receptor negativity providing prognostic information. Clin Med Insights Oncol 2014;8:107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yanai A, Inoue N, Yagi T, et al. Activation of mTOR/S6K but not MAPK pathways might be associated with high Ki-67, ER(+), and HER2(-) breast cancer. Clin Breast Cancer 2015;15:197–203. [DOI] [PubMed] [Google Scholar]
- 150.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 2003;100:3983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009;138:592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kotiyal S, Bhattacharya S. Breast cancer stem cells, EMT and therapeutic targets. Biochem Biophys Res Commun 2014;453:112–6. [DOI] [PubMed] [Google Scholar]
- 153.Martello G, Rosato A, Ferrari F, et al. A MicroRNA targeting dicer for metastasis control. Cell 2010;141:1195–207. [DOI] [PubMed] [Google Scholar]
- 154.Kovalchuk O, Filkowski J, Meservy J, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther 2008;7:2152–9. [DOI] [PubMed] [Google Scholar]
- 155.Liang Z, Wu H, Xia J, et al. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol 2010;79:817–24. [DOI] [PubMed] [Google Scholar]
- 156.Ma MT, He M, Wang Y, et al. MiR-487a resensitizes mitoxantrone (MX)-resistant breast cancer cells (MCF-7/MX) to MX by targeting breast cancer resistance protein (BCRP/ABCG2). Cancer Lett 2013;339:107–15. [DOI] [PubMed] [Google Scholar]
- 157.Rao X, Di Leva G, Li M, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 2011;30:1082–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Taylor DD, Gercel-Taylor C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front Genet 2013;4:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2011;2:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Putz U, Howitt J, Doan A, et al. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Sci Signal 2012;5:ra70. [DOI] [PubMed] [Google Scholar]
- 161.Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–9. [DOI] [PubMed] [Google Scholar]
- 162.Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014;25:501–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012;18:883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Luga V, Zhang L, Viloria-Petit AM, et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012;151:1542–56. [DOI] [PubMed] [Google Scholar]
- 165.Lim PK, Bliss SA, Patel SA, et al. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res 2011;71:1550–60. [DOI] [PubMed] [Google Scholar]
- 166.Yang M, Chen J, Su F, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer 2011;10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen WX, Liu XM, Lv MM, et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One 2014;9:e95240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li Q, Eades G, Yao Y, et al. Characterization of a stem-like subpopulation in basal-like ductal carcinoma in situ (DCIS) lesions. J Biol Chem 2014;289:1303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang H, Ren Y, Xu H, et al. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg Oncol 2013;22:217–23. [DOI] [PubMed] [Google Scholar]
- 170.Hashim A, Rizzo F, Marchese G, et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget 2014;5:9901–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fu A, Jacobs DI, Hoffman AE, et al. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis 2015;36:1094–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shore AN, Herschkowitz JI, Rosen JM. Noncoding RNAs involved in mammary gland development and tumorigenesis: there's a long way to go. J Mammary Gland Biol Neoplasia 2012;17:43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vikram R, Ramachandran R, Abdul KS. Functional significance of long non-coding RNAs in breast cancer. Breast Cancer 2014;21:515–21. [DOI] [PubMed] [Google Scholar]
- 174.Hansji H, Leung EY, Baguley BC, et al. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front Genet 2014;5:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Front Biosci (Schol Ed) 2015;7:94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010;329:689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang L, Zeng X, Chen S, et al. BRCA1 is a negative modulator of the PRC2 complex. EMBO J 2011;32:1584–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Xue X, Yang YA, Zhang A, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang YL, Overstreet AM, Chen MS, et al. Combined inhibition of EGFR and c-ABL suppresses the growth of triple-negative breast cancer growth through inhibition of HOTAIR. Oncotarget 2015;6:11150–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Padua Alves C, Fonseca AS, Muys BR, et al. Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells 2013;31:2827–32. [DOI] [PubMed] [Google Scholar]
- 182.Iacoangeli A, Lin Y, Morley EJ, et al. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis 2004;25:2125–33. [DOI] [PubMed] [Google Scholar]
- 183.Kondrashov AV, Kiefmann M, Ebnet K, et al. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP). J Mol Biol 2005;353:88–103. [DOI] [PubMed] [Google Scholar]
- 184.Gumireddy K, Li A, Yan J, et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J 2013;32:2672–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huang J, Zhou N, Watabe K, et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis 2014;5:e1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mourtada-Maarabouni M, Pickard MR, Hedge VL, et al. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009;28:195–208. [DOI] [PubMed] [Google Scholar]
- 187.Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 2010;3:ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lanz RB, McKenna NJ, Onate SA, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 1999;97:17–27. [DOI] [PubMed] [Google Scholar]
- 189.Colley SM, Leedman PJ. Steroid Receptor RNA Activator - A nuclear receptor coregulator with multiple partners: insights and challenges. Biochimie 2011;93:1966–72. [DOI] [PubMed] [Google Scholar]
- 190.Leygue E, Dotzlaw H, Watson PH, et al. Expression of the steroid receptor RNA activator in human breast tumors. Cancer Res 1999;59:4190–3. [PubMed] [Google Scholar]
- 191.Lanz RB, Chua SS, Barron N, et al. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol Cell Biol 2003;23:7163–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chujo T, Yamazaki T, Hirose T. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim Biophys Acta 2015. [DOI] [PubMed] [Google Scholar]
- 193.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031–41. [DOI] [PubMed] [Google Scholar]
- 194.Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013;73:1180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007;8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet 2013;9:e1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gutschner T, Hammerle M, Diederichs S. MALAT1—a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. [DOI] [PubMed] [Google Scholar]
- 198.Sun Y, Wu J, Wu SH, et al. Expression profile of microRNAs in c-Myc induced mouse mammary tumors. Breast Cancer Res Treat 2009;118:185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Guffanti A, Iacono M, Pelucchi P, et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics 2009;10:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol 2013;10:456–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Chakravarty D, Sboner A, Nair SS, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun 2014;5:5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Standaert L, Adriaens C, Radaelli E, et al. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA 2014;20:1844–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Choudhry H, Albukhari A, Morotti M, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 2015;34:4482–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yang J, Li T, Gao C, et al. FOXO1 3'UTR functions as a ceRNA in repressing the metastases of breast cancer cells via regulating miRNA activity. FEBS Lett 2014;588:3218–24. [DOI] [PubMed] [Google Scholar]
- 205.Paci P, Colombo T, Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst Biol 2014;8:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Heneghan HM, Miller N, Kelly R, et al. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist 2010;15:673–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Asaga S, Kuo C, Nguyen T, et al. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem 2011;57:84–91. [DOI] [PubMed] [Google Scholar]
- 208.Schwarzenbach H, Milde-Langosch K, Steinbach B, et al. Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res Treat 2012;134:933–41. [DOI] [PubMed] [Google Scholar]
- 209.Wu Q, Wang C, Lu Z, et al. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin Chim Acta 2012;413:1058–65. [DOI] [PubMed] [Google Scholar]
- 210.Guo LJ, Zhang QY. Decreased serum miR-181a is a potential new tool for breast cancer screening. Int J Mol Med 2012;30:680–6. [DOI] [PubMed] [Google Scholar]
- 211.Zhao FL, Hu GD, Wang XF, et al. Serum overexpression of microRNA-10b in patients with bone metastatic primary breast cancer. J Int Med Res 2012;40:859–66. [DOI] [PubMed] [Google Scholar]
- 212.Mar-Aguilar F, Mendoza-Ramirez JA, Malagon-Santiago I, et al. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers 2013;34:163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Si H, Sun X, Chen Y, et al. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol 2013;139:223–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chan M, Liaw CS, Ji SM, et al. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res 2013;19:4477–87. [DOI] [PubMed] [Google Scholar]
- 215.Cuk K, Zucknick M, Madhavan D, et al. Plasma microRNA panel for minimally –invasive detection of breast cancer. PLoS One 2013;8:e76729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Chen W, Cai F, Zhang B, et al. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: potential biomarkers. Tumour Biol 2013;34:455–62. [DOI] [PubMed] [Google Scholar]
- 217.Kumar S, Keerthana R, Pazhanimuthu A, et al. Overexpression of circulating miRNA-21 and miRNA-146a in plasma samples of breast cancer patients. Indian J Biochem Biophys 2013;50:210–4. [PubMed] [Google Scholar]
- 218.Zeng RC, Zhang W, Yan XQ, et al. Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Med Oncol 2013;30:477. [DOI] [PubMed] [Google Scholar]
- 219.Ng EK, Li R, Shin VY, et al. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One 2013;8:e53141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Gao J, Zhang Q, Xu J, et al. Clinical significance of serum miR-21 in breast cancer compared with CA153 and CEA. Chin J Cancer Res 2013;25:743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Shen J, Hu Q, Schrauder M, et al. Circulating miR-148b and miR-133a as biomarkers for breast cancer detection. Oncotarget 2014;5:528494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Erbes T, Hirschfeld M, Rucker G, et al. Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer 2015;15:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Shaker O, Maher M, Nassar Y, et al. Role of microRNAs -29b-2, -155, -197 and -205 as diagnostic biomarkers in serum of breast cancer females. Gene 2015;560:77–82. [DOI] [PubMed] [Google Scholar]
- 224.Zhang K, Zhao S, Wang Q, et al. Identification of microRNAs in nipple discharge as potential diagnostic biomarkers for breast cancer. Ann Surg Oncol 2015. [DOI] [PubMed] [Google Scholar]
- 225.Camps C, Buffa FM, Colella S, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 2008;14:1340–8. [DOI] [PubMed] [Google Scholar]
- 226.Quesne JL, Jones J, Warren J, et al. Biological and prognostic associations of miR-205 and let-7b in breast cancer revealed by in situ hybridization analysis of micro-RNA expression in arrays of archival tumour tissue. J Pathol 2012;227:306–14. [DOI] [PubMed] [Google Scholar]
- 227.Cheng CW, Wang HW, Chang CW, et al. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res Treat 2012;134:1081–93. [DOI] [PubMed] [Google Scholar]
- 228.Madhavan D, Zucknick M, Wallwiener M, et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin Cancer Res 2012;18:5972–82. [DOI] [PubMed] [Google Scholar]
- 229.Mulrane L, Madden SF, Brennan DJ, et al. miR-187 is an independent prognostic factor in breast cancer and confers increased invasive potential in vitro. Clin Cancer Res 2012;18:6702–13. [DOI] [PubMed] [Google Scholar]
- 230.Li Y, Ma X, Zhao J, et al. microRNA-210 as a prognostic factor in patients with breast cancer: meta-analysis. Cancer Biomark 2013;13:471–81. [DOI] [PubMed] [Google Scholar]
- 231.Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc Natl Acad Sci USA 2013;110:7413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Markou A, Yousef GM, Stathopoulos E, et al. Prognostic significance of metastasis-related microRNAs in early breast cancer patients with a long follow-up. Clin Chem 2014;60:197–205. [DOI] [PubMed] [Google Scholar]
- 233.Ma L, Li GZ, Wu ZS, et al. Prognostic significance of let-7b expression in breast cancer and correlation to its target gene of BSG expression. Med Oncol 2014;31:773. [DOI] [PubMed] [Google Scholar]
- 234.Lee SJ, Seo JW, Chae YS, et al. Genetic polymorphism of miR-196a as a prognostic biomarker for early breast cancer. Anticancer Res 2014;34:2943–9. [PubMed] [Google Scholar]
- 235.Chen B, Tang H, Liu X, et al. miR-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Cancer Lett 2015;356:410–7. [DOI] [PubMed] [Google Scholar]
- 236.Agarwal S, Hanna J, Sherman ME, et al. Quantitative assessment of miR34a as an independent prognostic marker in breast cancer. Br J Cancer 2015;112:61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wang S, Li H, Wang J, et al. Prognostic and biological significance of microRNA-127 expression in human breast cancer. Dis Markers 2014;2014:401986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zheng R, Pan L, Gao J, et al. Prognostic value of miR-106b expression in breast cancer patients. J Surg Res 2015;195:158–65. [DOI] [PubMed] [Google Scholar]
- 239.Yao Y, Hu J, Shen Z, et al. MiR-200b expression in breast cancer: a prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. J Cell Mol Med 2015;19: 760–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.D'Aiuto F, Callari M, Dugo M, et al. miR-30e* is an independent subtype-specific prognostic marker in breast cancer. Br J Cancer 2015;113:290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sorensen KP, Thomassen M, Tan Q, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat 2013;142:529–36. [DOI] [PubMed] [Google Scholar]
- 242.Kaboli PJ, Rahmat A, Ismail P, et al. MicroRNA-based therapy and breast cancer: a comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacol Res 2015;97:104–21. [DOI] [PubMed] [Google Scholar]
- 243.Lennox KA, Owczarzy R, Thomas DM, et al. Improved performance of anti-miRNA oligonucleotides using a novel non-nucleotide modifier. Mol Ther Nucleic Acids 2013;2:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Shen H, Mittal V, Ferrari M, et al. Delivery of gene silencing agents for breast cancer therapy. Breast Cancer Res 2013;15:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Baluk P, Hashizume H, McDonald DM. Cellular abnormalities of blood vessels as targets in cancer. Curr Opin Genet Dev 2005;15:102–11. [DOI] [PubMed] [Google Scholar]
- 246.Babar IA, Cheng CJ, Booth CJ, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA 2012;109:E1695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Mlotshwa S, Pruss GJ, MacArthur JL, et al. A novel chemopreventive strategy based on therapeutic microRNAs produced in plants. Cell Res 2015;25:521–4. [DOI] [PMC free article] [PubMed] [Google Scholar]