Summary
Multicellular organisms are composed of many cell types that acquire their specific fate through a precisely controlled pattern of gene expression in time and space dictated in part by cell type-specific promoter activity. Understanding the contribution of highly specialized cell types in the development of a whole organism requires the ability to isolate or analyze different cell types separately. We have characterized and validated a large collection of root cell type-specific promoters and have generated cell type-specific marker lines. These benchmarked promoters can be readily used to evaluate cell type-specific complementation of mutant phenotypes, or to knockdown gene expression using targeted expression of artificial miRNA. We also generated vectors and characterized transgenic lines for cell type-specific induction of gene expression and cell type-specific isolation of nuclei for RNA and chromatin profiling. Vectors and seeds from transgenic Arabidopsis plants will be freely available, and will promote rapid progress in cell type-specific functional genomics. We demonstrate the power of this promoter set for analysis of complex biological processes by investigating the contribution of root cell types in the IRT1-dependent root iron uptake. Our findings revealed the complex spatial expression pattern of IRT1 in both root epidermis and phloem companion cells and the requirement for IRT1 to be expressed in both cell types for proper iron homeostasis.
Keywords: SAND lines, RED TIDE lines, BREAK lines, INTACT, LINE UP lines
Introduction
During the development of multicellular organisms, each cell acquires its specific fate through a precisely controlled pattern of gene expression dictated in part by cell type-specific promoter activity. Such spatial distribution of biological functions is mirrored by differential gene expression. Yet, the function of a given gene is usually evaluated by generating genetically modified organisms using strong viral promoter elements from cytomegalovirus (CMV) in mammalian cells or the cauliflower mosaic virus in plants (CaMV35S). These promoters drive high levels of expression in most cell types, but prevent the analysis of organ-specific or cell type-specific effects. The complexity of biological processes inherent to the existence of multiple cell types therefore highlights the necessity to increase our resolution to the cell type level to deconstruct these processes.
Among the different organs of higher plants, the primary root is an excellent developmental model because of its simple and stereotyped organization of cell types (Dolan et al. 1993, Sarkar et al. 2007, Wildwater et al. 2005). Several root cell type-specific promoters have been identified over the past two decades in Arabidopsis thaliana, following the characterization of one’s favorite genes. More powerful were the gene trap strategies looking for transgenic lines where reporter genes are cell- or tissue-specific, temporally or conditionally regulated (Springer 2000). These specific promoters have been used to drive expression of a given gene in discrete territories of the root or in response to a particular signal/condition to evaluate its local contribution in a process involving other expression territories or to shed light on its ability to function as a mobile signal (Duan et al. 2013, Fridman et al. 2014, Gonzalez-Garcia et al. 2011, Hacham et al. 2011). Cell type-specific promoters have also been used to investigate transcriptomes, translatomes, proteomes or to profile chromatin changes in different root cell types (Birnbaum et al. 2003, Brady et al. 2007, Deal and Henikoff 2010, Fridman et al. 2014, Mustroph et al. 2009, Petricka et al. 2012, Vragovic et al. 2015), dramatically increasing our resolution on the cellular organization of higher plants. Large-scale analyses also characterized cell type-specific responses to endogenous signals like auxin and to changing environmental conditions such as nutrient availability and salt stress (Bargmann et al. 2013, Dinneny et al. 2008, Gifford et al. 2008, Iyer-Pascuzzi et al. 2011), and therefore provided a wealth of information on the regulatory networks driving complex developmental and environmental responses with a cellular resolution.
The necessity to generalize the deconstruction of biological processes in higher plants requires the establishment of a set of validated and integrated tools for cell type-specific functional genomics. This led us to revisit known Arabidopsis promoters to generate a collection of robust cell type-specific promoters driving expression in different developmental zones. These promoters have been used to establish several cell type-specific lines expressing cytosolic or nuclear-localized fluorescent proteins. These lines not only allowed us to ascertain promoter spatial specificity, but also represent new marker lines in different colors and subcellular localization for the different cell types of the root. In addition, these promoters can be used to drive cell type-specific expression of any Arabidopsis genes and evaluate cell type-specific complementation of mutant phenotypes, or to knock down gene expression using targeted expression of artificial miRNA for example. Taking advantage of a specific set of promoters, we established vectors and transgenic lines allowing cell type-specific nuclei purification for RNA and chromatin profiling, as well as for cell type-specific inducible expression. Altogether, these promoters, constructs and transgenic line collection represent a comprehensive toolbox to investigate with high precision gene function in Arabidopsis.
Results
Identification of root cell type-specific promoters
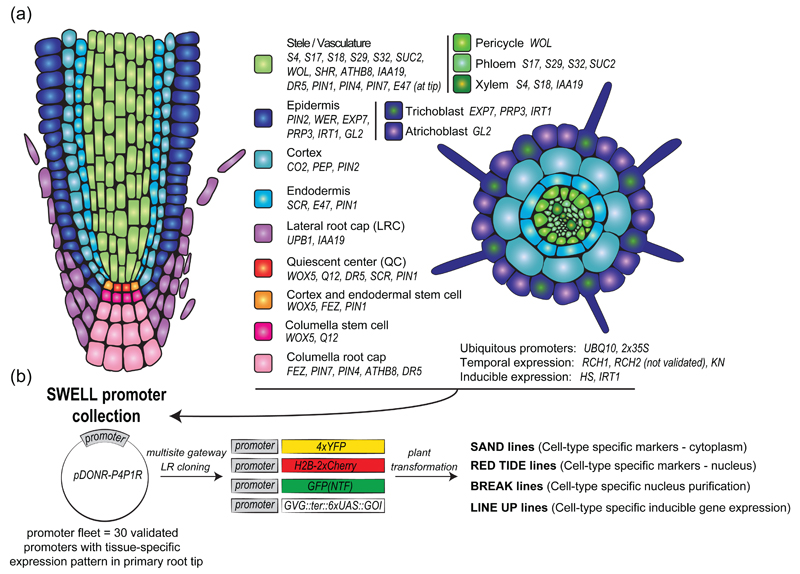
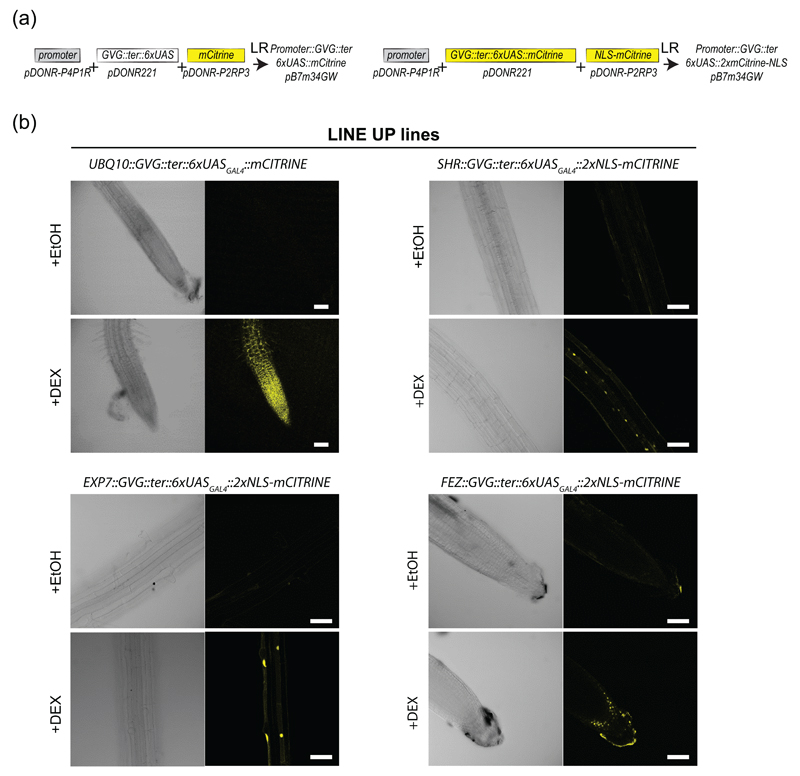
We generated a list of known promoters that, based on the literature, are active in specific root cell types or regulated by endogenous/exogenous cues but spatially restricted (Figure 1a). These promoters were cloned into the pDONR-P4P1R Multisite Gateway compatible entry vector and sequenced (Figure 1b). Care was taken to clone the exact same published promoter fragments, unless stated otherwise (Table S1). To validate the cell type-specificity of these promoters and monitor with the greatest sensitivity their exact expression territories, they were recombined in a destination vector containing four tandem copies of the Yellow Fluorescent Protein (YFP) (Figure 1b and 2a). The resulting vectors were transformed into Arabidopsis plant by floral dipping. The ratio of primary transformants showing fluorescence and the expected expression profile was scored (Table S1). The six brightest T1 plants exhibiting a fluorescence pattern consistent with previously published one were transferred to soil. Segregation analyses were performed at the T2 and T3 stages to isolate monoinsertional homozygous transgenic plants.
Figure 1. Strategy for generation of the SWELL promoter collection and derived transgenic lines.
(a) Schematic representations of the primary root tip (left, longitudinal section, right, transversal section) with the different promoter used in this study.
(b) Cloning strategy for generation of the different cell type-specific reporter line generated in this study.
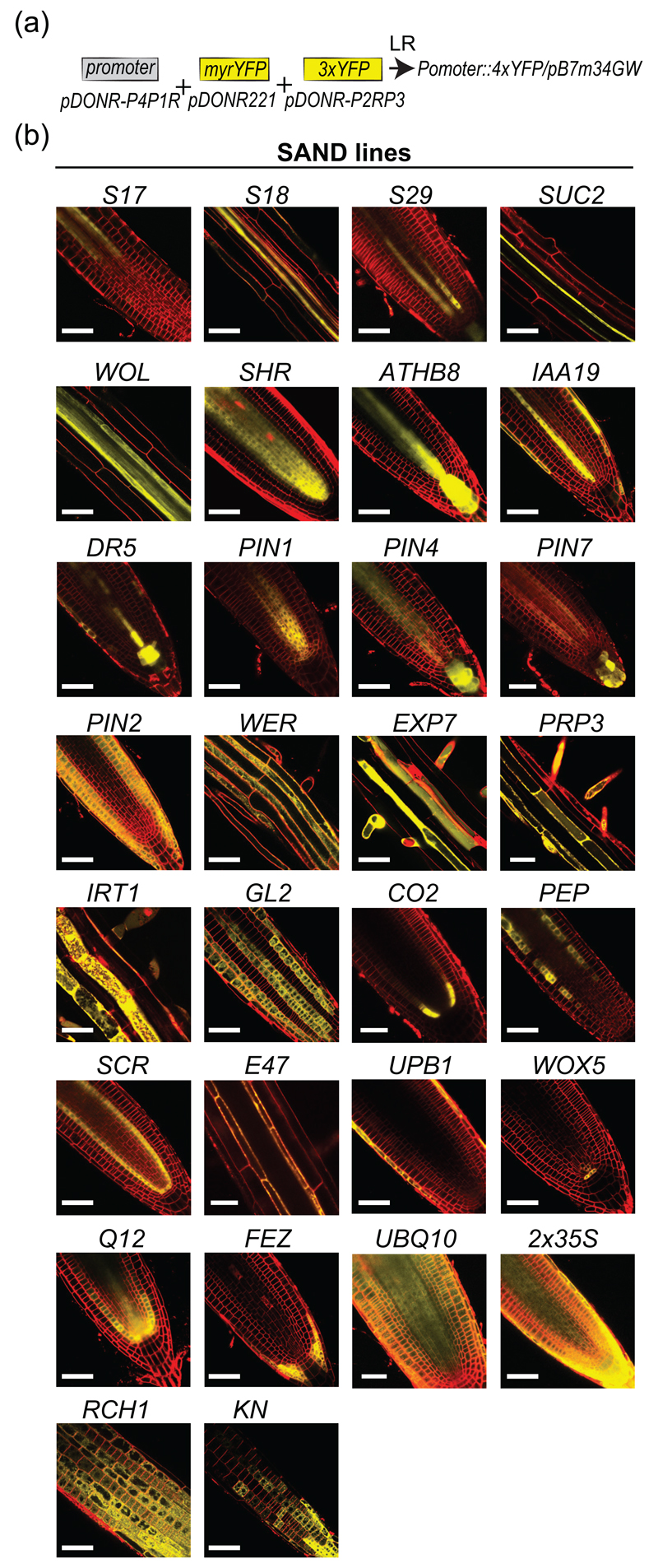
Figure 2. Expression pattern of the SAND reporter lines.
(a) Cloning strategy to generate the SAND lines.
(b) Images of the primary root for the SAND reporter lines. Depending on the lines, images of the root tip, elongation zone or differentiation zone is provided. The plasma membrane is counterstained with FM4-64 (red). Scale bars: 50 μm.
For each promoter in the collection, the expression pattern of the corresponding 4xYFP line was monitored at T2 and T3 generation (Figure 2b and S1). We used the plasma membrane red fluorescent dye FM4-64 to highlight root architecture. Most lines matched previously reported expression patterns (Figure 2b and S1). Table S1 summarizes the observed versus expected expression pattern. We also scored the number of T1 plants with the observed reported pattern as opposed to expression in other tissues. We noticed that some promoters drove very robust expression patterns, such as the promoters of SCR, PRP3 or CO2, which show similar expression in 100% of the T1s that we analyzed (22 out of 22, 12 out of 12 and 14 out of 14, respectively; Table S1). Conversely, some promoters showed the expected expression domain in only a portion of the T1 that we analyzed (for example the S4 promoter showed xylem expression in only 16 out of 37 T1 analyzed; Table S1). These variations in expression might be due to a stronger susceptibility of the regulatory regions to genomic position effects. In any case, this also highlights the need to systematically check individual transgenic lines for their respective expression, rather than to assume tissue specificity solely based on the promoter used in the construct.
In addition, the extreme sensitivity of our lines expressing 4xYFP revealed further complexity in the transcriptional activity of some previously published cell type specific promoters. This is notably the case for the xylem-specific S4 and S18 promoters (At3g25710 and At5g12870), respectively (Lee et al. 2006), and the pericycle-specific S17 promoter (At2g22850) (Lee et al. 2006), which all tend to show also expression, albeit weaker, in differentiated epidermal cells (Table S1). Similarly, the PIN2 promoter fragment used in our study is active in the epidermis and cortex not only at the root meristem but also in differentiated part of the root (Figure S1). This expression pattern is consistent with the broad activity of the PIN2 promoter along the entire root compared to the relative narrow expression of the PIN2 protein in the root meristem (Sieberer et al. 2000). However, the comparative analysis of PIN2 promoter versus PIN2 protein expression domain was done using GUS as a reporter protein and concluded that PIN2 promoter by itself does not confer tissue specificity (Sieberer et al. 2000). Our analysis of PIN2 promoter activity using the 4xYFP reporter suggests that it is specific for the epidermis and cortex cell files and that protein stability likely restricts PIN2 protein to the meristem in these tissues. This interpretation is consistent with the observation of PIN2 protein in mature trichoblasts and atrichoblast cells (Jones et al. 2009). In addition, we also noticed that the PIN2 promoter was active in the lateral root cap, but not in the columella (Figure 2b), which confirmed previous observations with either anti-PIN2 antibodies or in PIN2::PIN2-GFP lines (Abas et al. 2006, Men et al. 2008). The PIN1 promoter was active in the stele and endodermis, as expected from published immunolocalization (Blilou et al. 2005, Friml et al. 2002), however our promoter was also active in the QC and surrounding initials (Figure 2b and S1). This PIN1 promoter activity matches previously published in situ hybridization (Friml et al. 2002) and the expected broader expression domain of PIN1 transcript as compared to the PIN1 protein (Simaskova et al. 2015). We also noticed PIN1 promoter activity in few cells above the QC in LRC, epidermis and cortex, although this expression pattern was not consistently observed. The PIN7 promoter showed activity in the columella and stele, as previously reported (Vieten et al. 2005). However unlike previously characterized expression pattern, we noticed that PIN7 expression in the stele was extremely weak in the differentiated part of the root (Figure S1). This is likely a specific feature of this particular line, since our other PIN7 lines (BREAK line, see below) shows clear expression in the stele. FEZ mRNA and FEZ-GFP under the control of FEZ promoter were found in the root cap of 5 day-old seedlings (Willemsen et al. 2008). Our analysis of FEZ promoter activity confirmed that it is specific of the root cap (including columella stem cells and columella in half of the lines analyzed). However, we found in several independent lines (7 out of 18 lines tested) that it was not active in the columella and columella stem cells (Figure 2). The WOX5 promoter, described as quiescent center-specific (Sarkar et al. 2007), was also active in stem cells and endodermal cells, although to a lesser extent in the majority of the lines analyzed. The WOL promoter, reported to be stele-active (Bonke et al. 2003, Mustroph et al. 2009), also drove weak expression of YFP in cortical and epidermal cells in the differentiated zone of the root (6 lines out of 23; Table S1) (Figure 2b and S1). In the case of the trichoblast-specific WER promoter (Lee and Schiefelbein 1999), we observed expression in both trichoblast and atrichoblast cell files at the root tip, consistent with recent reports (Bruex et al. 2012), as well as in the lateral root cap (Figure 2b and S1). The broader expression pattern of our WOX5, WOL and WER promoter fragment are likely explained by the use of a shorter promoter fragment than the ones previously reported (Table S1), or to the extreme sensitivity to position effects of the transgene. Nonetheless, the shorter WER promoter may represent a good tool to drive gene expression in root epidermal cells, regardless of their hair or non-hair fate. Finally, we found that the E47 promoter was expressed in the endodermis in the mature part of the root but was expressed in the protoxylem at the root tip (Figure S1).
Overall, a large number of cell type-specific promoters or promoters with interesting expression profiles were isolated (Figure 2b). When possible, we selected two different promoters for a given cell type, with different expression during development. This is notably the case for the trichoblast-specific PRP3 and EXP7 promoters (Bernhardt and Tierney 2000, Cho and Cosgrove 2002), or the cortex-specific CO2 and PEP promoters (Heidstra et al. 2004, Mustroph et al. 2009). We observed that EXP7 and CO2 were expressed in younger root trichoblastic and cortical cells, respectively, compared to PRP3 and PEP. Altogether, the 30 different lines generated not only validate the specificity of promoter activity but also serve as marker lines for the corresponding cell type. The characterized cell type-specific promoters cloned into the pDONR-P4P1R Gateway vector compatible with Multisite Gateway cloning are called the “SWELL” promoters. The sequences of all the constructs may be downloaded from http://www.ens-lyon.fr/RDP/SiCE/Swelline.html. The corresponding transgenic plants expressing 4xYFP, serving as cell type-specific marker lines, are named the “SAND” lines. All DNA constructs and transgenic lines have been deposited in the NASC stock center for rapid distribution of this resource to the community.
Nuclear cell type-specific marker lines
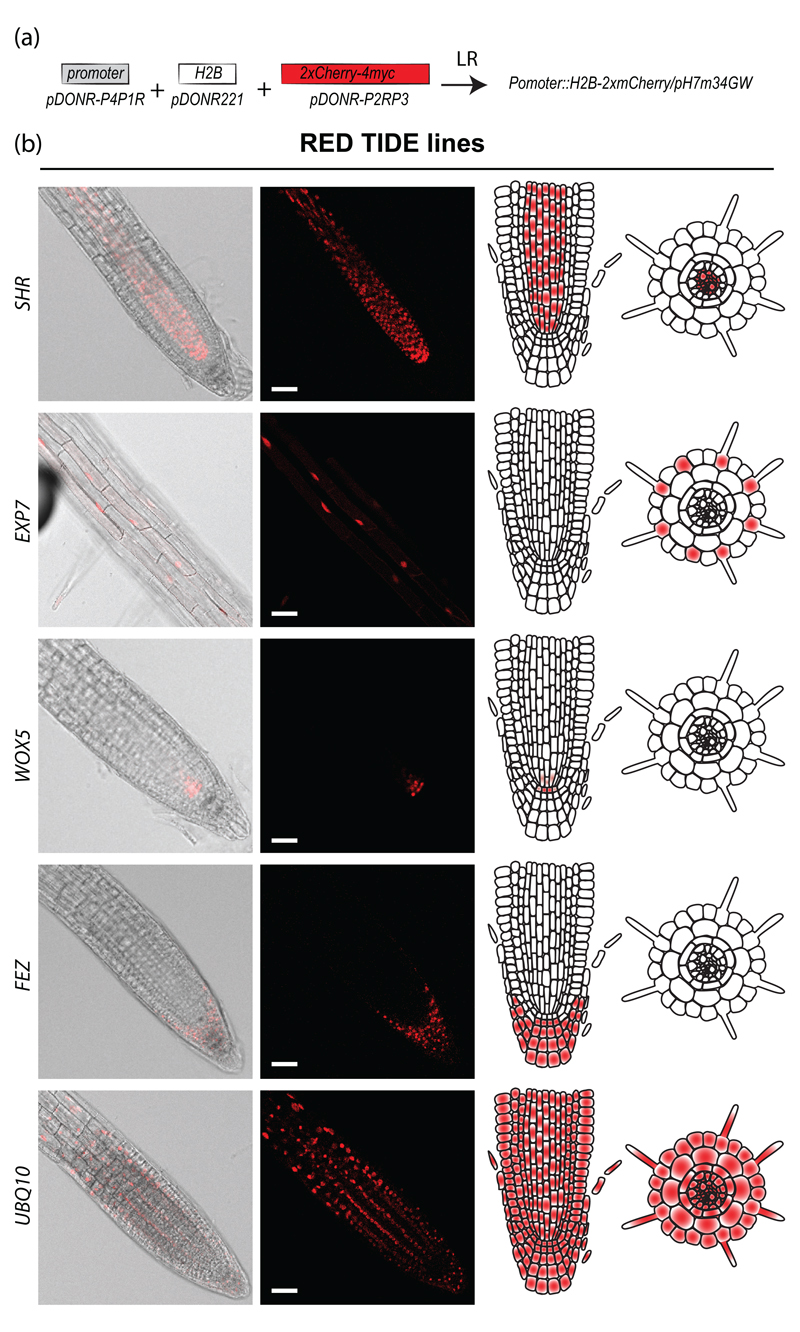
The 30 validated promoters were recombined with histone H2B fused to mCherry to generate cell type-specific marker lines expressing a nuclear red fluorescent reporter (Figure 1b and 3a). As previously mentioned, the six brightest primary transformants showing the expected expression profile were transferred to soil. Segregation analyses were performed at the T2 and T3 stages to isolate single insertion homozygous transgenic plants. Pictures were taken for each H2B-2xmCherry line (Figure 3b and S2). The expression patterns observed for the H2B-mCherry lines closely matched what was observed for the SAND lines. The H2B-2xmCherry lines may therefore serve as a nuclear cell type-specific marker line. They also offer a great alternative to DAPI staining, which is not cell type-specific and shows high toxicity in live imaging approaches and therefore may modify the processes being monitored. Alternatively, the cell type-specific H2B-2xmCherry lines can be used to purify nuclei from a cell type of interest by fluorescence-assisted cell sorting. These transgenic lines expressing a nuclear marker fused to mCherry in a cell type-specific fashion were named the “RED TIDE” lines.
Figure 3. Nuclei tagging with the RED TIDE reporter lines.
(a) Cloning strategy to generate the RED TIDE lines.
(b) Examples of RED-TIDE lines expression pattern for SHR∷H2B-2xmCherry; EXP7∷H2B-2xmCherry; WOX5∷H2B-2xmCherry, FEZ∷H2B-2xmCherry and UBQ10∷H2B-2xmCherry. Scale bars: 50 μm.
Cell type-specific purification of nuclei by INTACT
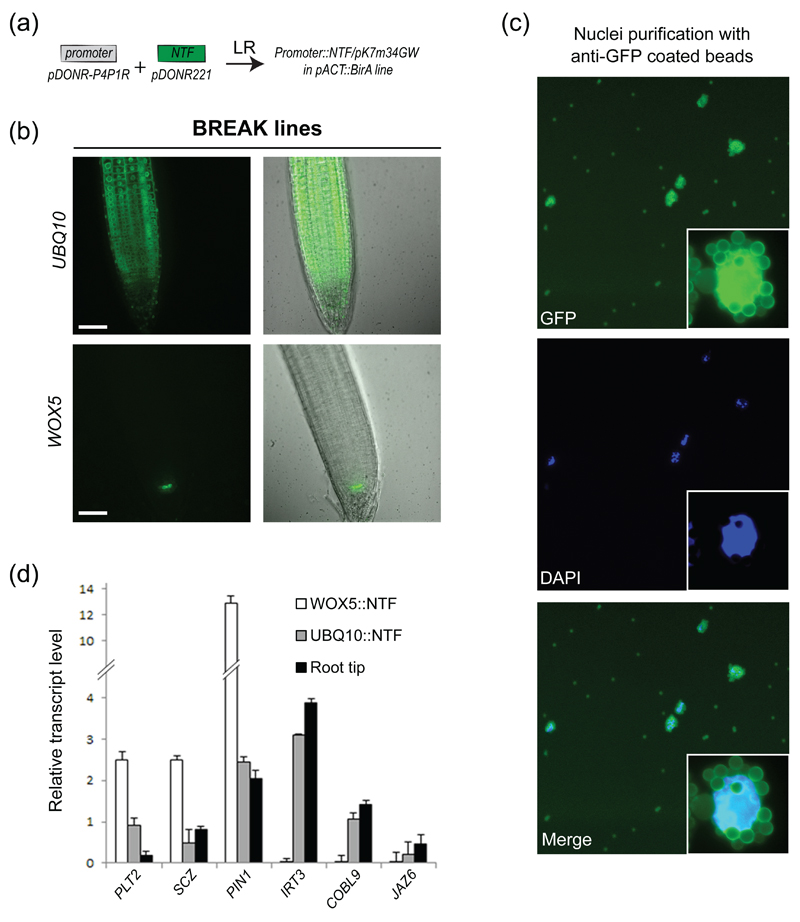
A method for studying gene expression and chromatin profiles in individual cell types was developed a few years ago and named INTACT (for Isolation of Nuclei TAgged in specific Cell Types) (Deal and Henikoff 2010). This method involves tagging nuclei in specific cell types and isolating these tagged nuclei using affinity purification. INTACT was used to study gene expression and chromatin marks in the two cell types of the Arabidopsis thaliana root epidermis (Deal and Henikoff 2010). To offer the possibility of generalizing the study of cell type-specific gene expression and chromatin modification associated, we recombined our high-confidence promoters to the INTACT cassette (NTF, Figure 1b and 4a), composed of WPP domain of Arabidopsis RanGAP1, which is necessary and sufficient for envelope association, GFP for visualization, and BLRP, which acts as a substrate for the E. coli biotin ligase BirA. The resulting binary vectors were transformed in Arabidopsis plants expressing constitutively the E. coli biotin ligase BirA (Deal and Henikoff 2010). The selection process followed exactly the same procedure as previously described for the SAND and RED TIDE lines. Importantly, expression profiles and the ability to purify nuclei were assessed. Globally, the expression of the INTACT cassette was driven in the expected root territories, as observed for the SAND and RED TIDE lines (Figure 4b and S3).
Figure 4. Nuclei purification using anti-GFP beads and a modified INTACT protocol.
(a) Cloning strategy to generate the BREAK lines.
(b) BREAK lines expression pattern for UBQ10∷NTF; WOX5∷NTF. Scale bars: 50 μm.
(c) Epifluorescence micrographs of affinity-captured nuclei. Note the autofluorescence of magnetic beads in the GFP channel. A higher magnification of a bead-bound nucleus is shown as an insert.
(d) RT-qPCR analyses of selected genes in INTACT-purified nuclei as well as root tips. Normalized transcript levels are given relative to AT5G60390 expression level arbitrarily set to 100. Error bars correspond to standard deviations from two biological replicates.
To capture nuclei expressing the biotinylated INTACT cassette, cell lysates were incubated with Streptavidin-coated magnetic beads and captured. We failed to recover bound nuclei for approximately 50% of the cell type-specific INTACT lines (Figure S3 and Table S2). We hypothesized that this could be due to poor biotinylation efficiency resulting from a weak expression of BirA in some cell types or from the silencing of the ACT2::BirA construct. To circumvent these limitations, we modified and streamlined the original INTACT protocol by capturing nuclei in batch rather than with a column, in a manner similar to what was recently proposed (Wang and Deal 2015), and by using magnetic beads coated with anti-GFP antibodies (Figure 4c, S3 and Table S2). This alternative, which has been reported in other systems (Henry et al. 2012, Ma and Weake 2014, Mo et al. 2015) renders optional the need of a two-component system that relies on the transgenic expression of the biotinylase. We found that this strategy offered a better yield since nuclei were purified in all transgenic lines except one (Figure S3 and Table S2), the latter likely because of very weak expression of the INTACT cassette.
As a proof of concept, we purified nuclei from the UBQ10 and WOX5 lines, expressing the NTF in most cell types or specifically in the quiescent center (QC), respectively. We determined transcript abundance of a selected set of genes expressed in (PLT2, At1g51190 ; SCZ, At1g46264 ; PIN1, At1g73590) or preferentially outside (IRT3, At1g60960 ; COBL9, At5g49270 ; JAZ6, At1g72450) the QC in these two cell populations as well as in the whole root tip (Brady et al. 2007, Nawy et al. 2005). Overall, transcript abundance was similar between the nuclear (UBQ10) and total (root tip) RNA samples irrespective of the expression level (Figure 4d). In addition, analysis of QC-expressed genes after nuclei purification clearly shows an enrichment for the WOX5:NTF line compared to UBQ10:NTF, illustrating the specificity and the sensitivity afforded by our lines and purification method. We named these cell type-specific INTACT transgenic lines the “BREAK” lines.
Cell type-specific induction of gene expression
The promoters characterized in this study offer the opportunity of expressing one’s favorite genes in a given cell type to deconstruct biological processes and to evaluate the contribution of different cell types. However, expressing genes in a highly controlled and timely fashion is often required to conclude on direct consequences of gene expression, especially for genes triggering detrimental phenotypes. To circumvent these issues, we engineered a single Gateway vector compatible with Multisite Gateway cloning for cell type-specific inducible gene expression (Figure 5a). This vector carries the glucocorticoid receptor-VP16-GAL4 DNA-binding (GVG) followed by a terminator and the 6xUASGAL4-CaM35Smini transcription units cloned into pDONR221 vector (P1-P2) (Figure 1). As a proof of concept, we generated a binary vector where the GVG cassette is under the control of the cell type-specific promoters and in which the 6xUASGAL4∷CaMV35Smini drives the expression of a fluorescent reporter (Figure 5a). Although the presence of two tandem mCitrine copies should greatly limit the diffusion of the fluorescent protein signal to neighboring cell types via plasmodesmata, we added the Nuclear Localization Signal (NLS) of the SV40 protein to sequester mCitrine in the nucleus (Vert and Chory 2006). We generated transgenic lines corresponding to the cell type-specific promoters UBQ10, EXP7, SHR and FEZ, and tested for dexamethasone-dependent induction of mCitrine or mCitrine-NLS. Plants treated with 30μM dexamethasone for 24h showed a clear induction of mCitrine expression compared to mock-treated plants (Figure 5b). In contrast to the widespread cell type distribution of mCitrine expression for UBQ10 promoter, the expression profiles of mCitrine-NLS appear restricted to the trichoblastic lineage, the vasculature and the columella for EXP7, SHR and FEZ, respectively (Figure 5b). This indicates that our dexamethasone-dependent induction of gene expression system occurred in a cell type-specific manner. The cell type-specific inducible transgenic lines were named the “LINE UP” lines.
Figure 5. Cell type-specific gene induction with the LINE UP reporter lines.
(a) Cloning strategy to generate the LINE UP lines.
(b) Images of the primary root for the LINE UP reporter lines driven by the UBQ10, EXP7, SHR and FEZ promoters, respectively, before and after induction with dexamethasone (DEX). Scale bars: 50 μm.
Deconstructing the cell type-specificity of root iron transport by IRT1
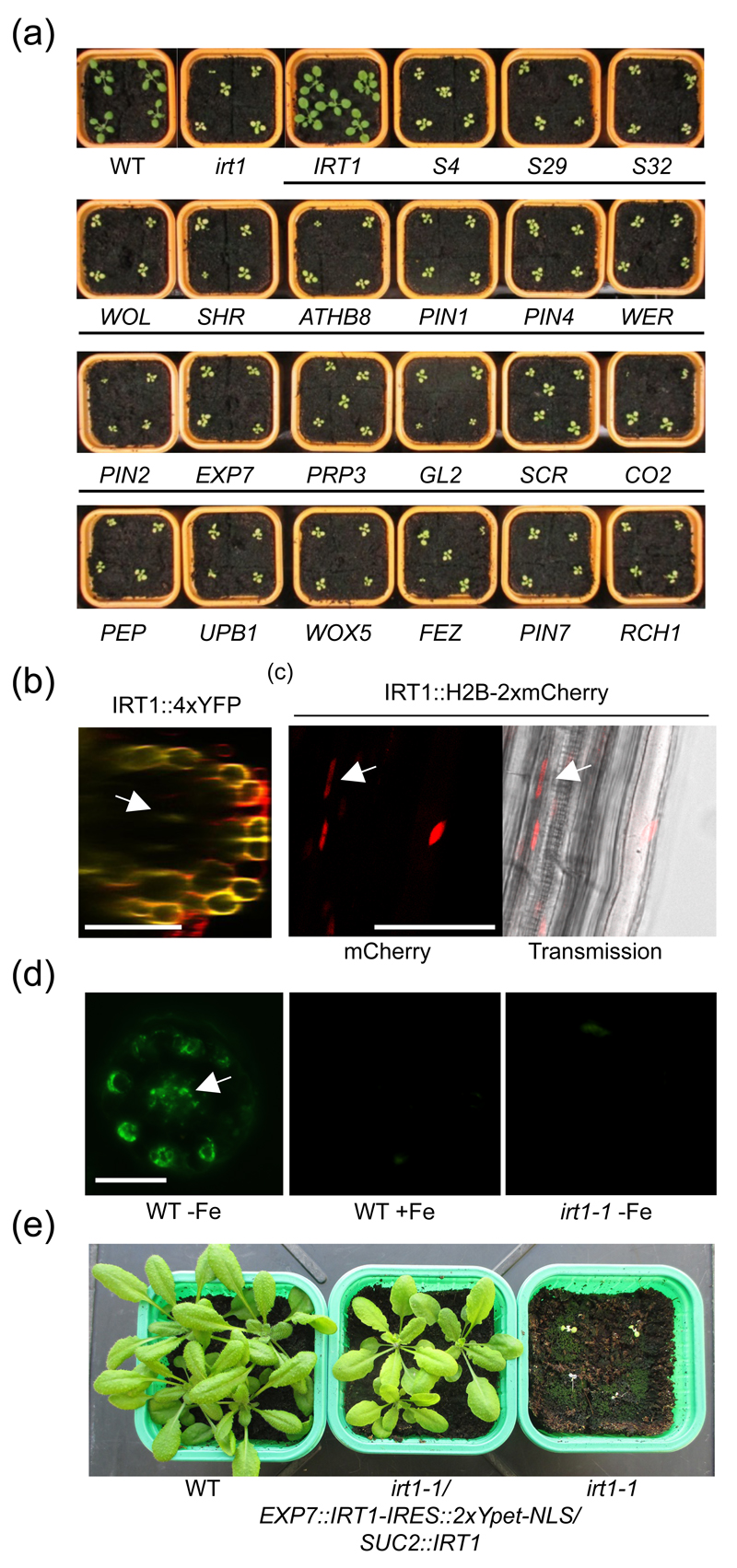
To demonstrate the usefulness of our cell type-specific SWELL promoter set, we sought to investigate further the contribution of root cell types to iron uptake from the soil. Nutrient diffusion from the soil to the vasculature can occur prior to deposition of the lignin-based casparian strip (Barberon and Geldner 2014). Once established, this diffusion barrier imposes nutrient uptake to occur in epidermal, cortical and the outer plasma membrane domain of endodermal cells, the latter being progressively insulated by deposition of suberin lamellae (Barberon and Geldner 2014). In Arabidopsis, iron is taken up by the root epidermis-expressed IRT1 transporter (Vert et al. 2002). An irt1-1 loss-of-function mutant for the IRT1 gene is severely chlorotic and dies early in development, unless massive amounts of iron are supplied to the shoots. To evaluate whether the absorption of iron from the soil could be achieved by other cell types than root epidermis, the irt1-1 mutant was transformed with the IRT1 gene under the control of representative SWELL promoters for different cell types. Because IRT1 protein cannot be tagged without impairing its transport activity, we took advantage of an internal ribosome entry site (IRES) (Ivanov et al. 1997). IRES allowed translation initiation of a nuclear-targeted fluorescent protein (2xYpet-NLS) in 3’ of the IRT1 mRNA, downstream of the IRT1 stop codon, thus creating a bicistronic transcript. The nuclear fluorescence 2xYpet-NLS therefore directly reflects the expression profile of IRT1 under the control of each promoter of interest. The ability of IRT1-IRES::2xYpet-NLS expressed under the control of our cell type-specific promoter collection to complement the chlorosis and restore the fertility of irt1-1 was used as a readout of iron uptake from the soil. Surprisingly, none of the SWELL promoters used led to complementation of irt1-1, except IRT1 promoter itself which drove expression of IRT1-IRES::2xYpet-NLS in the root epidermis (Figure 6a and S4). This first indicates that IRT1 protein expressed from the bicistronic transcript is functional and does not account for the overall lack of complementation of irt1-1. In addition, the lack of complementation resulting from expression driven by the root epidermis-specific WER, EXP7, PRP3 and GL2 promoters suggests that i) these different promoters do not induce sufficient expression of IRT1 or ii) IRT1 expression is not limited to the root periphery, in contrast to what has been previously reported (Vert et al. 2002), and expression in a different cell type is required for complementation of irt1-1. To decipher between these different scenarios, we reinvestigated the expression pattern of IRT1, taking advantage of the highly-sensitive IRT1 SAND line. To our surprise, we detected fluorescence in vascular tissues from older root zones, although to a lesser extent than what is observed in epidermal cells (Figure 6b). To facilitate the observation of IRT1 promoter activity in the vasculature, we used the IRT1 RED TIDE line, where the fluorescence of the H2B reporter is concentrated in nuclei. We observed a strong accumulation of H2B-2xmCherry in differentiated epidermal cells as well as in the vasculature, confirming that IRT1 expression is not restricted to root peripheral cell layer (Figure 6c). To ascertain that IRT1 is indeed expressed in the central cylinder, we performed immunolocalization using an anti-IRT1-specific antibody on root cross-sections. IRT1 protein was clearly detected in trichoblastic cells, as previously reported (Barberon et al. 2011), but was also observed in vascular tissues (Figure 6d). No signal was observed in iron-replete conditions, where IRT1 is not expressed, or in the irt1-1 mutant background, indicating that the signal observed in the central cylinder corresponds to IRT1 protein.
Figure 6. Spatial determinants of IRT1-dependent root iron uptake.
(a) Phenotype of 16 day-old wild-type (WT), irt1-1 and irt1-1 transformed with IRT1-IRES∷IRT1-2xYpet-NLS under the control of the indicated SWELL promoters.
(b) Confocal microscopy image of the IRT1 SAND line
(c) Confocal microscopy image of the IRT1 RED TIDE line
(d) Immunofluorescence analysis on root cross-section from iron-deficient (-Fe) and iron-sufficient (+Fe) wild-type (WT) plants and irt1-1, using anti-IRT1 antibodies.
(e) Phenotype of 4 week-old wild-type (WT), irt1-1 and irt1-1/EXP7∷IRT1-IRES∷IRT1-2xYpet-NLS/SUC2∷IRT1 plants grown on soil.
Scale bars: 50 μm.
To get a glimpse on where IRT1 is expressed in the vasculature, we analyzed the publicly available cell type-specific translatome datasets (Mustroph et al. 2009). In accordance with our observations that IRT1 is expressed in vascular tissues, IRT1 transcripts were strongly detected in phloem companion cells-specific translatome where the SUC2 promoter is active (Imlau et al. 1999). To examine whether the expression of IRT1 is required in both epidermal cells and phloem companion cells, we therefore transformed the irt1-1 mutant expressing the trichoblast-specific EXP7::IRT1-IRES::2xYpet-NLS with the phloem companion cell-specific SUC2::IRT1. The double transformants were selected and transferred to soil to evaluate the complementation of irt1-1 phenotypes. In contrast to lines expressing IRT1-IRES::2xYpet-NLS under the control of EXP7 or phloem-specific promoters alone that are severely chlorotic when grown in soil (Fig 6a), the double transgenic irt1-1/EXP7::IRT1-IRES::2xYpet-NLS/SUC2::IRT1 showed a clear restoration of the chlorosis defects and of the fertility (Figure 6e). These observations suggest that IRT1 is expressed in companion cells and that its activity in phloem cells is required for proper iron distribution in the plant. Altogether, the cell type-specific tools generated in this study provide unique tools to deconstruct complex biological processes and allowed to reveal additional functions for a theoretically well-characterized promoter.
Discussion
Multicellular organisms are composed of several specialized cell types that acquire particular fate and functions through a precisely established pattern of gene expression. Modern functional genomics therefore require thorough investigation of gene functions in a restricted number of cells to grasp their contribution in a biological process at stake in a whole organism. Here, we have generated a collection of cell type-specific or regulated promoters that are cloned in gateway compatible vectors. This set of tools allows to determine with great precision gene expression territories, to manipulate gene expression in a limited number of cells of interest, and to isolate biological information from relevant cell types.
As a proof of concept, we dissected further root iron uptake. Previous work demonstrated that the IRT1 is the major root iron transporter involved in iron uptake from the soil (Vert et al. 2002). This conclusion was drawn from the dramatic chlorosis displayed by the irt1-1 loss-of-function mutant. The restricted expression of IRT1 in root epidermal cells also indicates that iron is mostly taken up by the root epidermis from where it travels towards the vasculature. However, the ability of underlying cell types external to the casparian strip (i.e. cortex and outer polar domain of the endodermis) to transport iron remained unexplored. Driving IRT1 expression in different root cell types failed to functionally complement the chlorosis of irt1-1. More surprising is the fact that, among several root-epidermis specific promoters driving IRT1 expression, only IRT1 promoter allowed the complementation of irt1-1 growth defects. Several important conclusions can be drawn from such observations. First, IRT1-dependent iron uptake in cortical and endodermal cells appears not to be sufficient for complementation of irt1-1. Assuming that IRT1 protein is translated from the corresponding IRT1-IRES∷2xYpet-NLS in these cell types, the lack of complementation may reveal i) poor iron availability in the apoplasm surrounding cortical and the outer polar domain of endodermal cells due to the low mobility of iron from the soil, ii) the lack of reductase activity required for ferrous iron transport in the corresponding cell types, or iii) the necessity for interacting proteins required for IRT1 activity in inner cell types. The absence of reductase activity is unlikely since the Arabidopsis genome encodes many FRO2 ferric chelate reductase homologs that are expressed in the root. Moreover, mutants in cell-surface reductases such as frd1 display a relatively mild chlorosis compared to irt1 (Yi and Guerinot 1996), highlighting a somewhat minor role in iron uptake. IRT1 appears to be active in non-epidermal cells since 35S∷IRT1 plants expressing IRT1 close to endogenous levels in epidermal cells but also in other root cell types show increased iron accumulation and toxicity symptoms (Barberon et al. 2011). We therefore favor the hypothesis where iron availability is rather poor to sustain efficient IRT1-dependent iron uptake by cortical and endodermal cells. The second conclusion to be learned from the lack of complementation in irt1 plants expressing IRT1 in non-epidermal cells is that IRT1 expression territories are likely not restricted to the root epidermis. Careful re-examination of IRT1 expression pattern using the highly-sensitive IRT1 SAND and RED TIDE lines, and by immunolocalization revealed that IRT1 is also expressed in the root central cylinder. This observation parallels what has been shown for the direct IRT1 upstream regulator FIT that is also expressed in the vasculature (Jakoby et al. 2004). Interestingly, IRT1 expression in the root central cylinder, and more specifically in phloem companion cells, turns out to be critical for iron homeostasis. The simultaneous expression of IRT1 in trichoblast and phloem companion cells is indeed required for complementation of irt1 growth defects. The relevance for IRT1 expression in root phloem companion cells is still elusive, but we speculate that IRT1 may facilitate iron loading into root phloem for downward transport of iron or mediate the xylem-to-phloem transfer of iron, as previously suggested for the phloem companion cell-expressed NRT1.9 nitrate transporter (Wang and Tsay 2011). Further work will be necessary to elucidate the precise role of IRT1 in the phloem and further dissect how iron is distributed in the whole plant via IRT1-mediated transport.
Experimental procedures
Plant material and Constructs
Wild-type (Col0), irt1-1 (Vert et al. 2002), and the various transgenic plants generated in this study were grown at 21°C with 16 h light/8 h dark cycles.
Cell type-specific SWELL promoters were amplified from Col0 genomic DNA using the primers described in Table S3 and cloned into the Gateway pDONR-P4P1R vector (Life Technologies). UBQ10/pDONR-P4P1R; 2x35S/pDONR-P4P1R and mCitrine/pDONR-P3P2R; 2xmCherry-4xMyc/pDONR-P2RP3 and mCitrine-noSTOP/pDONR221 were previously described (Jaillais et al. 2011, Simon et al. 2014). The INTACT NTF cassette (gift from Roger Deal) was cloned into pDONR221 (see table S3 for primers). The myristoylation sequence and SV40 NLS sequence were added to mCitrine/pDONR221 and mCitrine/pDONR-P2RP3, respectively, using 5’phosphorylated primers followed by dpnI digestion and vector ligation. The GVG cassette (composed of GAL4 DNA binding domain, VP16 activation sequence from the Herpes virus and the GR rat glucocorticoid receptor), followed by the E9 terminator, the 6xUAS sequence and a minimal 35S promoter (35Smin) was synthesized with AttB1 and AttB2 sequences (Integrated DNA technologies), and recombined into pDONR221 to obtain GVG∷ter∷6xUAS/pDONR221. mCitrine was added to this vector using Gibson cloning (NEB) to generate GVG∷ter∷6xUAS∷mCitrine/pDONR221. 3xYpet was amplified from pBJ36 (gift from D.B. Long) and cloned into pDONR-P2RP3. IRES-2xYPet-NLS was synthesized by Integrated DNA technologies with AttB2r and AttB3 sequences and recombined into pDONR-P2RP3. IRT1 coding sequence was amplified by PCR and cloned in pDONR221. Final destination vectors were obtained by using three-fragment recombination system (Life Technologies), using the pB7m34GW/pH7m34GW/pK7m34GW destination vectors following the strategies described in Figure 2-6 (Karimi et al. 2007). A list of all the vectors generated and used in this work can be found in Table S3 along with their sequences, as well as at the following address: www.ens-lyon.fr/DRP/SICE/Swelline.html. Each SWELL promoter cloned into L4-L1R gateway compatible vectors (except the entry vectors for the following promoters: S4, S17, S18, S29, S32, IAA19, DR5, E47, UPB1 and Q12 that were provided by P. Benfey (Duke University, NC, USA)) were donated to the NASC (Set #N2106366), as well as the various tag used in this study cloned in their respective gateway compatible vectors (except the entry vectors for H2B that was provided by the F. Berger (GMI, Austria)).
The resulting constructs were transformed into wild-type plants (SAND, RED TIDE and LINE UP lines), birA-expressing plants (BREAK lines) or homozygous irt1-1 supplemented with iron. For all constructs, around 20 independent T1 lines were isolated and six representative mono-insertion lines were selected in T2. Independent lines homozygous for the transgene were selected in T3. A list of all the transgenic plants generated in this work can be found in Table S4.
The destination vectors corresponding to the SAND (NASC Set #N2106363), RED TIDE (NASC Set #N2106364), BREAK (NASC Set #N2106365) and LINE-UP collection as well as the corresponding transgenic lines (NASC SET #N2106367 for SAND lines, NASC SET #N2106368 for RED TIDE lines and NASC SET #N2106369 for BREAK lines) were also donated to the NASC (http://arabidopsis.info/CollectionInfo?id=156).
Confocal microscopy
Images of the SAND lines were performed on an inverted Zeiss LSM710 confocal microscope using a 40x Plan-apochromatic objective (numerical aperture 1.2, oil immersion). Dual-colour images were acquired by sequential line switching, allowing the separation of channels by both excitation and emission. Counter staining of plasma membranes were obtained by incubating roots with 1 μM FM4-64 solution (Lifesciences, stock solution at 3mM in H2O) for 10 minutes, followed by 15 to 45 minutes incubation in water prior to observation. mCitrine and FM4-64 were excited with a 515nm laser.
Images of the BREAK lines were performed on an inverted Zeiss microscope (AxioObserver Z1) equipped with a spinning disk module (CSU-W1-T3, Yokogawa) and a ProEM+ 1024B camera (Princeton Instrument) using a 40x C-Apochromat objective (numerical aperture 1.1, water immersion). GFP was excited with a 488nm laser.
Images of the RED TIDE and LINE UP lines, as well as irt1 complementation analyses were performed on an inverted Leica SP8 X microscope using a 40x HCX Plan-apochromatic objective (numerical aperture 1.3, oil immersion). Yellow fluorescent proteins (YFP, mCitrine and Ypet) and mCherry were excited with a white light laser at 514nm and 587 nm, respectively. Imaging of LINE UP lines was performed after induction with 30 μM dexamethasone (DEX) for 24 hours.
Quick assay to validate tagged-nuclei capture from BREAK lines
5-10 mm root tips from 7 day-old seedlings (50-300 plants, depending on the expected proportion of tagged nuclei) were chopped using a razor-blade in 1 mL of ice cold LB01 buffer (15 mM Tris, 2 mM Na2EDTA, 0.5 mM spermine tetrahydrochloride, 80 mM KCl, 20 mM NaCl, 0.1% Triton X-100, 15 mM b-mercaptoethanol, pH 7.5) supplemented with 4 μg/mL DAPI (Dolezel et al. 2007) and ground using a Dounce tissue homogenizer (Wheaton, USA). 1.5 μL of magnetic beads, Streptavidin-coated (M-280 Dynabeads, Life Technologies) or proteinA-G Dynabeads (Novex, Life Technologies) adsorbed with 0.1 μg anti-GFP antibodies (Abcam, ab290) were added to the nuclei suspension and incubated for 30 minutes at 4°C under gentle agitation. Bead-bound nuclei were then captured using a magnetic rack (Dynal, MPCS), washed three times with 1 mL of LB01 and resuspended in 20 μL of LB01 for epifluorescence imaging.
Nuclei purification from the BREAK lines
In vivo tagged-nuclei were purified from approximately 1 g of root tips using a modified version of the INTACT procedure described previously (Deal and Henikoff 2010). Root tips were hand-dissected from 7-d-old seedlings and ground in liquid nitrogen. All subsequent stages were performed at 4°C. The resulting fine powder was resuspended in 20 mL nuclei purification buffer (NPB: 20 mM MOPS, 40 mM NaCl, 90 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, 0.2 mM spermine, pH 7.0) supplemented with Complete protease inhibitors (Roche) and homogenized in a dounce tissue grinder (Wheaton, USA) using the loose and then tight pestle, in order to efficiently release the nuclei of all cell types. The obtained lysate was filtered through 70 and 40 μm nylon cell strainers (BD Falcon) and centrifuged for 5 min at 1000 g to pellet nuclei. Nuclei were resuspended in 1 mL NPB with 4 μg/mL DAPI, incubated on ice for 3 min, centrifuged for 5 min at 1000 g and resuspended in 1 mL NPB with 1% BSA. After estimating the number of nuclei using epifluorescence microscopy, nuclei were diluted in 7 mL NPB, 0.5% BSA, 0.1% Triton X-100 in a 15 mL Falcon tube and incubated for 30 min under gentle rotation with 1 μg of anti-GFP antibodies (Abcam, ab290) adsorbed on 10 mg of proteinA-G Dynabeads (Novex, Life Technologies). Bead-bound nuclei were selectively recovered using a Dynamag 15 magnet (Life Technologies) by carefully removing the supernatant. Bead-bound nuclei were gently resuspended in 10 mL NPB, 0.5% BSA, 0.1% Triton X-100 and magnetically re-captured. This wash step was repeated twice. Bead-bound nuclei were resuspended in 1 mL NPB, 0.5% BSA, 0.1% Triton X-100 and their purity and number was measured by epifluorescence microscopy.
Transcript analysis from INTACT-purified nuclei
Total RNA were isolated using the RNeasy Plus Micro Kit (Qiagen) and quantified using Ribogreen (Life Technologies) on a fluorospectrophotometer (ND3300, Thermo Scientific). 1 ng of total RNA was reverse transcribed and amplified using the Ovation RNA-Seq System V2 (NuGEN). Transcript abundance was measured by qPCR using amplified cDNA. The relative amount of each transcript was calculated with the 2-ΔCT method (Livak and Schmittgen 2001) using At5g60390 transcripts as endogenous control for data normalization. Each experiment was performed in at least two biological replicates. All primers used for RT-qPCR are listed in Table S3.
Immunolocalization
Immunolocalization on root cross-sections was performed using 7-d-old iron-deficient plants exactly as previously described (Barberon et al., 2011). Briefly, roots were fixed using paraformaldehyde (PFA), mounted in 3% (wt/vol) agarose, and cut with a Vibratome (Microm) in ~70-μm sections. Sections were blocked in BSA before incubation with anti-IRT1 antibodies. Sections were washed three times before incubation with the secondary (Alexa488–anti-rabbit goat antibody, MolecularProbes A11008; Invitrogen) and then tertiary (Alexa488–anti-goat donkey antibody, MolecularProbes A11055; Invitrogen) antibodies.
Supporting Information
Figure S1. Confocal imaging analyses of the SAND lines
Figure S2. Confocal imaging analyses of the RED TIDE lines
Figure S3. Confocal imaging analyses of the BREAK lines and nuclei purification validation
Figure S4. Expression pattern of irt1-1/IRT1∷IRT1-IRES∷2xYpet-NLS
Table S1. Promoter list, published expression patterns and references.
Table S2. Comparative nucleus purification analysis of INTACT lines using anti-strep and anti-GFP coated beads
Table S3. List of primers and entry gateway vectors used in this study
Table S4. List of transgenic lines generated in this study
Acknowledgments
We would like to thank Dr. P. Benfey (Duke University) for sharing previously published cell type-specific promoters, Dr. D.B. Long (UC Los Angeles) for the pBJ36-3xYpet vector, Dr. F. Berger (GMI Vienna) for H2BnoSTOP/pDONR-207, Dr. R. Deal (Emory University) for both the GFP(NTF) cassette and the ACT2∷birA transgenic line. The authors are indebted to Platim (UMS 3444 Biosciences Gerland-Lyon Sud) and the Imagerie-Gif imaging facility from I2BC for help with imaging, and J. Berger and A. Lacroix for plant care. Y.J. has received funding from the European Research Council - ERC Grant Agreement n. (3363360-APPL) and from the Marie Curie Action – CIG Grant Agreement (PCIG-GA-2011-303601) under the European Union's Seventh Framework Programme (FP/2007-2013). Work in the group of V. Colot was supported by the European Union Seventh Framework Programme Network of Excellence EpiGeneSys (HEALTH-F4-2010-257082), the CNRS and the Agence Nationale de la Recherche. A.K.M. is the recipient of a grant from the French Ministère de la Recherche et de l’Enseignement Supérieur. G.V. has received funding from Agence Nationale de la Recherche (ANR-13-JSV2-0004-01), and from the Marie Curie Action (PCIG12-GA-2012-334021) under the European Union's Seventh Framework Programme (FP/2007-2013).
Footnotes
Significance statement
In multicellular organisms, different cell types often perform discrete functions. Here we present a collection of benchmarked cell type-specific promoters for functional genomics of the Arabidopsis root, and use this resource as a test case to spatially deconstruct iron uptake.
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wisniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol. 2006;8:249–256. doi: 10.1038/ncb1369. [DOI] [PubMed] [Google Scholar]
- Barberon M, Geldner N. Radial transport of nutrients: the plant root as a polarized epithelium. Plant Physiol. 2014;166:528–537. doi: 10.1104/pp.114.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conejero G, Curie C, Friml J, Vert G. Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci U S A. 2011;108:E450–458. doi: 10.1073/pnas.1100659108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann BO, Vanneste S, Krouk G, Nawy T, Efroni I, Shani E, Choe G, Friml J, Bergmann DC, Estelle M, Birnbaum KD. A map of cell type-specific auxin responses. Mol Syst Biol. 2013;9:688. doi: 10.1038/msb.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C, Tierney ML. Expression of AtPRP3, a proline-rich structural cell wall protein from Arabidopsis, is regulated by cell-type-specific developmental pathways involved in root hair formation. Plant Physiol. 2000;122:705–714. doi: 10.1104/pp.122.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Bonke M, Thitamadee S, Mahonen AP, Hauser MT, Helariutta Y. APL regulates vascular tissue identity in Arabidopsis. Nature. 2003;426:181–186. doi: 10.1038/nature02100. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, Kang YH, Bernhardt C, Xia Y, Zheng X, Wang JY, Lee MM, Benfey P, Woolf PJ, et al. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 2012;8:e1002446. doi: 10.1371/journal.pgen.1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell. 2002;14:3237–3253. doi: 10.1105/tpc.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Dolezel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell. 2013;25:324–341. doi: 10.1105/tpc.112.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman Y, Elkouby L, Holland N, Vragovic K, Elbaum R, Savaldi-Goldstein S. Root growth is modulated by differential hormonal sensitivity in neighboring cells. Genes Dev. 2014;28:912–920. doi: 10.1101/gad.239335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benkova E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jurgens G, Palme K, et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell. 2002;108:661–673. doi: 10.1016/s0092-8674(02)00656-6. [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci U S A. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-Garcia S, Russinova E, Cano-Delgado AI. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development. 2011;138:849–859. doi: 10.1242/dev.057331. [DOI] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. Brassinosteroid perception in the epidermis controls root meristem size. Development. 2011;138:839–848. doi: 10.1242/dev.061804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 2004;18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry GL, Davis FP, Picard S, Eddy SR. Cell type-specific genomics of Drosophila neurons. Nucleic Acids Res. 2012;40:9691–9704. doi: 10.1093/nar/gks671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell. 1999;11:309–322. doi: 10.1105/tpc.11.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PA, Karpova OV, Skulachev MV, Tomashevskaya OL, Rodionova NP, Dorokhov Yu L, Atabekov JG. A tobamovirus genome that contains an internal ribosome entry site functional in vitro. Virology. 1997;232:32–43. doi: 10.1006/viro.1997.8525. [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN. Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell. 2011;21:770–782. doi: 10.1016/j.devcel.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Hothorn M, Belkhadir Y, Dabi T, Nimchuk ZL, Meyerowitz EM, Chory J. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25:232–237. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P. FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 2004;577:528–534. doi: 10.1016/j.febslet.2004.10.062. [DOI] [PubMed] [Google Scholar]
- Jones AR, Kramer EM, Knox K, Swarup R, Bennett MJ, Lazarus CM, Leyser HM, Grierson CS. Auxin transport through non-hair cells sustains root-hair development. Nat Cell Biol. 2009;11:78–84. doi: 10.1038/ncb1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P. Recombinational cloning with plant gateway vectors. Plant Physiol. 2007;145:1144–1154. doi: 10.1104/pp.107.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci U S A. 2006;103:6055–6060. doi: 10.1073/pnas.0510607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma J, Weake VM. Affinity-based isolation of tagged nuclei from Drosophila tissues for gene expression analysis. Journal of visualized experiments : JoVE. 2014 doi: 10.3791/51418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men S, Boutte Y, Ikeda Y, Li X, Palme K, Stierhof YD, Hartmann MA, Moritz T, Grebe M. Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nature cell biology. 2008;10:237–244. doi: 10.1038/ncb1686. [DOI] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, Eddy SR, et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:18843–18848. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17:1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Schauer MA, Megraw M, Breakfield NW, Thompson JW, Georgiev S, Soderblom EJ, Ohler U, Moseley MA, Grossniklaus U, Benfey PN. The protein expression landscape of the Arabidopsis root. Proc Natl Acad Sci U S A. 2012;109:6811–6818. doi: 10.1073/pnas.1202546109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Sieberer T, Seifert GJ, Hauser MT, Grisafi P, Fink GR, Luschnig C. Post-transcriptional control of the Arabidopsis auxin efflux carrier EIR1 requires AXR1. Curr Biol. 2000;10:1595–1598. doi: 10.1016/s0960-9822(00)00861-7. [DOI] [PubMed] [Google Scholar]
- Simaskova M, O'Brien JA, Khan M, Van Noorden G, Otvos K, Vieten A, De Clercq I, Van Haperen JM, Cuesta C, Hoyerova K, Vanneste S, et al. Cytokinin response factors regulate PIN-FORMED auxin transporters. Nat Commun. 2015;6:8717. doi: 10.1038/ncomms9717. [DOI] [PubMed] [Google Scholar]
- Simon MLA, Platre MP, Assil S, van Wijk R, Chen WY, Chory J, Dreux M, Munnik T, Jaillais Y. A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. The Plant Journal. 2014 doi: 10.1111/tpj.12358. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer PS. Gene traps: tools for plant development and genomics. Plant Cell. 2000;12:1007–1020. doi: 10.1105/tpc.12.7.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benkova E, Benjamins R, Beeckman T, Luschnig C, Friml J. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development. 2005;132:4521–4531. doi: 10.1242/dev.02027. [DOI] [PubMed] [Google Scholar]
- Vragovic K, Sela A, Friedlander-Shani L, Fridman Y, Hacham Y, Holland N, Bartom E, Mockler TC, Savaldi-Goldstein S. Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc Natl Acad Sci U S A. 2015;112:923–928. doi: 10.1073/pnas.1417947112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Deal RB. Epigenome profiling of specific plant cell types using a streamlined INTACT protocol and ChIP-seq. Methods Mol Biol. 2015;1284:3–25. doi: 10.1007/978-1-4939-2444-8_1. [DOI] [PubMed] [Google Scholar]
- Wang YY, Tsay YF. Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell. 2011;23:1945–1957. doi: 10.1105/tpc.111.083618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123:1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Willemsen V, Bauch M, Bennett T, Campilho A, Wolkenfelt H, Xu J, Haseloff J, Scheres B. The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev Cell. 2008;15:913–922. doi: 10.1016/j.devcel.2008.09.019. [DOI] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 1996;10:835–844. doi: 10.1046/j.1365-313x.1996.10050835.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.