We report the first comprehensive genomic analysis of a case of mixed conventional and fibrolamellar HCC (mFL-HCC). This study confirms the expression of DNAJB1:PRKACA, a fusion previously associated with pure FL-HCC but not conventional HCC, in mFL-HCC. These results indicate the DNAJB1:PRKACA fusion has diagnostic utility for both pure and mixed FL-HCC.
Keywords: mixed fibrolamellar hepatocellular carcinoma, genome analysis, fusion transcript, DNAJB1:PRKACA
Abstract
Background
Mixed fibrolamellar hepatocellular carcinoma (mFL-HCC) is a rare liver tumor defined by the presence of both pure FL-HCC and conventional HCC components, represents up to 25% of cases of FL-HCC, and has been associated with worse prognosis. Recent genomic characterization of pure FL-HCC identified a highly recurrent transcript fusion (DNAJB1:PRKACA) not found in conventional HCC.
Patients and Methods
We performed exome and transcriptome sequencing of a case of mFL-HCC. A novel BAC-capture approach was developed to identify a 400 kb deletion as the underlying genomic mechanism for a DNAJB1:PRKACA fusion in this case. A sensitive Nanostring Elements assay was used to screen for this transcript fusion in a second case of mFL-HCC, 112 additional HCC samples and 44 adjacent non-tumor liver samples.
Results
We report the first comprehensive genomic analysis of a case of mFL-HCC. No common HCC-associated mutations were identified. The very low mutation rate of this case, large number of mostly single-copy, long-range copy number variants, and high expression of ERBB2 were more consistent with previous reports of pure FL-HCC than conventional HCC. In particular, the DNAJB1:PRKACA fusion transcript specifically associated with pure FL-HCC was detected at very high expression levels. Subsequent analysis revealed the presence of this fusion in all primary and metastatic samples, including those with mixed or conventional HCC pathology. A second case of mFL-HCC confirmed our finding that the fusion was detectable in conventional components. An expanded screen identified a third case of fusion-positive HCC, which upon review, also had both conventional and fibrolamellar features. This screen confirmed the absence of the fusion in all conventional HCC and adjacent non-tumor liver samples.
Conclusion
These results indicate that mFL-HCC is similar to pure FL-HCC at the genomic level and the DNAJB1:PRKACA fusion can be used as a diagnostic tool for both pure and mFL-HCC.
introduction
patient history
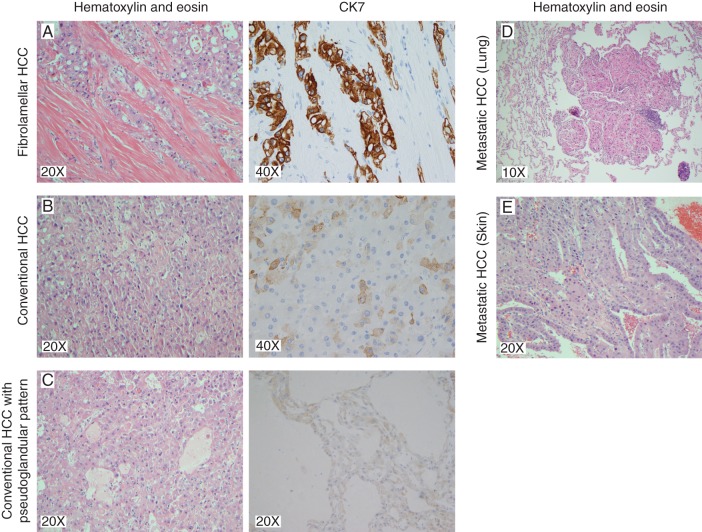
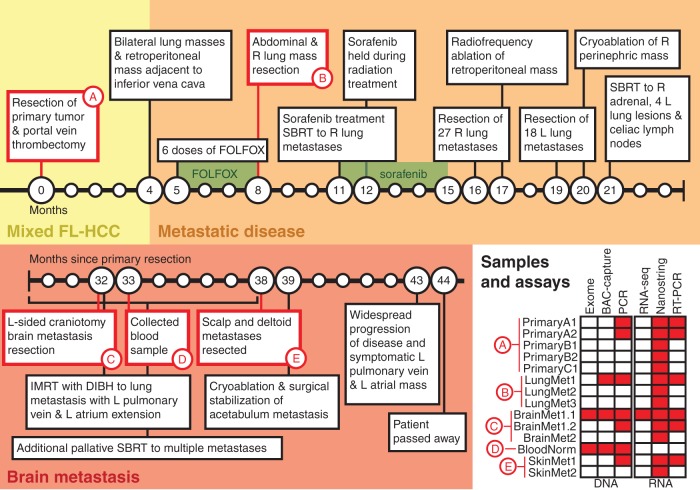
A woman in her 20s was diagnosed with mixed fibrolamellar and well-differentiated multifocal hepatocellular carcinoma (mFL-HCC) (Figure 1). She had no underlying liver disease, known cancer risk factors, or evidence of metastatic disease on imaging. A complete timeline is provided in Figure 2 and more detailed case presentation in supplementary Information, available at Annals of Oncology online. She underwent a right hepatic trisegmentectomy and portal vein thrombectomy for a pT4bN0M0 tumor with three large tumor nodules (supplementary Figure S1, available at Annals of Oncology online). Pathologic examination revealed that the largest nodule showed typical histologic features of FL-HCC (PrimaryA1, A2), the second largest showed only conventional HCC (PrimaryB1, B2) and the smallest showed a mix of FL-HCC and conventional HCC (PrimaryC1) (Figure 1A–C; supplementary Figure S2, available at Annals of Oncology online). Four months after surgery, the patient developed metastatic disease and received chemotherapy. At 8 months, surgical resection of metastatic nodules in the abdomen and lungs (LungMet1-3; Figure 1D) was performed, both of which had characteristics of FL-HCC on pathologic evaluation. Three months later she began a 4-month course of sorafenib for recurrent disease suspended during stereotactic body radiotherapy (SBRT) to metastatic lung lesions (supplementary Figure S3, available at Annals of Oncology online). She continued to have progressive disease on imaging and, over the next 5 months, she underwent surgical and locoregional therapy for recurrent disease in her torso. Subsequently, she did well until about 32 months when she developed a pulmonary metastasis and a solitary left temporal lobe brain metastasis (BrainMet1.1, 1.2, 2) that was surgically resected followed by SBRT. SBRT was also used for palliation of multiple recurrences. At month 38, symptomatic scalp (SkinMet1–2; Figure 1E) and left deltoid masses were resected followed by treatment of a left acetabulum lesion. Shortly before entering hospice, the patient developed symptomatic progression and increased tumor burden at multiple sites. For the last 28 months of her life, the patient declined all systemic therapies other than alternative ones. At no time did she develop recurrent liver lesions. She died about 44 months after the initial surgery and diagnosis.
Figure 1.
Tumor pathology images showing mixed FL-HCC characteristics. The primary tumor resection included three tumor nodules exhibiting pathology consistent with fibrolamellar HCC, conventional HCC and mixed fibrolamellar and conventional HCC. Representative images of H&E and immunohistochemical staining for CK7 are shown for each nodule. (A) Fibrolamellar HCC with diffusely positive CK7 staining in the tumor cells. (B) Conventional HCC with solid patterns primarily negative for CK7 staining with rare positive cells. (C) A region of the mixed nodule displaying conventional HCC with solid and pseudoglandular patterns negative for CK7 staining. Metastatic tissue exhibiting features of fibrolamellar HCC from (D) lung and (E) skin, resected 8 and 38 months after the primary tumor, respectively.
Figure 2.
A timeline depicts the clinical course of the patient in the months following resection of the primary mixed fibrolamellar hepatocellular carcinoma. Samples collected during the course of treatment and used for genomic analysis are identified. The table in the bottom right indicates specific assays used on samples from each time point. Additional sample details can be found in supplementary Table S1, available at Annals of Oncology online. FOLFOX, 5-fluorouracil, leucovorin, and oxaliplatin; R, right; L, left; SBRT, stereotactic body radiotherapy (Cyberknife™); IMRT, intensity modulated radiation therapy; DIBH, deep inspiration breath hold.
Approximately 30 months into her cancer course, the patient consented to genomic analyses to attempt to identify specific targeted gene therapies. The BrainMet1 sample was chosen for initial profiling and blood (BloodNorm) was collected for normal comparator in genomic analyses. Targeted validation was performed on additional RNA and DNA samples from the primary tumor and additional metastases (Figure 1; supplementary Table S1 and Methods, available at Annals of Oncology online). Ultimately, the patient declined additional treatments suggested by this analysis.
mixed and pure FL-HCC versus conventional HCC
FL-HCC is a rare liver tumor (0.02/100 000 age adjusted incidence; ∼1% of HCC cases) [1] typically affecting adolescents and young adults with no history of primary liver disease or cirrhosis [2]. mFL-HCC is defined by the presence of features of both conventional HCC and some but not all of the criteria for pure FL-HCC [3] as initially defined by Edmondson [4]. Patients with mFL-HCC may represent up to 25% of cases of FL-HCC, tend to be older than pure FL-HCC (but still younger than conventional HCC), and have been associated with worse prognosis [3]. Recent studies have also revealed distinct transcriptional signatures for pure and mixed FL-HCC [5]. Specifically, pure FL-HCC was characterized by overexpression of ERBB2, genes involved in glycolysis, and several neuroendocrine genes compared with conventional HCC or mFL-HCC. As such, mFL-HCC may represent a separate entity, or an intermediate entity on a continuum, from pure FL-HCC to conventional HCC.
Recent surveys of the genomic landscape of both conventional HCC (predominantly hepatitis-associated) and pure FL-HCC have differentiated the genomic profiles of these two subtypes. Conventional HCC is characterized by recurrent point mutations (including in TP53, CTNNB1, and TERT), HBV integration sites, structural variants, and copy number variants (CNVs) affecting chromatin regulators and the TERT promoter, among others [6–8]. Overall, FL-HCCs show fewer large-scale chromosomal alterations and less frequent methylation of tumor suppressor promoters with methylation changes primarily near liver developmental genes when compared with conventional HCC [9–11]. Most notably, a novel DNAJB1:PRKACA fusion has been identified in association with pure FL-HCC but not in conventional HCC [9, 11–13]. Although methylation and RNA expression profiling has been evaluated in a very small number of mFL-HCC cases, no comprehensive genomic profiles of mFL-HCC have been published.
results
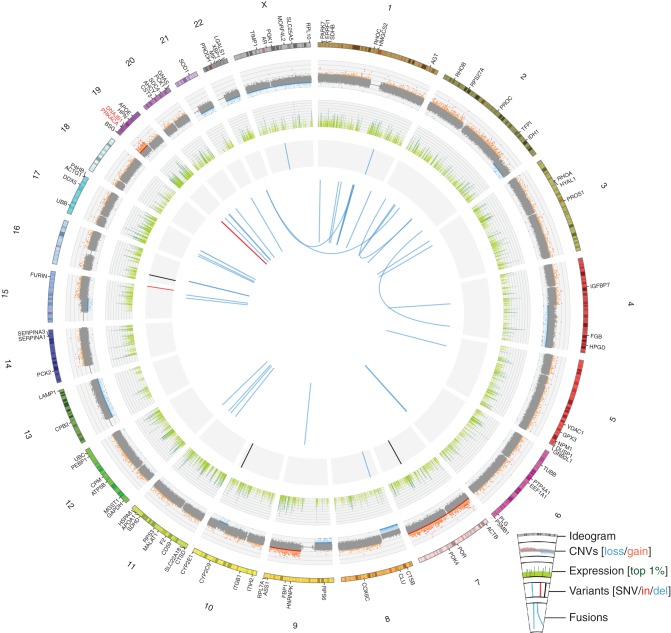
Exome sequencing with custom capture probes also targeting HBV, HCV and the TERT promoter region (supplementary Table S2, available at Annals of Oncology online) produced coverage of 99.1% of targeted regions at a minimum 20× depth (mean 347×) for BrainMet1.1 DNA and 96.1% at 20× (mean 114×) for BloodNorm DNA. Figure 3 depicts the key findings of exome and transcriptome sequencing. No evidence of HCV or HBV expression or integration, or TERT promoter mutations was observed (supplementary Figure S4, available at Annals of Oncology online). Similar to previous reports for pure FL-HCC, a very low somatic mutation rate was observed compared with the median of 45 nonsynonymous exome mutations (∼2 mutations/Mb) previously reported for conventional HCC [13, 14]. In fact, only two somatic exome SNVs (TACC2, LOC81691) and zero indels were unambiguously somatic variants. None of the previously reported, recurrently mutated genes in HCC were mutated in this case, nor were the few mutations observed obvious cancer-driving genes.
Figure 3.
A Circos plot displays genomic and transcriptomic alterations observed in the BrainMet1.1 sample from the index case of mixed fibrolamellar HCC. For the outermost ring, a chromosome ideogram is shown with selected genes labeled. The first data track displays copy number variants (CNVs) from genotype array data with tumor-normal logR ratios plotted and segments called by cnv-hmm (unpublished software). Copy-altered segments are indicated as dark red (gain) or dark blue (loss). The next data track plots RNA-seq gene-level fragments per kilobase of transcript per million mapped reads (FPKM) values on a log2 scale in light green with the top 1% most highly expressed genes indicated in dark green. Next, single-nucleotide variants (black), small insertions (red), and small deletions (blue) from exome data are indicated. These include two somatic and five ambiguous variants (supplementary Results, available at Annals of Oncology online). Finally, the centermost ring plots RNA-seq fusions (ChimeraScan) in light blue with the intrachromosomal fusion between DNAJB1 and PRKACA highlighted in red.
A high number of primarily single-copy, long-range CNVs were observed (Figure 3), including previously described, recurrent deletions affecting chr 4q (q22.3–q35.2) and 22 as well as amplification of 19p [9]. RNA-sequencing data analysis identified a large number of highly expressed cancer genes (top 2%) including ERBB2 and EGFR. Immunohistochemistry determined that the tumor was androgen receptor negative, ERBB2 score of 2+ and EGFR (membranous staining) intermediate at 2+. Note that ERBB2 was highly expressed in this case as recently reported for pure FL-HCC but not for other mixed FL-HCC [5].
Only three fusion transcripts were predicted by both ChimeraScan and Integrate: DNAJB1:PRKACA, TTC33:PRKAA1, and BCAS3:CLTC (supplementary Table S3A and B and Results, available at Annals of Oncology online). By far the most highly expressed fusion was DNAJB1:PRKACA (supplementary Appendix 1, available at Annals of Oncology online), the product of combining exon1 of DNAJB1 to exon2 of PRKACA in a head-to-tail fashion (supplementary Figures S5 and S6, available at Annals of Oncology online). The fusion appears to be upregulated and expressed in an allele-specific manner (supplementary Results, available at Annals of Oncology online). RT-PCR followed by sequencing of the amplified RNA products was used to validate the predicted DNAJB1:PRKACA fusion (supplementary Table S4, Figures S7–S9 and Methods and Results, available at Annals of Oncology online). No evidence for the DNAJB1:PRKACA fusion was identified in RNA-seq data from 30 conventional HCC cases or 30 adjacent nontumor liver tissues.
To determine whether this fusion was unique to the fibrolamellar components of this tumor, we extended our analysis to additional samples from this individual (Figure 2, samples and assays; supplementary Table S1, available at Annals of Oncology online). This included bulk formalin-fixed paraffin-embedded samples from each nodule of the primary tumor with conventional, mixed and fibrolamellar HCC pathology and metastatic sites (brain, lung, skin) with FL-HCC pathology. The presence of the DNAJB1:PRKACA fusion was assayed using custom Nanostring probes (nCounter Elements™ TagSets) specific to the fusion (supplementary Table S5, available at Annals of Oncology online). All primary tumor samples, regardless of pathology, tested positive for the fusion. Of the metastatic sites, only a single sample (LungMet1) was negative for the fusion. This same RNA sample resulted in RT-PCR bands of unexpected size but upon sequencing showed evidence of the fusion. Two additional RNA samples from this same metastatic site (LungMet2-3) were evaluated and tested positive for the fusion (supplementary Figure S10, available at Annals of Oncology online). We repeated this experiment in a second individual (Case2) with mFL-HCC that presented with fibrolamellar HCC and later developed a primary liver recurrence and lymph node metastasis with conventional HCC pathology (supplementary Information and Figure S11, available at Annals of Oncology online). The fusion was readily detected in the lymph node metastasis (Case2_LymphMet) with conventional HCC pathology but not in the primary (Case2_Primary1-2) or liver recurrence (Case2_LiverRec) samples. As no other conventional HCC samples have tested positive for the fusion [9, 12, 13], the most likely explanation is that the fusion was present in the fibrolamellar primary and retained in the metastasis but that low available RNA quantities and poor sample quality from archival tissue prevented detection. An additional 112 HCC samples and 44 adjacent matched normal tissues were also analyzed for the presence of the fusion (supplementary Methods and Results, Figure S10, available at Annals of Oncology online). A single HCC sample, but no normal samples, tested positive for presence of the fusion. Evaluation of the pathology reports from this sample indicated this tumor had regions of both fibrolamellar and conventional HCC consistent with mFL-HCC (Case3). No additional samples (N = 111) had descriptions consistent with mixed or pure FL-HCC upon review.
We used the comprehensive data available for the index patient to further characterize the nature of the genomic alteration leading to the DNAJB1:PRKACA fusion. Analysis of genotyping array data (BrainMet1.1) for the genomic region between DNAJB1 and PRKACA revealed a focal-deletion of ∼400 kb on chr19 (supplementary Figure S12, available at Annals of Oncology online). The exact genomic breakpoints responsible for the DNAJB1:PRKACA fusion were determined by a novel custom capture-based sequencing approach. Three BAC clones spanning the region from DNAJB1 to PRKACA were identified (supplementary Table S6, Figure S13 and Methods, available at Annals of Oncology online), used to create a BAC-clone capture reagent, and then captured libraries were sequenced either on Illumina or Pacific Biosciences instruments (supplementary Figures S14–S18, Tables S7–S9, and Appendix 2, available at Annals of Oncology online). Both methods supported a deletion of 406 272 bases between the first intron of DNAJB1 and first intron of PRKACA (chr19: 14 221 615–14 627 887, hg19) in the BrainMet1.1 sample. Support for the fusion was also observed for the LungMet1 sample (Illumina) but not for the BloodNorm sample (either platform). PCR validation yielded products of the expected size in all primary and metastatic (but not normal) samples from the patient (supplementary Tables S10 and S11 and Figure S19, available at Annals of Oncology online). Analysis of variant allele fractions in this region also revealed an expected loss of heterozygosity event within the deletion breakpoints (supplementary Figure S20, available at Annals of Oncology online).
As mentioned above, there was also evidence of amplification of the entire chr 19p arm (Figure 3). It is difficult to know for certain whether the focal-deletion occurred first and then was subsequently amplified. However, this would be parsimonious with an evolutionary process promoting further activation of PRKACA. Recurrent 19p amplifications have been previously reported in small comparative genomic hybridization studies of pure FL-HCC [10]. More recently, Graham et al. observed aneuploidy affecting the PRKACA locus, both involving the fusion and the wild-type allele, in a large percentage of cells from pure FL-HCC patients using fluorescence in situ hybridization (FISH) probes [12]. Taken together, the likely explanation for upregulation of PRKACA is allele-specific expression driven by the DNAJB1 promoter-swap and/or amplification of the fusion allele.
Analysis of microarray data from Malouf et al. [5] for 39 HCCs (17 pure FL-HCC, 5 mixed, 7 noncirrhotic HCC, 10 adjacent livers) also demonstrated significant upregulation of PRKACA in pure FL compared with conventional HCC (216234_s_at: P = 0.009; 202801_at: P = 2.31e−05) and normal liver tissues (216234_s_at: P = 0.003; 202801_at: P = 2.3e−05; supplementary Figure S21, available at Annals of Oncology online). mFL-HCC expression levels for PRKACA were intermediate and significantly higher than conventional HCC for one probe set (202801_at: P = 0.030) but not otherwise significantly different from other disease types. DNAJB1 was significantly down-regulated in pure FL compared with nontumor liver tissues (200664_s_at: P = 0.005; 200666_s_at: P = 0.011; supplementary Figure S22, available at Annals of Oncology online). In general, DNAJB1 showed the inverse pattern of PRKACA with the lowest DNAJB1 expression in pure FL-HCC, followed by intermediate levels in mixed, and then progressively higher levels in conventional and normal samples. Normal RNA-seq data were not obtained from this mFL-HCC case for direct comparison; however, the level of PRKACA expression in the mFL-HCC was higher (3.97 standard deviations or 4.11-fold) and a statistical outlier when compared with data from 30 conventional HCC samples (Grubb's test, P = 3.56e−05) (supplementary Figure S23, available at Annals of Oncology online). DNAJB1 expression in the mFL-HCC was comparable with the mean of the conventional HCC cases.
discussion
We present here the first comprehensive exome and transcriptome analysis of a mixed fibrolamellar HCC (mFL-HCC). We observed a remarkably low mutation rate with no strong cancer-driving SNV or indel candidates consistent with previous reports of pure FL-HCC [9, 13]. However, RNA-seq analysis identified high expression of many cancer-relevant and potentially druggable genes and the DNAJB1:PRKACA fusion. Shortly after completion of our initial analysis, Honeyman et al. [13] reported an identical chimeric RNA transcript in a cohort of pure FL-HCC patients. Subsequent reports have independently validated the high recurrence of this fusion in pure FL, but not conventional, HCC [9, 11, 12, 15]. Consistent with our observations, Honeyman et al. reported the PRKACA transcript expression was significantly upregulated in relation to normal tissue [13]. Identification of this fusion in mFL-HCC is important to our understanding of the etiology of this HCC subtype.
A small body of literature suggests that mFL-HCC is a subtype of disease clinically distinct from conventional HCC and pure FL-HCC. However, in this case, it appears to share the recently discovered and most important genetic feature of FL-HCC reported to date and thus could be grouped together with FL-HCC as a candidate for PRKACA targeted therapy (if one can be developed). FL-HCC and by extension mFL-HCC diagnosis remains challenging [5]. This is especially true for cases in which the disease occurs at an atypical age or in the context of underlying cirrhotic liver and is magnified by tumor heterogeneity and poor reproducibility of pathologic examination of primary liver tumors with fibrous stroma [16]. The studies referenced show that, while FL-HCC had better interobserver reproducibility than other types of HCC there was still substantial disagreement with a real possibility of both false-positive and false-negative diagnoses. A highly specific and easily detected event such as the DNAJB1:PRKACA fusion could represent a much more accurate diagnostic tool.
Recent studies evaluating the presence of the DNAJB1:PRKACA fusion in pure FL-HCC have reported detection in 79%–100% of samples [9, 11, 12]. One group also evaluated this fusion in five cases of mFL-HCC but failed to identify this fusion in any sample [11]. We not only identified the fusion in all portions of the primary tumor, regardless of pathology, but in all subsequent metastases studied in our initial mFL-HCC patient. Expansion of this analysis to an additional patient with temporally separated presentations of FL and conventional HCC, confirmed this finding. Finally, screening a large cohort of HCC with a Nanostring assay identified a third case with mixed fibrolamellar characteristics. Graham et al. reported the use of a FISH assay to identify the fusion in FL-HCC samples that failed in their initial round of RT-PCR detection. Together these studies indicate that more sensitive methods should be considered when evaluating the presence of this fusion in both mixed and pure FL-HCC [12]. Although more mFL-HCC samples should be evaluated for this fusion, we present evidence that it can be detected in at least a subset of these very rare cases.
The increased kinase activity observed as a result of the recurrent DNAJB1:PRKACA fusion presents the obvious strategy of PRKACA inhibition as a potential treatment modality for both mixed and pure FL-HCC [13]. A search for PRKACA-specific inhibitors using the Drug-Gene Interaction Database (DGIdb) [17] identified a small number of potential compounds; however, they generally lack potency or specificity (supplementary Table S12A–E and Results, available at Annals of Oncology online) [18]. Given the finding in this and other recent studies of the likely importance of PRKACA activation in mixed and pure FL-HCC, renewed efforts are warranted for the development of inhibitors specific to PRKACA or perhaps the DNAJB1:PRKACA fusion protein itself. Lacking specific inhibitors for PRKACA, we considered other targets, potentially upregulated by PRKACA activation by comparing expression levels in pure FL versus conventional HCC and nontumor tissues as described above. We identified five potentially druggable targets, including ERBB2, one of the most highly expressed genes in the mFL-HCC of this study (supplementary Results, Figure S24, and Table S13, available at Annals of Oncology online). Drugs such as lapatinib, which targets ERBB2, have good phase III safety and efficacy data in breast cancer. Lapatinib was investigated in a phase II clinical trial in HCC and, although well tolerated, was discontinued due to lack of efficacy in advanced patients [19]. However, the status of ERBB2 overexpression was not evaluated in that study and to our knowledge ERBB2 inhibitors have not been systematically evaluated in FL-HCC, the subtype most associated with overexpression. In the index case for this study, the patient favored palliative locoregional therapy and chose to not pursue systemic therapy with limited data. This is likely to be a common outcome of such genome-driven approaches. In many cases, even highly relevant events such as the DNAJB1:PRKACA fusion are not actionable, and other genomic events are not sufficiently compelling to alter treatment decisions.
conclusion
This study extends the recent discovery of DNAJB1:PRKACA fusions in pure fibrolamellar HCC to mixed FL-HCC. It seems likely that this highly recurrent and unique genomic and transcriptomic event may be a useful diagnostic tool for distinguishing pure and mixed FL-HCC from conventional HCC even when fibrolamellar characteristics are absent at pathology. We also add to evidence that FL-HCC may be driven by structural variants and fusion transcripts far more than SNVs/Indels. We present a novel strategy for identifying DNA breakpoints in the absence of whole-genome sequence data by using BAC-clone DNA to create custom hybrid-capture reagents to enrich for and sequence relatively large genomic regions. This approach represents a convenient strategy for discovery and validation of structural variations in small sample sets for large genomic regions that are challenging for PCR validation but would not be cost-effective for conventional capture-based approaches using commercial probes. Similarly, the Nanostring Elements technology represents a convenient and rapid approach for sensitive detection of fusions in large sample sets. While the DNAJB1:PRKACA fusion certainly stood out as interesting in our initial analysis, it was not deemed clinically actionable. Its likely importance as a driving event was only illuminated upon publication of the Honeyman et al. analysis of 11 FL-HCCs. This illustrates a key challenge of personalized medicine in that individual tumor genomes must be interpreted in the context of a constantly accumulating literature. Finally, this case presents a reminder of how far targeted therapeutics still have to go. PRKACA seems a clear candidate for drug development. However, clinically useful inhibitors remain elusive. Efforts should be redoubled given the new significance of this kinase in both mixed and pure fibrolamellar HCC.
funding
MG was supported by the National Human Genome Research Institute of the National Institutes of Health (NIH) under award number K99HG007940. OLG was supported by the National Cancer Institute of the NIH under award number K22CA188163.
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
acknowledgements
We gratefully acknowledge Kay Washington for her assistance in the identification, coordination and pathologic imaging of Case2; Jason Waligorski, Tracie Miner, Chris Markovic, Matt Cordes, Catrina Fronick, Shelly O'Laughlin and Robert Fulton for assistance with data production; Adam Coffman, Thomas Mooney, Eddie Belter, Jim Weible, Anthony Brummet, Mark Burnett and Ian Ferguson for technical assistance with automated analysis pipelines; Stephanie Jackson and Elizabeth Applebaum for sample management support; and Richard K. Wilson for his overall mentorship and support. We would also acknowledge the care provided by the physicians and surgeons in Boston, St Louis, Nashville, and Europe.
references
- 1.El-Serag HB, Davila JA. Is fibrolamellar carcinoma different from hepatocellular carcinoma? A US population-based study. Hepatology 2004; 39: 798–803. [DOI] [PubMed] [Google Scholar]
- 2.Torbenson M. Review of the clinicopathologic features of fibrolamellar carcinoma. Adv Anat Pathol 2007; 14: 217–223. [DOI] [PubMed] [Google Scholar]
- 3.Malouf GG, Brugieres L, Le Deley MC et al. . Pure and mixed fibrolamellar hepatocellular carcinomas differ in natural history and prognosis after complete surgical resection. Cancer 2012; 118: 4981–4990. [DOI] [PubMed] [Google Scholar]
- 4.Edmondson HA. Differential diagnosis of tumors and tumor-like lesions of liver in infancy and childhood. AMA J Dis Child 1956; 91: 168–186. [DOI] [PubMed] [Google Scholar]
- 5.Malouf GG, Job S, Paradis V et al. . Transcriptional profiling of pure fibrolamellar hepatocellular carcinoma reveals an endocrine signature. Hepatology 2014; 59: 2228–2237. [DOI] [PubMed] [Google Scholar]
- 6.Fujimoto A, Totoki Y, Abe T et al. . Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012; 44: 760–764. [DOI] [PubMed] [Google Scholar]
- 7.Jhunjhunwala S, Jiang Z, Stawiski EW et al. . Diverse modes of genomic alterations in hepatocellular carcinoma. Genome Biol 2014; 15: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nault JC, Mallet M, Pilati C et al. . High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun 2013; 4: 2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornella H, Alsinet C, Sayols S et al. . Unique genomic profile of fibrolamellar hepatocellular carcinoma. Gastroenterology 2015; 148: 806–818.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakar S, Chen X, Ho C et al. . Chromosomal changes in fibrolamellar hepatocellular carcinoma detected by array comparative genomic hybridization. Mod Pathol 2009; 22: 134–141. [DOI] [PubMed] [Google Scholar]
- 11.Malouf GG, Tahara T, Paradis V et al. . Methylome sequencing for fibrolamellar hepatocellular carcinoma depicts distinctive features. Epigenetics 2015; 10: 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham RP, Jin L, Knutson DL et al. . DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Mod Pathol 2015; 28: 822–829. [DOI] [PubMed] [Google Scholar]
- 13.Honeyman JN, Simon EP, Robine N et al. . Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 2014; 343: 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cleary SP, Jeck WR, Zhao X et al. . Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 2013; 58: 1693–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu L, Hazard FK, Zmoos AF et al. . Genomic analysis of fibrolamellar hepatocellular carcinoma. Hum Mol Genet 2015; 24: 50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malouf G, Falissard B, Azoulay D et al. . Is histological diagnosis of primary liver carcinomas with fibrous stroma reproducible among experts? J Clin Pathol 2009; 62: 519–524. [DOI] [PubMed] [Google Scholar]
- 17.Wagner AH, Coffman AC, Ainscough BJ et al. . DGIdb 2.0: mining clinically relevant drug-gene interactions. Nucleic Acids Res 2016; 44: D1036–D1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pawson AJ, Sharman JL, Benson HE et al. . The IUPHAR/BPS Guide to PHARMACOLOGY: an expert-driven knowledgebase of drug targets and their ligands. Nucleic Acids Res 2014; 42: D1098–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramanathan RK, Belani CP, Singh DA et al. . A phase II study of lapatinib in patients with advanced biliary tree and hepatocellular cancer. Cancer Chemother Pharmacol 2009; 64: 777–783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.