Abstract
Schizophrenia is currently diagnosed by physicians through clinical assessment and their evaluation of patient’s self-reported experiences over the longitudinal course of the illness. There is great interest in identifying biologically based markers at the onset of illness, rather than relying on the evolution of symptoms across time. Functional network connectivity shows promise in providing individual subject predictive power. The majority of previous studies considered the analysis of functional connectivity during resting-state using only fMRI. However, exclusive reliance on fMRI to generate such networks, may limit inference on dysfunctional connectivity, which is hypothesized to underlie patient symptoms. In this work, we propose a framework for classification of schizophrenia patients and healthy control subjects based on using both fMRI and band limited envelope correlation metrics in MEG to interrogate functional network components in the resting state. Our results show that the combination of these two methods provide valuable information that captures fundamental characteristics of brain network connectivity in schizophrenia. Such information is useful for prediction of schizophrenia patients. Classification accuracy performance was improved significantly (up to ≈ 7%) relative to only the fMRI method and (up to ≈ 21%) relative to only the MEG method.
I. Introduction
The general approach for the diagnosis of schizophrenia is based on a patients self-reported experiences and observed behavior over the longitudinal course of the illness. There is great interest in identifying biologically based marker of illness, rather than relying on symptom assessment because the current approach may postpone the diagnosis of the disorder, whereas early diagnosis can improve treatment response and reduce associated costs [1]. But small numbers of training subjects and high dimensional datasets make it challenging to design robust and accurate classifiers for schizophrenia. Functional connectivity shows promise in providing individual subject predictive power. Seed-based functional connectivity approaches assess the temporal correlation between a seed region and individual brain voxels [2], [3]. Independent component analysis (ICA) based functional network connectivity (FNC) is a data-driven approach that summarizes the overall connection between independent brain maps over time [4], [5]. Therefore, the FNC feature provides a picture of the connectivity pattern over time between independent components.
Most of the previous FNC studies have focused only on the performance of a resting-state with fMRI method. However, exclusive reliance on fMRI to generate such networks may limit inference on dysfunctional connectivity which is hypothesized to underlie patient symptoms [7]. Whilst the blood oxygenation-level dependent (BOLD) response measured by fMRI allows high spatial resolution maps, it is limited by being an indirect and slow physiological signal [9]. Neural oscillatory activity, which comprises rhythmic electrical activity in cell assemblies, is thought to underlie BOLD responses. This occurs in the ~1–900Hz band; such rapid electrical signals cannot be assessed using fMRI but can be measured directly by techniques such as MEG [10]. Measurement of the resting state brain activity using both fMRI and MEG, within a common sample of subjects, combines the strengths of each modality by allowing comparison of hemodynamic and electrophysiological effects. In this way we provide significant insight into FNC, with special relevance for the study of schizophrenia and similar conditions.
Significant progress towards integrating MEG and fMRI has been made in the past decade. A recent study [11] used intrinsic connectivity networks (ICNs) in MEG in a similar way to that typically used in fMRI [11]. Also, our recent study [12] used a method based on group spatial ICA, for the first time estimating FNC networks from both MEG and fMRI. The purpose of the present study is to use both fMRI and band limited envelope correlation metrics in MEG to interrogate FNC in the resting state in a sample of healthy normal volunteers and schizophrenia patients to improve the classification accuracy of schizophrenia patients.
II. Materials and Methods
A. Participants
This study combined existing data from 91 subjects. Inclusion criteria for patient selection included diagnosis of schizophrenia or schizoaffective disorder between 18 to 65 years of age. TABLE I. provides demographic characteristics and clinical variables of the participants.
TABLE I.
Demographic Clinical Variables for SPs and HCs.
| Demographics | Mean (SD) | t or x2 | |
|---|---|---|---|
| Patients (n=44) | Controls (n=47) | (p-value) | |
| Age | 37. 3 (13.9) | 35.2(11.8) | 0.78 (0.44) |
| Gender (M/F) | 37/7 | 34/13 | 0.27 (0.78) |
| Ethnicity (H/NH) | 23/21 | 26/21 |
Abbreviations: M=Male, F=Female, H=Hispanic, NH= Non-Hispanic
B. fMRI Data Acquisition and Preprocessing
fMRI data was collected on a single 3-Tesla Siemens Trio scanner with a 12-channel radio frequency coil with a repetition time of 2 sec. High-resolution T1-weighted structural images were acquired with a five-echo MPRAGE sequence. Resting-state scans consisted of 149 volumes per run. After initial standard preprocessing [4], [13] the imaging data was decomposed into functionally homogeneous cortical and sub-cortical regions exhibiting temporally coherent activity using a high model order (100) group-level spatial independent component analysis (GICA).
We used a relatively high model order ICA (number of components, IC = 75), since such models yield refined components that correspond to known anatomical and functional segmentation. Of the 75 components returned by the GICA, 39 were identified as non-artifactual independent components (IC) using a combination of two methods [14] for fMRI method. In the first method we examined the power spectra with two criteria in mind: dynamic range and low frequency/high frequency ratio. Dynamic range refers to the difference between the peak power and minimum power at frequencies to the right of the peak in the power spectra. Low frequency to high frequency power ratio is the ratio of the integral of spectral power below 0.10 Hz to the integral of power between 0.15 and 0.25 Hz. To verify the results, three expert reviewers evaluated the components for functional relevance. In this evaluation, if a component exhibited 1) peak activation in gray matter, 2) low spatial overlap with known vascular, ventricular, motion, and susceptibility artifacts, and 3) Time Courses (TCs) dominated by low frequency fluctuations, it was classified as a non-artifactual component.
Subject specific time courses (TCs) and spatial maps (SMs) were obtained using back reconstruction [15]. The FNC for each subject was estimated from the TC matrix as a C×C sample covariance matrix by using a cosine similarity measure. For tasks in the analysis, we isolated activations related to particular tasks within an fMRI scanning session. Task-related component time courses for separate components within a task were then correlated with one another exclusively over non-zero areas of the hemodynamic predictor function using a cosine similarity measure to yield task-related FNC scores for pairs of components.
C. MEG Data Acquisition and Preprocessing
MEG data were collected in a magnetically and electrically shielded room using a whole-cortex 306-channel MEG array. MEG data were sampled at a rate of 1000 Hz, with a bandpass filter of 0.10 to 330 Hz. Head position was monitored continuously throughout the MEG session. Raw data were collected and stored. Participants were instructed to keep their eyes open and maintain fixation during the 6-minute scan to minimize occipital alpha rhythm [10]. Artifact removal, correction for head movement, and down sampling to 250 Hz were conducted offline using Elekta Maxfilter software, with 123 basis vectors, a spatiotemporal buffer of 10 s, and a correlation limit of 0.95.
Covariance matrices were generated independently for each subject and frequency band, using all recorded data. Covariance matrices were regularized using a value of 4 times the minimum singular value of the unregularized matrix. Source orientation at each voxel was based on a nonlinear search for maximum projected signal-to-noise ratio. The forward solution was based on a dipole model [16] and a single-shell boundary element model [17]. Beamformer projection was performed separately for each subject and frequency range. Then source-space signals were normalized by an estimate of projected noise [18] and transformed to standard (MNI) space using FLIRT in FSL. A Hilbert transform was applied to the time course at each voxel time to derive the analytic signal. The Hilbert envelope at each voxel was down sampled to an effective sampling rate of 1 Hz [11]. Source space envelope data were smoothed spatially (6 mm at full-width half-maximum), and the voxel size was resampled to 3×3×3 mm to facilitate comparison with the fMRI data.
Group spatial ICA was applied to the individual subject data using the GIFT toolbox (http://mialab.mrn.org/software/gift). Each frequency range was treated as a session in GIFT to permit analysis of each band, as well as the mean across bands. For MEG ICA processing and non-artifactual components selection, we follow the procedures that are applied to the fMRI as mentioned in previous section. Of the 75 components requested from the group ICA, 29 were retained as non-artifactual components for MEG method.
III. Classification
Determining a reliable biological feature for a mental disorder is an important step for developing a more accurate and reliable framework for diagnosis, and ultimately treatment [19]. Resting-state fMRI connectivity has been used in determining the differences based on biological features of mental disorders including schizophrenia. [20]–[23]. Using methods based on group spatial ICA, we estimate networks from both MEG and fMRI and findings from these two neuroimaging modalities, with the hypotheses that using both MEG and fMRI measures of among-network connectivity would show improvement to classification of schizophrenia patients. In this chapter, we used dynamic FNC to determine reliable differences based on dynamic FNC differences of schizophrenia.
A. Dynamic Functional Network Connectivity and Clustering
Recent studies [13], [21]–[24] show that connectivity dynamics can capture repetitive patterns of interactions among intrinsic networks during a rest or task related experiments that cannot be detected with FNC (static functional connectivity analyses). These repetitive patterns of interactions contain valuable information for individual prediction of schizophrenia patients. Such information is useful for training and replicates in testing.
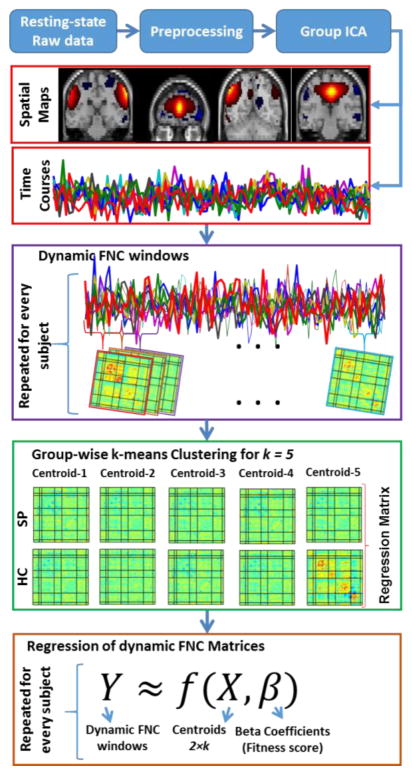
In order to obtain connectivity dynamics we follow the steps of a previous study [24]. First, we computed correlations between non-artifactual components’ time courses using a sliding window approach with a rectangular window of 25 TR (in steps of 1TR) convolved with Gaussian of sigma 3 TRs to obtain tapering along the edges. To characterize the full covariance matrix, we estimated covariance from regularized inverse covariance matrix (ICOV) [25] by using the graphical LASSO framework [26]. Then we placed a penalty on the L1 norm of the precision matrix to enforce sparsity. The regularization parameter was optimized for each subject separately by evaluating the log-likelihood of unseen data of the subject in a cross-validation framework. Second, we selected group centrotypes by using k-means clustering algorithm from all of the dynamic windowed FNC matrices for each group. Then for each FNC time point, we regressed out the dynamic FNC matrix against these 2×k states and obtained the corresponding beta coefficients. We used the mean of these beta coefficients and finalized 2×k features for each subject for classification. See Figure 1. for schematic description of dynamic FNC, clustering and regression of dynamic FNC matrices
Figure 1.
Schematic description of dynamic FNC, clustering and regression of dynamic FNC matrices
In order to compute the optimal accuracy score, we define most efficient cluster number by using elbow criterion of the cluster validity index, which is computed as the ratio between within-cluster distances to between-cluster distance.
IV. Results
We evaluated the performance improvement of classification based on dynamic FNC and combination estimated networks from both MEG and fMRI methods. Our main focus was to extract reliable features from the dynamic FNC matrices and combine these features to perform the best classification results.
First, we used fMRI dynamic FNC matrixes and MEG dynamic FNC matrixes separately (for each frequency) for classification (See TABLE II. ) then we combined fMRI (subject×time×FNC=91×119×703) and MEG (frequency× subject×time×FNC=5×91×270×496) dynamic FNC matrixes as a data set for classification (See TABLE III. ). And we compared results to show the improvement of combining fMRI and MEG methods for classification.
TABLE II.
Classification accuracy obtained from fMRI data, MEG data for each frequency and combination of all MEG data frequencies by using majority voting method
| Acc % | fMRI | MEG A | MEG B | MEG D | MEG G | MEG T | MEG MJ Vt |
|---|---|---|---|---|---|---|---|
| NBC | 82.42 | 65.93 | 71.43 | 71.43 | 51.65 | 53.85 | 65.93 |
| nSVM | 83.52 | 69.23 | 72.53 | 69.23 | 51.65 | 58.24 | 69.23 |
| LDF | 82.42 | 65.93 | 68.13 | 71.43 | 52.75 | 53.85 | 65.93 |
TABLE III.
Classification accuracy obtained from the combination of fMRI data and MEG data for each frequency and the combination of all by using majority voting method
| Acc % | fMRI | fMRI | fMRI | fMRI | fMRI | MJ Vt |
|---|---|---|---|---|---|---|
| MEG A | MEG B | MEG D | MEG G | MEG T | ||
| NBC | 83.52 | 87.91 | 86.81 | 83.52 | 85.71 | 90.11 |
| nSVM | 82.42 | 85.71 | 84.62 | 82.42 | 81.32 | 87.91 |
| LDF | 82.42 | 83.52 | 83.52 | 82.42 | 83.52 | 85.71 |
We used leave-one-out cross validation method. One subject for testing and the rest of the data (90 subjects) were used as a training data set. And this process is repeated for each subject. In each cross-validation run, we obtained 5 cluster centroids for each group and regressed out the dynamic FNC matrix against these 10 centroids (5 centroids for each group) and computed the corresponding beta coefficients for all dynamic FNC for each subject. Then, we used the mean of these beta coefficients across the subjects and finalized 10 features for each subject for classification.
We performed the leave one out method with three well known classification algorithms; linear discriminant classifier (LDC), Naïve Bayes classifier (NBC) and non-linear SVM (nSVM) with Gaussian radial bases function kernel to test the hypothesis.
TABLE II. reports the classification results that were obtained from fMRI data, MEG data for each frequency and combination of all MEG data frequencies by using majority voting method. Results show that the classification accuracy obtained from fMRI data (nSVM-83.52%) provides better classification performance than MEG data for all frequencies and combination of all MEG data frequencies by using majority voting method. Comparison of internal MEG frequencies shows that beta (nSVM-72.53%) frequency has better performance than other frequencies and combination of all MEG data frequencies. Similarly, FDR-corrected group differences of MEG-beta and MEG-delta frequencies show more significant differences than other frequencies.
TABLE III. summarizes the classification accuracy obtained from the combination of fMRI data and MEG data for each frequency and the combination of all by using majority voting method. Combination of features obtained from dynamic FNC of fMRI and MEG-Beta frequency provided better results (NBC-87.91%) than other frequencies. The best performance is provided by the combination of all by using majority voting method (NBC-90.11%).
We repeated the clustering method by using different distance functions such as Euclidian, correlation, cosine similarities. We did not find any performance differences.
V. Conclusion
Our results provided evidence that the combination of fMRI and MEG modalities captures important information for classification that is missed by using only one modality. This suggests that the combination of these two methods provides valuable information that captures fundamental characteristics of brain network connectivity in schizophrenia. These results may help to design an objective biological marker-based diagnostic test for schizophrenia for early diagnosis.
Acknowledgments
Research supported by NIH grants P20GM103472/1R01EB006841 to VDC and NIH P20RR021938, P20GM103472, & K01AA021431.
Contributor Information
Mustafa S Cetin, The Mind Research Network, Albuquerque, NM 87106 USA.
Jon M. Houck, Department of Psychology, University of New Mexico, & the Mind Research Network, Albuquerque, NM 87106 USA
Victor M. Vergara, The Mind Research Network, Albuquerque, NM 87106 USA
Robyn L. Miller, The Mind Research Network, Albuquerque, NM 87106 USA
Vince Calhoun, The Mind Research Network, Albuquerque, NM 87106 and The Department of Electrical & Computer Engineering, University of New Mexico, Albuquerque, NM 87131 USA.
References
- 1.Kubicki M, Mccarley R, Westin C-F, Park H-J, Maier S, Kikinis R, Jolesz FA, Shenton ME. A review of diffusion tensor imaging studies in schizophrenia. J Psychiatr Res. 2007:15–30. doi: 10.1016/j.jpsychires.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anti correlated functional networks. Proc Natl Acad Sci USA. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordes D, Haughton V, Carew JD, Arfanakis K, Maravilla K. Hierarchical clustering to measure connectivity in fMRI resting-state data. MagnReson Imaging. 2002;4(20):305–317. doi: 10.1016/s0730-725x(02)00503-9. [DOI] [PubMed] [Google Scholar]
- 4.Cetin MS, Christensen F, Abbott CC, Stephen JM, Mayer AR, Cañive JM, Calhoun VD. Thalamus and posterior temporal lobe show greater inter-network connectivity at rest and across sensory paradigms in schizophrenia. NeuroImage. 2014;97:117–126. doi: 10.1016/j.neuroimage.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jafri MJ, Pearlson GD, Stevens M, Calhoun VD. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. Neuroimage. 2008:1666–1681. doi: 10.1016/j.neuroimage.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullmore ET, Frangou S, Murray RM. The dysplastic net hypothesis: an integration of developmental and dysconnectivity theories of schizophrenia. Schizophr Res. 1997;28:143–156. doi: 10.1016/s0920-9964(97)00114-x. [DOI] [PubMed] [Google Scholar]
- 7.Stephan KE, Baldeweg T, Friston KJ. Synaptic Plasticity and Dysconnection in Schizophrenia. Biol Psychiatry. 2006;59:929–939. doi: 10.1016/j.biopsych.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Bandettini PA, Jesmanowicz A, Wong EC, Hyde JS. Processing strategies for time-course data sets in functional MRI of the human brain. Magn Reson Med J Soc Magn Reson Med Soc Magn Reson Med. 1993;30:161–173. doi: 10.1002/mrm.1910300204. [DOI] [PubMed] [Google Scholar]
- 9.Kim SG, Richter W, Uğurbil K. Limitations of temporal resolution in functional MRI. Magn Reson Med J Soc Magn Reson Med Soc Magn Reson Med. 1997;37(4):631–636. doi: 10.1002/mrm.1910370427. [DOI] [PubMed] [Google Scholar]
- 10.Cohen D. Magnetoencephalography: evidence of magnetic fields produced by alpha rhythm currents. Science. 1968;161:784–6. doi: 10.1126/science.161.3843.784. [DOI] [PubMed] [Google Scholar]
- 11.Brookesa MJ, Woolrichb M, Luckhoob H, Pricea D, Halea JR, Stephensona MC, Barnesc GR, Smithd SM, Morrisa PG. Investigating the electrophysiological basis of resting state networks using magnetoencephalography. Neuroimage. 2011:302–308. doi: 10.1073/pnas.1112685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houck JM, Cetin MS, Mayer AR, Bustillo JR, Stephen JM, Aine CJ, Cañive JM, et al. Magnetoencephalographic and functional MRI connectomics in schizophrenia via intra- and inter-network connectivity. NeuroImaging. 2015 doi: 10.1016/j.neuroimage.2016.10.011. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen EA, Damaraju E, Plis SM, Erhardt EB, Calhoun VD. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex. 2012 Nov;24(3):663–676. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen EA, Erhardt EB, Wei Y, Eichele T, Calhoun VD. Capturing inter-subject variability with group independent component analysis of fMRI data: a simulation study. NeuroImage. 2011:4141–4159. doi: 10.1016/j.neuroimage.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erhardt EB, Rachakonda S, Bedrick EJ, Allen EA, Adali T, Calhoun VD. Comparison of multi-subject ICA methods for analysis of fMRI data. Hum Brain Mapp. 2011;32:2075–2095. doi: 10.1002/hbm.21170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarvas J. Basic mathematical and electromagnetic concepts of the biomagnetic inverse problem. Phys Med Biol. 1987;32:11. doi: 10.1088/0031-9155/32/1/004. [DOI] [PubMed] [Google Scholar]
- 17.Hamalainen MS, Sarvas J. Realistic conductivity geometry model of the human head for interpretation of neuromagnetic data. IEEE Trans Biomed Eng. 1989;36:165–171. doi: 10.1109/10.16463. [DOI] [PubMed] [Google Scholar]
- 18.Hall EL, Woolrich MW, Thomaz CE, Morris PG, Brookes MJ. Using variance information in magnetoencephalography measures of functional connectivity. NeuroImage. 2013;67:203–212. doi: 10.1016/j.neuroimage.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Keshavan MS, Clementz BA, Pearlson GD, Sweeney JA, Tamminga CA. Reimagining psychoses: an agnostic approach to diagnosis. Schizophr Res. 2013;146:10–16. doi: 10.1016/j.schres.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 20.Arbabshirani MR, Kiehl KA, Pearlson GD, Calhoun VD. Classification of schizophrenia patients based on resting-state functional network connectivity. Front Brain Imaging Methods. 2013 doi: 10.3389/fnins.2013.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rashid Barnaly, Damaraju Eswar, Pearlson Godfrey D, Calhoun Vince D. Dynamic connectivity states estimated from resting fMRI Identify differences among Schizophrenia, bipolar disorder, and healthy control subjects. Front Hum Neurosci. 2014;8(897) doi: 10.3389/fnhum.2014.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rashid Barnaly, Arbabshirani Mohammad Reza, Damaraju Eswar, Miller Robyn, Cetin Mustafa S, Pearlson Godfrey, Calhoun Vince. Classification of Schizophrenia and Bipolar Patients Using Static and Time-Varying Resting-State Fmri Brain Connectivity. presented at the IEEE International Symposium on Biomedical Imaging; 2015. [Google Scholar]
- 23.Ünal Sakoğlu, Pearlson Godfrey D, Kiehl Kent A, Michelle Wang Y, Michael Andrew M, Calhoun Vince D. A method for evaluating dynamic functional network connectivity and task-modulation: application to schizophrenia. Magn Reson Mater Phys Biol Med. 2010;23(5–6):351–366. doi: 10.1007/s10334-010-0197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damaraju E, Allen EA, Belger A, Ford JM, McEwen S, Mathalon DH, Mueller BA, et al. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. NeuroImage Clin. 2014;5:298–308. doi: 10.1016/j.nicl.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. Network modelling methods for FMRI. Neuroimage. 2011:875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- 26.Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008:432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]