Abstract
Estrogen exerts cellular effects through both nuclear (ESR1 and ESR2) and membrane-bound estrogen receptors (G-protein coupled estrogen receptor, GPER); however, it is unclear if they act independently or engage in crosstalk to influence hormonal responses. To investigate each receptor’s role in proliferation, transcriptional activation, and protein phosphorylation in breast cancer cells (MCF-7), we employed selective agonists for ESR1 propyl-pyrazole-triol (PPT), ESR2 diarylpropionitrile (DPN), and GPER (G-1) and also determined the impact of xenoestrogens bisphenol-A (BPA) and genistein on these effects. As anticipated, 17β-estradiol (E2), PPT, DPN, BPA, and genistein each enhanced proliferation and activation of an ERE-driven reporter gene whereas G-1 had no significant impact. However, G-1 significantly reduced E2-, PPT-, DPN-, BPA-, and genistein-induced proliferation and ERE activation at doses greater than 500 nM indicating that G-1 mediated inhibition is not ESR isotype specific. As membrane receptors initiate cascades of phosphorylation events, we performed a global phosphoproteomic analysis on cells exposed to E2 or G-1 to identify potential targets of receptor crosstalk via downstream protein phosphorylation targets. Of the 211 phosphorylated proteins identified, 40 and 13 phosphoproteins were specifically modified by E2 and G-1, respectively. Subnetwork enrichment analysis revealed several processes related to cell cycle were specifically enriched by G-1 compared with E2. Further there existed a number of newly identified proteins that were specifically phosphorylated by G-1. These phosphorylation networks highlight specific proteins that may modulate the inhibitory effects of G-1 and suggest a novel role for interference with nuclear receptor activity driven by E2 and xenoestrogens.
Keywords: estrogen, estrogen receptor, xenoestrogens, GPER, G-1, phosphoproteomic
The predominant female sex hormone, 17β-Estradiol (E2), is essential for reproductive development and function in males and females (Hess, 2003; Prossnitz and Barton, 2011) and plays critical roles in the physiology of the nervous, immune, vascular, muscular, skeletal, and endocrine systems (Prossnitz and Barton, 2011). Disruption of estrogen signaling contributes to multiple disorders such as reproductive abnormalities, cardiovascular disease, hypertension, metabolic diseases, immune disorders, and reproductive cancers, among others (Deroo and Korach, 2006). As such, it is important to expand our knowledge of the molecular mechanisms of estrogen-receptor ligands to increase our understanding of potential pharmacological and toxicological targets. However, attempts to understand physiological and pathophysiological molecular responses of estrogens are complicated by the existence of multiple types of estrogen receptors (ESRs) capable of signaling through genomic and non-genomic mechanisms (Heldring et al., 2007; Prossnitz and Barton, 2011) and the diverse array of estrogen active chemicals in the environment (Lorand et al., 2010).
The classical, genomic-based mode of action of E2 and other known estrogen active chemicals such as bisphenol-A (BPA) and genistein is well established to involve binding and modulation of nuclear ESRs, of which 2 primary subtypes have been identified in mammals (ESR1 and ESR2) (Kuiper et al., 1997). Once bound to a ligand, the ESRs adopt a conformational change that promotes dimerization and recruitment of various co-regulatory proteins to the transcriptional complex (McKenna and O’Malley, 2001). This ESR transcriptional complex subsequently interacts directly or through tethering with select response elements in the promoters of responsive genes, thereby initiating or inhibiting downstream gene expression (Hall et al., 2001).
In addition to nuclear ESRs, a more recently identified putative membrane-bound ESR, G-protein coupled estrogen receptor (GPER), has been implicated in mediating non-genomic effects of E2 (Revankar et al., 2005a). The GPER has been found to mediate E2-induced rapid responses that include activation of ERK-1/-2, mobilization of intracellular calcium stores, and stimulation of intracellular cAMP production (Prossnitz et al., 2008; Revankar et al., 2005b). It is known that multiple well-established ESR ligands, such as E2, ICI 182,780 and tamoxifen display similar binding affinities (Hall et al., 2001; Korach et al., 2003; Revankar et al., 2005a) for nuclear and membrane ESRs. However, a more unusual observation is that the latter compounds which act as antagonists for ESRs, agonize the GPER.
These results have led to speculation that ESRs and GPER may engage in crosstalk, and possibly modulate compensatory or opposing cell signaling pathways. For example, GPER was able to mediate proliferative effects of E2 in breast cancer cells lacking ESRs suggesting compensatory mechanisms (Pandey et al., 2009) whereas GPER inhibited the proliferation of ESR positive cells suggesting opposing mechanisms are activated (Ariazi et al., 2010). Further, the observation that ESR1 and GPER are able to activate kinases such as ERK1/2 (Filardo et al., 2000; Kang et al., 2010) and PI3K (Revankar et al., 2005a) suggests that activation of signaling cascades by protein phosphorylation may be a point of convergent or divergent cellular signaling pathways that are potentially targets of modulation by xenoestrogens.
Separating cellular signaling networks specifically modulated by GPER and ESRs has been facilitated by the identification of a substituted dihydroquinoline, termed G-1, that was proposed as a selective GPER agonist (Bologa et al., 2006). Availability of this ligand has allowed for studies aimed at investigating a GPER-specific role in non-genomic biological responses to E2. However, a number of studies that have employed G-1, primarily in cell-based experiments, have produced mixed results. For example, in assessing proliferation, some studies show that G-1 enhances while others find it inhibits proliferation in breast cancer cells both expressing and lacking ESRs (Albanito et al., 2007, 2008; Ariazi et al., 2010; Lubig et al., 2012; Lucki and Sewer, 2011). Interestingly, it has been more recently suggested that G-1 may signal independently of GPER at high doses (Wang et al., 2012). Moreover, only one study to date has assessed the role of GPER in mediating estrogen-driven effects that include direct action on ESR activity (Gao et al., 2011).
Based on the current body of knowledge and gaps in our understanding of GPER signaling by E2 and xenoestrogens and potential for ESR crosstalk, the overall goal of this research was 2-fold. First, we sought to determine the impact of GPER activation on classic E2- and xenoestrogen-driven cellular responses in breast cancer cells, including proliferation and ESR isotype specific reporter gene activity. Second, we sought to identify novel non-genomic signaling pathways perturbed by E2 and G-1 using a global phosphoproteomic approach.
MATERIALS AND METHODS
Chemicals
All xenoestrogens and other test substances used in the studies were dissolved in 0.1% DMSO and included 17β-estradiol (Sigma Aldrich, St Louis, Missouri), BPA (supplied by NIEHS), Genistein (Acros Organics, Morris Plains, New Jersey), (propyl-pyrazole-triol (PPT, Tocris, Ellisville, Missouri), diarylpropionitrile (DPN, Tocris), (±)-1-[(3aR*,4S*,9bS*)-4-(6-Bromo-1,3-benzodioxol-5-yl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinolin-8-yl]- ethanone (G-1, Santa Cruz Biotechnology, Santa Cruz, CA), and (3aS*,4R*,9bR*)-4-(6-Bromo-1,3-benzodioxol-5-yl)-3a,4,5,9b-3H-cyclopenta[c]quinolone (G-15, Tocris).
Cell culture
Human breast cancer cells (MCF-7), a gift from Dr Nasser Chegini previously at University of Florida College of Medicine, were cultured in Minimum Essential Medium (MEM) without phenol red (Cellgro 17-305-CV, Manassas, Virginia) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, Penstrep, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, and 1.5 g/l sodium bicarbonate. Cells were maintained at 37 °C in humidified 5% CO2 atmosphere. All exposures were performed in phenol red free MEM with 10% charcoal-stripped FBS and the supplements listed earlier.
Proliferation assays
MCF-7 cells were seeded at a concentration of 4 × 103 cells per well of a 96-well Tissue Culture Treated Microplate (Costar Corning Incorporated) in 100 µl cell culture medium. The cells were allowed to adhere overnight then were switched to cell culture medium containing 10% charcoal-stripped FBS for 24 h. Cells were then exposed to various concentrations of test substances dissolved in DMSO (final DMSO ≤ 0.2%) or vehicle control for 24, 48, and 72 h. Proliferation was measured by modified MTT assay (Promega Corp., Wisconsin). Briefly, proliferation was measured every 24 h by incubating cells in 15 µl Dye Solution for 4 h followed by addition of 100 µl Solubilization/Stop buffer. After 1 h, absorbance measurements (570 nm) were acquired on a BioTek Synergy H1 plate reader. Exposure media was changed every 48 h throughout the time-course.
Reporter gene activity assays
MCF-7 cells were seeded at a concentration of 2 × 105 cells per well of a 24-well Tissue Culture Treated Plates (Multiwell, Falcon) in 1.0 ml of complete cell culture medium. The cells were allowed to adhere overnight then were switched to 10% charcoal-stripped FBS-containing medium without Penstrep. Cells were transiently transfected with 500 ng reporter plasmid containing a 2X estrogen response element (2XERE) upstream of the luciferase gene and 50 ng Renilla pRL-TK using Lipofectamine 2000 (Invitrogen). After 24 h, cells were exposed to chemicals dissolved in DMSO for 24 h. Cells were washed with 500 µl 1X Phosphate Buffered Saline without Magnesium and Calcium (Corning cellgro, Manassas, Virginia) and collected in 100 µl 1X Passive Lysis Buffer (Promega). To facilitate lysis, cells were rocked on a shaking platform at 50 rpm for 20 min at room temperature. Cell lysates were collected and placed in 1.5 ml microcentrifuge tubes and centrifuged at 12 000 × g at 4 °C. Thereafter, luciferase activity was measured using the Dual Luciferase Reporter Kit (Promega Corp., Wisconsin) on a BioTek Synergy H1 plate reader. To each well of a 96-well LUMITRAC 200 white immunology plate (USA Scientific), 20 µl of each lysate was added followed by 50 µl of the Firefly Luciferase Reagent. The activity in each well was read, followed by the addition of 50 µl of Stop & Glo Substrate. Firefly luciferase luminescence was normalized to Renilla luminescence and reported as either fold change over control or percent maximal response.
Phosphoproteomic enrichment and LC-MS/MS
MCF-7 cells were exposed to 10 nM E2, 1 µM G-1 or vehicle control (n = 1) for 30 min and total protein was isolated from cells by washing 3 times with 2 ml ice-cold PBS and harvested in 100 ul ice-cold whole cell lysis buffer (20 mM Tris pH 7.6, 1% Triton-X 100, 137 mM NaCl, 0.1 mM EDTA, 1X EDTA Free Protease Inhibitor Tablet [Pierce 88 661]). Lysates were passed through a 26-gauge needle and incubated on ice for 30 min, then centrifuged at 14 000×g for 15 min at 4 °C. Supernatant was collected and quantified by the Bradford Protein Assay (BioRad). Protein phosphorylation was assessed using a quantitative, label-free approach at Duke University by the Proteomics Core Facility (Soderblom et al., 2011). Protein extracts were digested with trypsin and subjected to phosphopeptide enrichment using TiO2-packed spin columns. After elution, phosphopeptides were identified using DIA MSE HPLC-MS/MS. All MS/MS samples were analyzed for protein identity using Mascot (Matrix Science, London, UK). Mascot was set to search the Swiss Prot_2012× database (selected for Homo sapiens, unknown version, 20321 entries) assuming the digestion enzyme trypsin. Mascot was searched with a fragment ion mass tolerance of 0.040 Da and a parent ion tolerance of 10.0 PPM. Iodoacetamide derivative of cysteine was specified in Mascot as a fixed modification. Deamidation of asparagine and glutamine, oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were specified in Mascot as variable modifications. Scaffold (version Scaffold_4.4.1.1, Proteome Software Inc., Portland, Oregon) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at >52.0% probability to achieve an false discovery rate (FDR) <1.0% by the Peptide Prophet algorithm (Beausoleil et al., 2006) with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at >32.0% probability to achieve an FDR < 1.0% and contained at least 1 identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm (Olsen et al., 2006). The lower protein probability scores were allowed due to identification of proteins by only the phosphorylated peptides. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Scaffold Post-Translational Modification (PTM) (Proteome Software, Portland, Oregon) was used to annotate PTM sites contained in MS/MS spectra. The program utilizes the ASCORE’s probabilistic approach and scoring technique developed by Beausoleil et al. (2006). Phosphopeptides with ASCORE values above 13 (≥ 95% certainty) were considered unambiguously assigned.
Pathway analysis
Subnetwork enrichment analysis of proteins/chemicals regulating cell processes was performed using Pathway Studio Version 9.0 (Elsevier) using all identified phosphorylated proteins. For the analysis, 2 entities (proteins) had to be present in a subnetwork for inclusion and results were limited to 200 subnetworks with best p-value < .05 for enrichment cut-off. Pathways were generated in pathway studio using only proteins differentially phosphorylated among E2 and G-1 experimental groups ie, excluding proteins similarly phosphorylated in all 3 exposure groups. The shortest path algorithm was used and the filter parameters were set to include cell process and protein entities, and the relation type was designated as regulation.
Statistical analysis
Statistically significant differences in the proliferation experiments were determined by 2-tailed Mann-Whitney test using a 95% confidence interval. Differences were considered significant with a P-value < 0.05. Data obtained from the gene reporter and qRT-PCR assays were analyzed by 1-way ANOVA followed by Newman-Keuls Multiple Comparisons post-hoc test to determine statistical differences between treatments. Differences were considered significant with a P-value < .05.
RESULTS
G-1 Suppresses Nuclear ESR-Driven Cell Proliferation
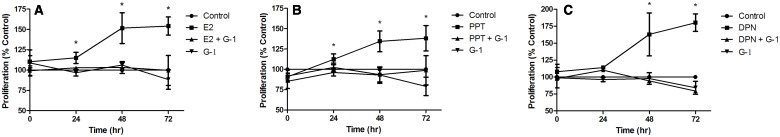
We utilized a modified MTT assay to explore the effect of GPER activation by the putative selective agonist, G-1, on the proliferation of breast cancer cells. MCF-7 cells were exposed to 10 nM E2 alone or in combination with 1 µM G-1, and proliferation was measured every 24 h over a 72-h time period. The dose of G-1 was chosen based on a previous study showing that no substantial binding of this compound to the ESRs occurred at concentrations up to 1 μM. The known Ki values of E2 and G-1 for GPER are reported as 5.7 and 11 nM, respectively (Bologa et al., 2006). As expected, E2 significantly increased cellular proliferation compared with the untreated control group (P < .05); however, exposure to 10 nM E2 in the presence of 1 μM G-1 eliminated proliferation, which was undistinguishable from the control cells (Figure 1A). Co-exposure of E2 with a lower dose of G-1 (10 nM), which is close to the calculated Ki of G-1 for GPER, had no effect on E2-induced proliferation (data not shown). Importantly, the observed reduction of E2-induced proliferation by G-1 was not due to a decrease in cell viability in G-1 exposed cells (Supplementary Figure S1). Of note, individual exposures to low (10 nM, data not shown) and high (1 µM) doses of G-1 individually had no effect on proliferation (Figure 1A).
FIG. 1.
A high dose (1 μM) of the GPER agonist (G-1) inhibits ESR1 and ESR2 induced proliferation. MCF-7 cells were exposed to the ESR agonist (10 nM E2, A), the ESR1 specific agonist (100 nM PPT, B), or the ESR2 specific agonist (100 nM DPN, C), individually and in the presence of 1 μM G-1. Absorbance at 570 nm was measured and expressed as percent of control. Bars are Mean ± SEM of at least 1 experiment. Asterisk (*) indicates significant differences from control (P < .05).
Next, cells were exposed to 100 nM PPT (ESR1 specific agonist), or 100 nM DPN (ESR2 specific agonist) individually and in combination with 1 µM G-1 in order to determine the ESR isoform specificity of the inhibitory effect of G-1 on MCF-7 proliferation. Although exposure of MCF-7 cells to both 100 nM PPT and 100 nM DPN significantly increased proliferation over the untreated control group (P < .05), co-exposure with 1 µM G-1 decreased proliferation to the level of control cells for both treatments (Figures 1B and C).
G-1 Inhibits Xenoestrogen-Induced Cellular Proliferation
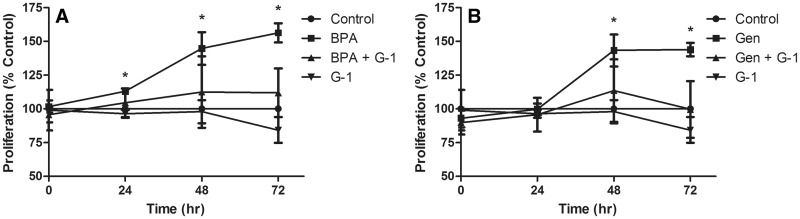
MCF-7 cells were exposed to 10 µM BPA or 1 µM genistein individually and in combination with a high dose of G-1 (1 µM) to determine whether G-1 could also suppress xenoestrogen-driven cellular proliferation. Cell proliferation was assessed with a modified MTT assay every 24 h throughout a 72-h time period. In agreement with previous reports (Hsieh et al., 1998; Nakaya et al., 2007; Schafer et al., 1999), exposure to BPA and genistein significantly increased proliferation of MCF-7 cells compared with the untreated control group over the course of 72 h (P < .05) (Figures 2A and B). Similar to results obtained with E2, co-exposure with the high dose of G-1 (1 µM) decreased BPA- and genistein-driven proliferation to the level of the untreated control group (Figures 2A and B). This inhibition was not observed when the low dose (10 nM) of G-1 was used (data not shown).
FIG. 2.
A high dose (1 μM) of G-1 inhibits xenoestrogen induced proliferation. MCF-7 cells were exposed to BPA (10 μM BPA, A) or genistein (1 μM Gen, B) individually and in the presence of 1 μM G-1. Absorbance at 570 nm was measured and expressed as percent of control. Bars are Mean ± SEM of 2 experiments. Asterisk (*) indicates significant differences from control (P < .05).
G-1 Suppresses ESR-Mediated Transcriptional Activation
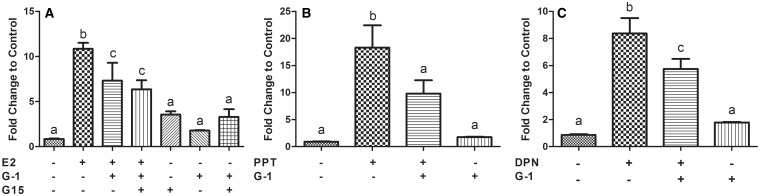
We examined the ability of G-1 to modulate ESR activity by utilizing a 2XERE-driven luciferase reporter gene assay in order to further examine a plausible mechanism for the G-1 mediated inhibition of E2 and xenoestrogen-driven cell proliferation. In addition to E2, ESR isoform specific agonists (PPT, DPN) were used individually and in combination with G-1 to examine effects on ESR activity. MCF-7 cells were transfected with a 2XERE-Luciferase reporter gene, and then exposed to E2 or the ESR isoform specific agonists individually and in combination with 1 µM G-1 for 24 h. Data from these experiments revealed that individual exposures to 10 nM E2, 100 nM PPT, and 100 nM DPN significantly increased (P < .05) ERE activation 12, 17, and 8-fold, respectively, over the untreated control group (Figures 3A and C). These results indicated that both ESR1 and ESR2 activated the 2XERE reporter in our model cell line. Exposing MCF-7 cells to 1 µM G-1 did not have any effect on ERE activation. In agreement with results from the proliferation assays, exposure to 10 nM E2, 100 nM PPT, or 100 nM DPN in combination with 1 µM G-1 significantly decreased (P < .05) ERE activation compared with individual exposures with 10 nM E2, 100 nM PPT, and 100 nM DPN, (37.5, 44, and 25%, respectively) (Figures 3A and C). Surprisingly, attempts to rescue this inhibition by G-1 in E2 exposed cells by pretreating cells with a GPER selective antagonist (20 µM G-15) were unsuccessful (Figure 3A).
FIG. 3.
A high dose (1 μM) of G-1 inhibits ESR1- and ESR2-induced transcriptional activation at an ERE. MCF-7 cells were exposed to E2 (A), PPT (B), or DPN (C) individually and in the presence of 1 μM G-1. Fold changes are expressed in Relative Luciferase Units (RLU) compared with control. Bars are Mean ± SEM of at least 3 experiments. Letters indicate statistically significant differences (P < .05).
G-1 Mediated Suppression of E2 and Xenoestrogen-Driven ESR Activity Is Effective at High Doses
After establishing that a high dose of G-1 inhibited ESR-mediated activity, we next sought to determine whether the inhibition was dose-dependent and whether xenoestrogen-induced transcriptional activation was similarly inhibited. For these experiments, MCF-7 cells were transfected with a 2XERE-Luciferase reporter gene and exposed to 10 nM E2 individually and in combination with increasing concentrations of G-1 (10, 100, 500 nM, or 1 µM) for 24 h. Results revealed that only the high dose of G-1 (1 µM) caused a statistically significant (P < .05) reduction (60%) in ERE activation compared with cells exposed only to 10 nM E2 (Figure 4A). Because 1 µM G-1 also inhibited xenoestrogen-induced proliferation, we next examined the potential inhibitory effects of low and high doses of G-1 on xenoestrogen-induced ERE activation. For these experiments, EC50 values for BPA and genistein were calculated based on a dose-response assessment of ERE activity and were determined to be 640 nM and 284 nM, respectively (data not shown). MCF-7 cells were exposed to these concentrations individually and in combination with increasing concentrations of G-1 (10, 100, 500 nM, or 1 µM) for 24 h. Similar to results observed with E2, low doses of G-1 (10–500 nM) had no statistically significant effect on BPA-induced ESR activity whereas co-exposure to 1 µM G-1 resulted in a statistically significant (P < .05) reduction (37%) in ERE activation compared with cells exposed to BPA (640 nM) individually (Figure 4B). Co-exposure to both 500 nM G-1 and 1 μM G-1 significantly reduced (P < .05) genistein (284 nM)-induced ESR activity by 45 and 33%, respectively.
FIG. 4.
G-1 mediated reduction in (xeno)estrogen induced transcriptional activation at an ERE is most effective at a high dose. MCF-7 cells were exposed to the ESR agonist (10 nM E2, A), BPA (640 nM, B), or genistein (284 nM Gen, C) individually and in the presence of increasing concentrations of G-1. Fold changes in RLU compared with control were calculated and graphed as percent of individual exposure. Bars are Mean ± SEM of 3 experiments. Letters indicate statistically significant differences (P < .05).
G-1 and E2 Exhibit Overlapping and Unique Phosphorylation Profiles
We utilized a global, non-gel-based phosphoproteomic strategy to identify rapid signaling pathways modulated by E2 and G-1. MCF-7 cells were exposed to vehicle control, 10 nM E2 or 1 µM G-1 for 30 minutes in order to identify early targets of G-1 and E2 -induced non-genomic signaling networks. Phosphopeptides were identified using LC-MS/MS strategies and matched to proteins using MASCOT. Results revealed the presence of 299 phosphorylated peptides derived from 211 proteins. Most peptides were phosphorylated on only one residue (239), 55 were phosphorylated at 2 sites, 3 peptides were phosphorylated at 3 sites, and 2 peptides were phosphorylated at 4 sites. The distribution of the 336 unique phosphorylation sites on the 299 peptides identified was 90.8% serine, 8.9% threonine, and 0.3% tyrosine which is consistent with other studies investigating phosphorylation induced by membrane receptors which identified 90% serine, 10% threonine, and 0.5% tyrosine phosphorylation events (Olsen et al., 2006).
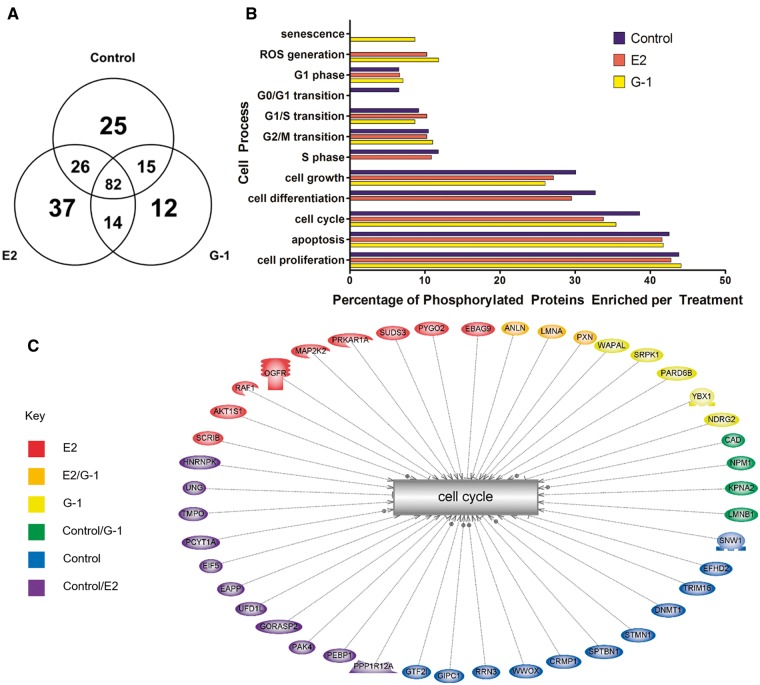
The distribution of phosphoproteins by treatment group is depicted in Figure 5A. Overall, 37 and 12 phosphoproteins were differentially phosphorylated by E2 and G-1, respectively, and 14 were similarly phosphorylated by both. Gene symbols for each protein are provided in parenthesis throughout this report for reference to the Subnetwork Enrichment and Pathway analyses. As expected a number of proteins involved in signal transduction and protein phosphorylation were phosphorylated after exposure to E2 such as RAF proto-oncogene serine/threonine-protein kinase (RAF1), epidermal growth factor receptor substrate 15-like 1 (EPS15L1), and proline-rich AKT1 substrate 1 (AKT1S1). G-1 induced phosphorylation of serine/arginine-rich protein-specific kinase 1 (SRPK1), and both treatments caused phosphorylation of ras GTPase-activating protein-binding protein 1 (G3BP1) at the same sites (S149 and S232). Other proteins that were phosphorylated by both treatments were phosphorylated on distinct residues by each such as La-related protein 1 (LARP1) which was phosphorylated at S774 by E2 and T526 by G-1 while S627 and S631 were phosphorylated after each treatment including control (Table 1).
FIG. 5.
E2 and G-1 exhibit unique and overlapping phosphorylation profiles. MCF-7 cells were exposed to 10 nM E2, 1 μM G-1, or vehicle control for 30 min to capture early targets of non-genomic signaling pathways mediated by phosphorylation cascades. A, Venn diagram organizing all identified phosphorylated proteins among the exposure groups. B, Histogram representing the number of phosphorylated proteins sorted by Subnetwork Enrichment Analysis into cell processes using Pathway Studio as a percentage of the total number of phosphorylated proteins identified in each exposure group. C, Pathway depicting the differentially phosphorylated proteins ie, those not found to be similarly phosphorylated in all 3 exposure groups that were involved in the cell process of cell cycle.
TABLE 1.
Identified Phosphorylated Proteins Involved in Cell Cycle
| Gene Symbol | Protein Name | Sequence |
Phosphorylation Sites by Treatment |
||
|---|---|---|---|---|---|
| Control | E2 | G-1 | |||
| NDRG2 | Protein NDRG2 | TAsLTSAASVDGNR | S332 | ||
| PARD6B | Partitioning defective 6 homolog beta | HGAGsGCLGTMEVK | S11 | ||
| SRPK1 | Serine/threonine-protein kinase SRPK1 | GSAPHSESDLPEQEEEILGsDDDEQEDPNDYCK | S51 | ||
| WAPAL | Wings apart-like protein homolog | RPESPSEIsPIKGSVR | S226 | ||
| VEEESTGDPFGFDsDDESLPVSSK | S77 | ||||
| YBX1 | Nuclease-sensitive element-binding protein 1 | NYQQNYQNsESGEKNEGSESAPEGQAQQR | S165 | ||
| ANLN | Actin-binding protein anillin | ATsPVKSTTSITDAK | S295 | S295 | |
| G3BP1 | Ras GTPase-activating protein-binding protein 1 | SSsPAPADIAQTVQEDLR | S232 | S232 | |
| YQDEVFGGFVTEPQEEsEEEVEEPEER | S149 | S149 | |||
| LMNA | Prelamin-A/C | SGAQASSTPLsPTR | S22 | S22 | |
| PXN | Paxillin | FIHQQPQSSsPVYGSSAK | S85 | S85 | |
| EAPP | E2F-associated phosphoprotein | YYDDIYFDsDsEDEDRAVQVTK | S109 | S109,S111 | |
| GORASP2 | Golgi reassembly-stacking protein 2 | VGDSTPVSEKPVSAAVDANASEsP | S451 | S451 | |
| HNRPK | Heterogeneous nuclear ribonucleoprotein K | DYDDMsPR | S284 | S284 | |
| EIF5 | Eukaryotic translation initiation factor 5 | WLKEAEEEssGGEEEDEDENIEVVYSK | S389,S390 | S389,S390 | |
| PPP1R12A | Protein phosphatase 1 regulatory subunit 12A | DKKsPLIESTANMDNNQSQK | S299 | S299 | |
| PAK4 | Serine/threonine-protein kinase PAK 4 | DKRPLsGPDVGTPQPAGLASGAK | S181 | S181 | |
| PCYT1A | Choline-phosphate cytidylyltransferase A | MLQAIsPK | S315 | ||
| WPFSGKTSPPCsPANLSR | S347 | ||||
| PEBP1 | Phosphatidylethanolamine-binding protein 1 | NRPTsISWDGLDSGK | S52 | S52 | |
| TMPO | Lamina-associated polypeptide 2, isoform alpha | GPPDFssDEEREPtPVLGSGAAAAGR | T74, S66, S67 | S66,S67 | |
| UFD1L | Ubiquitin fusion degradation protein 1 homolog | GVEPSPsPIKPGDIKR | S247 | S247 | |
| UNG | Uracil-DNA glycosylase | HAPsPEPAVQGTGVAGVPEESGDAAAIPAK | S243 | S243 | |
| KAPAGQEEPGtPPSSPLSAEQLDR | T60 | ||||
| AKT1S1 | Proline-rich AKT1 substrate 1 | AATAARPPAPPPAPQPPsPTPsPPRPTLAR | S88,S92 | ||
| PRKAR1A | cAMP-dependent protein kinase type I-alpha regulatory subunit | TDSREDEIsPPPPNPVVK | S83 | ||
| MAP2K2 | Dual specificity mitogen-activated protein kinase 2 | LNQPGtPTR | T394 | ||
| OGFR | Opioid growth factor receptor | SQGDEAGGHGEDRPEPLsPK | S378 | ||
| PYGO2 | Pygopus homolog 2 | GGGtPDANSLAPPGK | T302 | ||
| RAF1 | RAF proto-oncogene serine/threonine-protein kinase | STsTPNVHMVSTTLPVDSR | S258 | ||
| EBAG9 | Receptor-binding cancer antigen expressed on SiSo cells | KLsGDQITLPTTVDYSSVPK | S36 | ||
| SCRIB | Protein scribble homolog | MAESPCSPSGQQPPsPPsPDELPANVK | S1306,S1309 | ||
| SUDS3 | Sin3 histone deacetylase corepressor complex component SDS3 | RPAsPSSPEHLPATPAESPAQR | S234 | ||
| DNMT1 | DNA (cytosine-5)-methyltransferase 1 | EADDDEEVDDNIPEMPsPK | S714 | ||
| CRMP1 | Dihydropyrimidinase-related protein 1 | sIPHITSDR | S8 | ||
| EFHD2 | EF-hand domain-containing protein D2 | RADLNQGIGEPQsPSRR | S74 | ||
| GIPC1 | PDZ domain-containing protein GIPC1 | SAGGRPGsGPQLGTGR | S232 | ||
| GTF2I | General transcription factor II-I | SPGSNsKVPEIEVTVEGPNNNNPQTSAVR | S679 | ||
| RRN3 | RNA polymerase I-specific transcription initiation factor RRN3 | VIIKEGDVDVsDsDDEDDNLPANFDTCHR | S170,S172 | ||
| SNW1 | SNW domain-containing protein 1 | GPPsPPAPVMHSPSRK | S224 | ||
| SPTBN1 | Spectrin beta chain, brain 1 | AQTLPTSVVTITSESsPGKR | S2341 | ||
| STMN1 | Stathmin | DLsLEEIQK | S46 | ||
| ESVPEFPLsPPKKK | S38 | ||||
| TRIM16 | Tripartite motif-containing protein 16 | ETEEQDsDSAEQGDPAGEGKEVLCDFCLDDTR | S60 | ||
| WWOX | WW domain-containing oxidoreductase | YAGLDDtDsEDELPPGWEER | T12,S14 | ||
| KPNA2 | Importin subunit alpha-2 | NVSSFPDDATsPLQENR | S62 | S62 | |
| LMNB1 | Lamin-B1 | LKLsPsPSSR | S391,S393 | S391,S393 | |
| NPM1 | Nucleophosmin | CGSGPVHISGQHLVAVEEDAEsEDEEEEDVK | S125 | S125 | |
| CAD | CAD protein | IHRAsDPGLPAEEPK | S1859 | S1859 | |
| DPYSL2 | Dihydropyrimidinase-related protein 2 | TVtPASSAKtSPAKQQAPPVR | T514 | T514,T521 | T514,T521 |
| EEF1B2 | Elongation factor 1-beta | YGPADVEDTTGSGATDsKDDDDIDLFGsDDEEESEEAKR | S95,S106 | S106 | S95,S106 |
| EIF3B | Eukaryotic translation initiation factor 3 subunit B | ALENGDADEPSFsDPEDFVDDVsEEELLGDVLK | S154,S164 | S164 | S154 |
| EIF4G1 | Eukaryotic translation initiation factor 4 gamma 1 | sFsKEVEER | S1185, S1187 | S1185, S1187 | S1185,S1187 |
| TASTPtPPQTGGGLEPQANGETPQVAVIVRPDDR | T207 | T207 | |||
| LARP1 | La-related protein 1 | AVtPVPTKTEEVSNLK | T526 | ||
| KNTFTAWsDEEsDYEIDDRDVNK | S627,S631 | S627,S631 | S627,S631 | ||
| SLPTTVPEsPNYR | S774 | ||||
| NCL | Nucleolin | AAAAAPAsEDEDDEDDEDDEDDDDDEEDDsEEEAMETTPAK | S184,S206 | S206 | |
| KEDsDEEEDDDsEEDEEDDEDEDEDEDEIEPAAMK | S145,S153 | S145,S153 | S145,S153 | ||
| KVVVSPtKK | T69 | ||||
| LELQGPRGsPNAR | S563 | S563 | S563 | ||
| MAPPPKEVEEDsEDEEMsEDEEDDssGEEVVIPQKK | S34,S41 | S28,S34,S41,S42 | S41,S42 | ||
| NAKKEDsDEEEDDDsEEDEEDDEDEDEDEDEIEPAAMK | S145,S153 | S145 | |||
| MCM2 | DNA replication licensing factor MCM2 | GLLYDsDEEDEERPAR | S139 | S139 | S139 |
| GNDPLTSsPGR | S27 | S27 | |||
| RGLLYDsDEEDEERPAR | S139 | S139 | |||
| MAPT | Microtubule-associated protein tau | AKTDHGAEIVYKsPVVsGDTsPR | S713 | S713,S717,S721 | S713,S717 |
| SGYSSPGsPGTPGSR | S520 | S520 | |||
| VAVVRtPPKsPSSAK | T548,S552 | T548,S552 | |||
| ACLY | ATP-citrate synthase | AKPAMPQDSVPsPR | S481 | S481 | S481 |
| TAsFSESRADEVAPAKK | S455 | S455 | S455 | ||
| AKT1 | RAC-alpha serine/threonine-protein kinase | SGsPsDNsGAEEMEVSLAKPK | S124,S129 | S126,S129 | S124,S129 |
| CCNY | Cyclin-Y | SAsADNLTLPR | S326 | S326 | S326 |
| CFL1 | Cofilin-1 | AsGVAVSDGVIK | S3 | S3 | S3 |
| GSK3A | Glycogen synthase kinase-3 alpha | GEPNVSyICSR | Y279 | Y279 | Y279 |
| HDAC2 | Histone deacetylase 2 | IACDEEFsDsEDEGEGGRR | S422,S424 | S422,S424 | S422,S424 |
| HDGF | Hepatoma-derived growth factor | AGDLLEDsPKRPK | S165 | S165 | S165 |
| HSP90AA1 | Heat shock protein HSP 90-alpha | ESEDKPEIEDVGsDEEEEKK | S263 | S263 | S263 |
| HSP90AB1 | Heat shock protein HSP 90-beta | IEDVGsDEEDDSGKDKK | S255 | S255 | S255 |
| HSPB1 | Heat shock protein beta-1 | QLsSGVSEIR | S82 | S82 | S82 |
| HUWE1 | E3 ubiquitin-protein ligase HUWE1 | GSGTAsDDEFENLR | S1907 | S1907 | S1907 |
| GSKsPAKVSDGGSSSTDFK | S3555 | S3555 | S3555 | ||
| CLNS1A | Methylosome subunit pICln | FEEESKEPVADEEEEDsDDDVEPITEFR | S102 | S102 | S102 |
| IRS1 | Insulin receptor substrate 1 | VNLsPNRNQSAK | S1078 | S1078 | S1078 |
| MAP4 | Microtubule-associated protein 4 | DMEsPTKLDVTLAK | S280 | S280 | S280 |
| NSUN2 | tRNA (cytosine-5-)-methyltransferase NSUN2 | AGEPNsPDAEEANsPDVTAGCDPAGVHPPR | S743,S751 | S743,S751 | S743,S751 |
| PDCD5 | Programmed cell death protein 5 | KVMDsDEDDDY | S119 | S119 | S119 |
| PEA15 | Astrocytic phosphoprotein PEA-15 | DIIRQPsEEEIIK | S116 | S116 | S116 |
| PPP6R3 | Serine/threonine-protein phosphatase 6 regulatory subunit 3 | IQQFDDGGsDEEDIWEEK | S617 | S617 | S617 |
| PREX1 | PtdIns(3,4,5)-dependent Rac exchanger 1 | SNSSYLGSDEMGsGDELPCDMR | S1200 | S1200 | S1200 |
| SEPT2 | Septin-2 | IYHLPDAEsDEDEDFKEQTR | S218 | S218 | S218 |
| SL9A1 | Sodium/hydrogen exchanger 1 | SKETSSPGTDDVFTPAPSDsPSSQR | S785 | S785 | S785 |
| CTTN | Src substrate cortactin | AKTQtPPVsPAPQPTEERLPSSPVYEDAASFK | T401,S405 | T401,S405 | T401,S405 |
| TPI1 | Triosephosphate isomerase | KQsLGELIGTLNAAK | S21 | S21 | S21 |
Lower case letters denote phosphorylated amino acids.
A Subnetwork Enrichment Analysis was performed in Pathway Studio to organize the 211 phosphorylated proteins into functional groups according to their Gene Ontology (GO) terms. A large number of proteins were assigned to the categories of cell proliferation, apoptosis, cell cycle, cell growth, G2/M transition, G1/S transition, G1 phase, cell differentiation, G0/G1 transition, S phase, ROS generation, and senescence (Figure 5B). The phosphorylated proteins involved in cell cycle are listed in more detail in Table 1. Of note, although a similar percentage of phosphorylated proteins were involved in cell cycle, the suite of proteins phosphorylated varied among the treatments (Figure 5C). Among the proteins involved in cell cycle, E2 specifically induced phosphorylation of 9 proteins, G-1 specifically induced phosphorylation of 5 proteins, and both E2 and G-1 induced the phosphorylation of 3 proteins that were not phosphorylated in the control group. Proteins that shared phosphorylation profiles between E2 and G-1 included actin-binding protein anillin (ANLN), DNA (cytosine-5)-methyltransferase 1 (DNMT1), dihydropyrimidinase-related protein 1 (CRMP1), dihydropyrimidinase-related protein 2 (DPYSL2), ras GTPase-activating protein-binding protein 1 (G3BP1) PDZ domain-containing protein GIPC1 (GIPC1), prelamin-A/C (LMNA), DNA replication licensing factor MCM2 (MCM2), and microtubule-associated protein tau (MAPT). Noteworthy were 3 proteins specifically phosphorylated by G-1; N-Myc downstream regulated gene 2 (NDRG2), wings apart-like protein homolog (WAPL), and partitioning defective 6 homolog beta (PAR6B). Other proteins conspicuously lacked phosphorylation in treatments dosed with G-1 (PEBP1, UNG, RLA2, UFD1, EAPP, GORS2, HNRPK, IF5, MYPT1, PAK4) compared with control and E2 treatments, which induced phosphorylation of these proteins. Other trends included proteins that were phosphorylated only by E2 or showed shared phosphorylation patterns by E2 and G-1 as previously described. Pathways were constructed in Pathway Studio to highlight the aforementioned differential phosphorylation patterns (Figure 6). The proteins that were differentially phosphorylated between the treatments and involved in cell cycle are enlarged for emphasis.
FIG. 6.
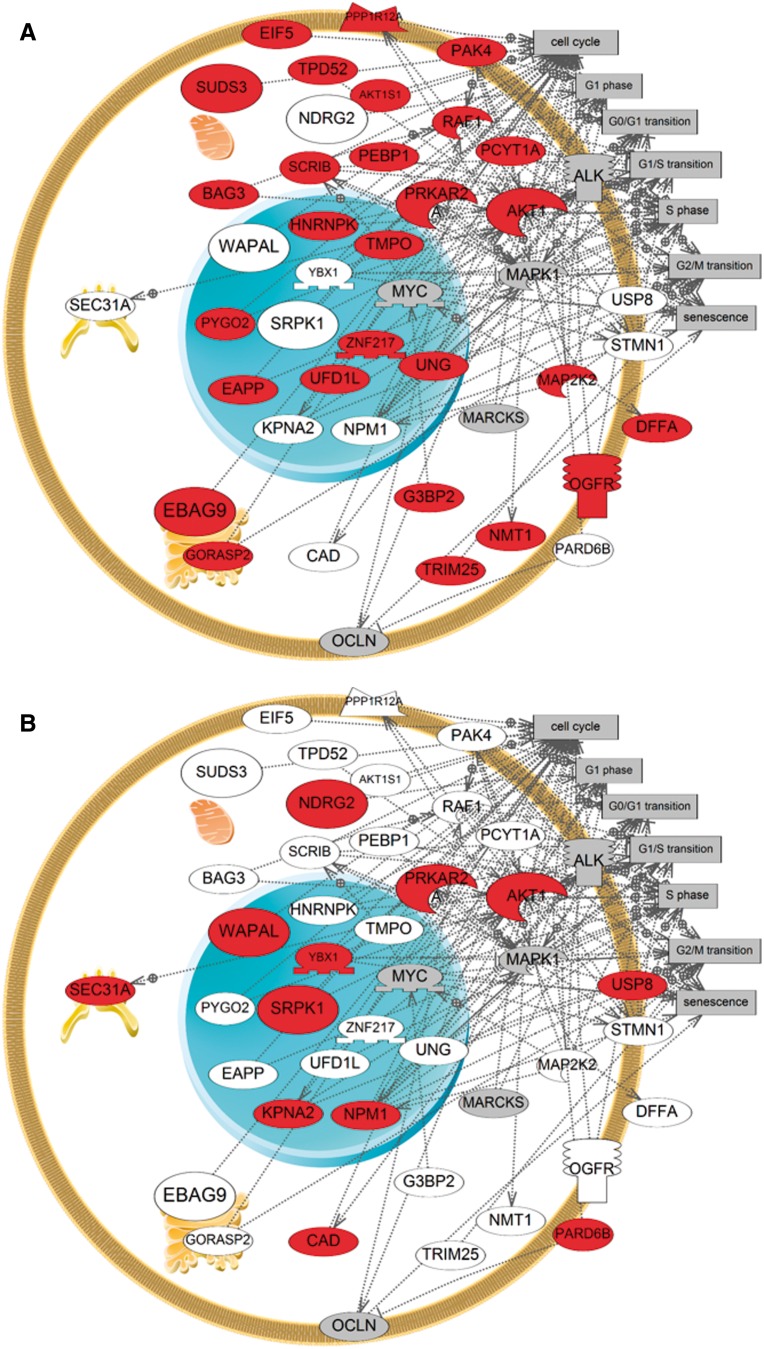
Exposure of MCF-7 cells to 10 nM E2 (A) and 1 μM G-1 (B) induced phosphorylation of proteins involved in cellular processes associated with cell cycle although the specific phosphorylation profiles varied. Proteins in red were identified in respective treatments while proteins in white were not identified in that exposure group. Proteins in gray were not identified in our analysis in any exposure group but were added using the shortest path algorithm in Pathway Studio. Cell Processes (gray boxes) included in the pathways were limited to those found to be significantly enriched by Subnetwork Enrichment Analysis (P < .05). Proteins involved in cell cycle that were differentially phosphorylated between the treatments are shown larger for emphasis.
DISCUSSION
The biochemistry and molecular biology of nuclear ESRs has been a focus of study for decades, but the recently discovered, putative membrane receptor for estrogens, GPER, has been found to contribute to rapid non-genomic actions of E2 (Revankar et al., 2005c; Thomas et al., 2005). Some suggest that GPER and ESRs may signal through both convergent and divergent pathways, which complicates attempts to fully understand their role in E2 signaling.
Several studies report that G-1 inhibits proliferation in a variety of cell types (Ariazi et al., 2010; Wang et al., 2012; Wei et al., 2014; Weißenborn et al., 2014) and attempts to elucidate responsible mechanisms have shown this compound can produce off-target effects (Wang et al., 2012). Although our studies similarly show repression of both E2-induced proliferation and ERE activation by G-1 at doses >1 μM without a reduction in cell viability, we further suggest that this effect is due to inhibition of ESR activity that is not receptor isotype specific. Further, G-1 doses below 1 μM had no effect on ESR induced proliferation (data not shown) or ERE activation (Figure 4). This finding is in agreement with other reports that show G-1 at doses above 1µM inhibited proliferation of certain breast and ovarian granulosa cells while a lower dose did not have a statistically significant effect suggesting that the inhibition has a dose threshold (Ariazi et al., 2010; Gertz et al., 2012; Routledge et al., 2000; Wang et al., 2012). Of note, low doses of G-1 (10–100 nM) also failed to inhibit ERE activation driven by lower doses of E2 (10–100 pM) which is not consistent with assertions that failure of low doses of G-1 to inhibit ESR function is due to saturation of the ESRs (data not shown). These data suggest that one plausible mechanism by which G-1 inhibits E2 induced proliferation is through interference of transcriptional activation at an ERE.
We further report the novel observation that the inhibitory effect of G-1 is not ESR isotype specific as proliferation and activation of a 2XERE reporter gene induced by both ESR-specific agonists PPT (ESR1) and DPN (ESR2) was inhibited by G-1. PPT induced greater induction at an ERE which coincides with previous reports that show ESR1 is more highly expressed in MCF-7 cells (Mollerup et al., 2002). The repression of proliferation without a complete loss of ERE activation suggests that G-1 is targeting other mechanisms of ESR induced proliferation which perhaps involves phosphorylation of proteins.
Because previous reports indicated that BPA and genistein can bind ESRs (Kuiper et al., 1997) and more recently GPER with weak affinity (GPER RBA for genistein = 13.41; BPA = 2.83, (Thomas and Dong, 2006), and are able to stimulate proliferation of MCF-7 cells (Hsieh et al., 1998; Nakaya et al., 2007; Recchia et al., 2004; Schafer et al., 1999), we examined the ability of G-1 to inhibit xenoestrogen-induced cellular proliferation and ERE activation by these compounds. We found that 1 µM G-1 inhibited proliferation and activation at an ERE stimulated by BPA and genistein (Figures 2A and B 4B and C) while lower doses (≤ 500 nM) of G-1 were less effective. Interestingly, co-exposure to 500 nM G-1 significantly reduced genistein induced ERE activation by about 2 fold (P < .05). Although a similar trend was observed for BPA, repression at this dose was not significant. The exposure doses of BPA and genistein produced equivalent activation (based on EC50 of ERE activity) and therefore the more sensitive repression of genistein-induced ERE activity is not likely due to variable receptor binding affinities per se but is more likely a result of variable promoter activity due to distinct receptor conformation, co-regulatory recruitment or phosphorylation of interacting signaling proteins. It is also possible that lower doses of G-1 are more effective at inhibiting ESR2 induced transcriptional activation in this cell line, as it has been reported that genistein has a stronger affinity for ESR2 relative to ESR1 (Kuiper et al., 1998).
Only a handful of studies have investigated the precise non-genomic mechanisms involved in G-1-mediated inhibition of proliferation and most have primarily focused on a few select kinases (eg, MAPK, PI3K) and downstream genes involved in cell cycle progression (eg, cyclins, tumor suppressor proteins) and apoptosis (caspases) (Ariazi et al., 2010; Bologa et al., 2006; Lubig et al., 2012; Wang et al., 2012; Wei et al., 2014; Weißenborn et al., 2014). To our knowledge, no studies have assessed the global phosphoproteome which can facilitate the identification of biological pathways that are modulated by G-1. As the propagation of signaling pathways through kinases and other molecules largely relies on reversible post-translational modifications such as phosphorylation (Metodiev and Alldridge, 2008), global phosphoproteomic strategies are emerging as powerful discovery-based tools to decipher rapid and complex signaling networks (Harsha and Pandey, 2010; Sudhir et al., 2011).
Results of our phosphoproteomic analysis of MCF-7 cells exposed to E2 and G-1 revealed both distinct and overlapping profiles. We identified 211 proteins in total that were phosphorylated on specific serine, threonine, and tyrosine residues among the treatments. A similar percentage of proteins phosphorylated by E2 and G-1 were sorted into cell processes involved in cell growth, cell cycle, apoptosis, cell proliferation, and more specifically, G1 phase, G1/S transition, and G2/M transition, however the specific suite of proteins phosphorylated were different. As seen in Figure 5C, similar percentages of phosphorylated proteins were sorted into cell cycle; however, there were a number of distinct targets of phosphorylation in each exposure group. Further, G-1 specifically did not induce phosphorylation of proteins sorted into cell differentiation or S phase. These results are consistent with previous reports indicating 1 μM G-1 decreased the number of MCF-7 cells in S phase after 24 h (Weißenborn et al., 2014). A GPER independent mechanism is further suggested by another study where 1 μM G-1 decreased the number of GPER negative HEK293 cells in S phase (Wang et al., 2012). Our results highlight protein targets involved in S phase that could contribute to the observed effects, in addition to previously studied cyclins and tumor suppressor proteins, that were specifically not phosphorylated by G-1 but were phosphorylated in other treatments in our analysis such as RAF1, HNRNPK, PRKAR1A, PEBP1, UNG, TMPO, SCRIB, PCYT1A, UFD1L, and EAPP. It is possible that phosphorylation of these proteins facilitates progression of cells into S phase and that the absence of their phosphorylation in the G-1 group could contribute to the observed repression of E2-induced proliferation by G-1.
Our observation that a high dose of G-1 and E2 exhibited unique but overlapping phosphorylation profiles is intriguing because it substantiates the notion that G-1 may signal independently of GPER at high doses but through GPER at lower doses as studies have indicated that low doses of G-1 activated kinases analogous to E2 (Kato et al., 1995; Albanito et al., 2007; Revankar et al., 2005a). Certainly these data warrant future knockdown studies that would directly assess the involvement of GPER in the effects observed. Nonetheless, we identified a number of proteins that were similarly phosphorylated by E2 and G-1 that have been shown to be modulated by E2 such as ANLN which is involved in cell cycle progression and is highly expressed in diverse human tumors (Hall et al., 2005), and DNMT1 which has been shown to be down-regulated at the protein level by E2 in lung cancer cells (Lai et al., 2009) and CD4-positive T cells (Wu et al., 2014), and CRMP1 which has also been shown to be down-regulated at the protein level by E2 in rat brain mitochondria (Nilsen et al., 2007). Of note is the E2-specific phosphorylation of ESR binding site associated, antigen, 9 (EBAG9), a gene known to be transcriptionally regulated by ESR1 (Nakashima et al., 1999).
A number of the proteins that were specifically phosphorylated by G-1 compared with controls were involved in cell cycle (Figure 6) including N-Myc downstream regulated gene 2 (NDRG2), wings apart-like protein homolog (WAPL), and partitioning defective 6 homolog beta (PAR6B). Of particular interest is the G-1 specific phosphorylation of the candidate tumor suppressor protein NDRG2 which plays a role in cell growth inhibition of colon cancer cells when phosphorylated at the particular residue (S332) identified in our phosphoproteomic analysis (Kim et al., 2009). Neither the control nor E2 treatments resulted in S332 phosphorylation of this protein. Although the results of our phosphoproteomic analysis are qualitative, the data offer new insight regarding a role for G-1 in influencing ESR activity through differential phosphorylation of target proteins. These studies provide a solid foundation for more extensive quantitative analyses in future studies.
Overall, results of this work indicate that doses of G-1 > 500 nM inhibit E2 and xenoestrogen-induced, nuclear ESR-mediated proliferation of human breast cancer cells. Because the dose of G-1 used produced no substantial binding to ESR1 or ESR2 (Bologa et al., 2006), we hypothesize that the observed inhibition is not due to interference of receptor binding, but may be a result of suppression of downstream processes such as ESR-mediated transcriptional activation, or through modulation of signaling cascades targeting cell cycle proteins among others. Although the direct involvement of GPER cannot be definitively answered without knockdown studies, these data support the possibility that G-1 may act independently of GPER and are significant because they highlight a novel role for G-1 in inhibiting E2 and xenoestrogen-dependent proliferation. Additional experiments with a larger sample size, a range of doses, and additional time points are needed to increase our understanding of the role of G-1 modulated cell signaling pathways and phosphorylation profiles in inhibiting ESR function. Although these data do not necessarily support a role for xenoestrogens in activating GPER, elucidating pathways that inhibit their effects (through ESRs) have enormous implications for adjuvant therapies in the treatment of ESR-positive breast cancers or ESR-mediated endocrine disease.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
ACKNOWLEDGMENTS
This work was supported by the UF Research Foundation and by National Institute of Environmental Health Sciences (NIH) (1R21ES16617R21).
REFERENCES
- Albanito L., Madeo A., Lappano R., Vivacqua A., Rago V., Carpino A., Oprea T. I., Prossnitz E. R., Musti A. M., Ando S. (2007). G protein–coupled receptor 30 (GPR30) mediates gene expression changes and growth response to 17β-estradiol and selective GPR30 ligand G-1 in ovarian cancer cells. Cancer Research 67, 1859–1866. [DOI] [PubMed] [Google Scholar]
- Albanito L., Sisci D., Aquila S., Brunelli E., Vivacqua A., Madeo A., Lappano R., Pandey D. P., Picard D., Mauro L. (2008). Epidermal growth factor induces G protein-coupled receptor 30 expression in estrogen receptor-negative breast cancer cells. Endocrinology 149, 3799–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariazi E. A., Brailoiu E., Yerrum S., Shupp H. A., Slifker M. J., Cunliffe H. E., Black M. A., Donato A. L., Arterburn J. B., Oprea T. I., et al. (2010). The G protein–coupled receptor GPR30 inhibits proliferation of estrogen receptor–positive breast cancer cells. Cancer Research 70, 1184–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006). A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature B iotechnology 24, 1285–1292. [DOI] [PubMed] [Google Scholar]
- Bologa C. G., Revankar C. M., Young S. M., Edwards B. S., Arterburn J. B., Kiselyov A. S., Parker M. A., Tkachenko S. E., Savchuck N. P., Sklar L. A. (2006). Virtual and biomolecular screening converge on a selective agonist for GPR30. Nature Chemical Biology 2, 207–212. [DOI] [PubMed] [Google Scholar]
- Deroo B. J., Korach K. S. (2006). Estrogen receptors and human disease. The Journal of Clinical Investigation 116, 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardo E. J., Quinn J. A., Bland K. I., Frackelton A. R. (2000). Estrogen-induced activation of Erk-1 and Erk-2 requires the G protein-coupled receptor homolog, GPR30, and occurs via trans-activation of the epidermal growth factor receptor through release of HB-EGF. Molecular Endocrinology 14, 1649–1660. [DOI] [PubMed] [Google Scholar]
- Gao F., Ma X., Ostmann A. B., Das S. K. (2011). GPR30 activation opposes estrogen-dependent uterine growth via inhibition of stromal ERK1/2 and estrogen receptor alpha (ERα) phosphorylation signals. Endocrinology 152, 1434–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J., Reddy T. E., Varley K. E., Garabedian M. J., Myers R. M. (2012). Genistein and bisphenol A exposure cause estrogen receptor 1 to bind thousands of sites in a cell type-specific manner. Genome Research 22, 2153–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. M., Couse J. F., Korach K. S. (2001). The multifaceted mechanisms of estradiol and estrogen receptor signaling. Journal of Biological Chemistry 276, 36869–36872. [DOI] [PubMed] [Google Scholar]
- Hall P. A., Todd C. B., Hyland P. L., McDade S. S., Grabsch H., Dattani M., Hillan K. J., Russell S. H. (2005). The septin-binding protein anillin is overexpressed in diverse human tumors. Clinical Cancer Research 11, 6780–6786. [DOI] [PubMed] [Google Scholar]
- Harsha H., Pandey A. (2010). Phosphoproteomics in cancer. Molecular Oncology 4, 482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldring N., Pike A., Andersson S., Matthews J., Cheng G., Hartman J., Tujague M., Ström A., Treuter E., Warner M. (2007). Estrogen receptors: How do they signal and what are their targets. Physiological Reviews 87, 905–931. [DOI] [PubMed] [Google Scholar]
- Hess R. A. (2003). Estrogen in the adult male reproductive tract: A review. Reproductive Biology and Endocrinology 1, 52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C. Y., Santell R. C., Haslam S. Z., Helferich W. G. (1998). Estrogenic effects of genistein on the growth of estrogen receptor-positive human breast cancer (MCF-7) cells in vitro and in vivo. Cancer Research 58, 3833–3838. [PubMed] [Google Scholar]
- Kang L., Zhang X., Xie Y., Tu Y., Wang D., Liu Z., Wang Z. Y. (2010). Involvement of estrogen receptor variant ER-α36, not GPR30, in nongenomic estrogen signaling. Molecular Endocrinology 24, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S., Endoh H., Masuhiro Y., Kitamoto T., Uchiyama S., Sasaki H., Masushige S., Gotoh Y., Nishida E., Kawashima H. (1995). Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science (New York, NY) 270, 1491.. [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Yoon S. Y., Kim J. T., Choi S. C., Lim J. S., Kim J. H., Song E. Y., Lee H. G., Choi I., Kim J. W. (2009). NDRG2 suppresses cell proliferation through down-regulation of AP-1 activity in human colon carcinoma cells. International Journal of Cancer 124, 7–15. [DOI] [PubMed] [Google Scholar]
- Korach K. S., Emmen J., Walker V. R., Hewitt S. C., Yates M., Hall J. M., Swope D. L., Harrell J. C., Couse J. F. (2003). Update on animal models developed for analyses of estrogen receptor biological activity. The Journal of Steroid Biochemistry and Molecular Biology 86, 387–391. [DOI] [PubMed] [Google Scholar]
- Kuiper G. G., Carlsson B., Grandien K., Enmark E., Häggblad J., Nilsson S., Gustafsson J. (1997). Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology 138, 863–870. [DOI] [PubMed] [Google Scholar]
- Kuiper G. G., Lemmen J. G., Carlsson B., Corton J. C., Safe S. H., van der Saag P. T., van der Burg B., Gustafsson JAk. (1998). Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor β. Endocrinology 139, 4252–4263. [DOI] [PubMed] [Google Scholar]
- Lai J. C., Wu J. Y., Cheng Y. W., Yeh K. T., Wu T. C., Chen C. Y., Lee H. (2009). O6-methylguanine-DNA methyltransferase hypermethylation modulated by 17β-estradiol in lung cancer cells. Anticancer Research 29, 2535–2540. [PubMed] [Google Scholar]
- Lorand T., Vigh E., Garai J. (2010). Hormonal action of plant derived and anthropogenic non-steroidal estrogenic compounds: Phytoestrogens and xenoestrogens. Current Medicinal Chemistry 17, 3542–3574. [DOI] [PubMed] [Google Scholar]
- Lubig J., Lattrich C., Springwald A., Häring J., Schüler S., Ortmann O., Treeck O. (2012). Effects of a combined treatment with GPR30 agonist G-1 and herceptin on growth and gene expression of human breast cancer cell lines. Cancer Investigation 30, 372–379. [DOI] [PubMed] [Google Scholar]
- Lucki N. C., Sewer M. B. (2011). Genistein stimulates MCF-7 breast cancer cell growth by inducing acid ceramidase (ASAH1) gene expression. Journal of Biological Chemistry 286, 19399–19409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N., O’Malley B. W. (2001). Nuclear receptors, coregulators, ligands, and selective receptor modulators. Annals of the New York Academy of Sciences 949, 3–5. [DOI] [PubMed] [Google Scholar]
- Metodiev M., Alldridge L. (2008). Phosphoproteomics: A possible route to novel biomarkers of breast cancer. Proteomics Clinical Applications 2, 181–194. [DOI] [PubMed] [Google Scholar]
- Mollerup S., Jørgensen K., Berge G., Haugen A. (2002). Expression of estrogen receptors α and β in human lung tissue and cell lines. Lung Cancer 37, 153–159. [DOI] [PubMed] [Google Scholar]
- Nakashima M., Sonoda K., Watanabe T. (1999). Inhibition of cell growth and induction of apoptotic cell death by the human tumor-associated antigen RCAS1. Nature Medicine 5, 938–942. [DOI] [PubMed] [Google Scholar]
- Nakaya M., Onda H., Sasaki K., Yukiyoshi A., Tachibana H., Yamada K. (2007). Effect of royal jelly on bisphenol A-induced proliferation of human breast cancer cells. Bioscience, Biotechnology, and Biochemistry 71, 253–255. [DOI] [PubMed] [Google Scholar]
- Nilsen J., Irwin R. W., Gallaher T. K., Brinton R. D. (2007). Estradiol in vivo regulation of brain mitochondrial proteome. The Journal of Neuroscience 27, 14069–14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006). Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648. [DOI] [PubMed] [Google Scholar]
- Pandey D. P., Lappano R., Albanito L., Madeo A., Maggiolini M., Picard D. (2009). Estrogenic GPR30 signalling induces proliferation and migration of breast cancer cells through CTGF. The EMBO Journal 28, 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prossnitz E. R., Arterburn J. B., Smith H. O., Oprea T. I., Sklar L. A., Hathaway H. J. (2008). Estrogen signaling through the transmembrane G protein-coupled receptor GPR30. Annual Review of Physiology 70, 165–190. [DOI] [PubMed] [Google Scholar]
- Prossnitz E. R., Barton M. (2011). The G-protein-coupled estrogen receptor GPER in health and disease. Nature Reviews Endocrinology 7, 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchia A., Vivacqua A., Gabriele S., Carpino A., Fasanella G., Rago V., Bonofiglio D., Maggiolini M. (2004). Xenoestrogens and the induction of proliferative effects in breast cancer cells via direct activation of oestrogen receptor α. Food Additives and Contaminants 21, 134–144. [DOI] [PubMed] [Google Scholar]
- Revankar C. M., Cimino D. F., Sklar L. A., Arterburn J. B., Prossnitz E. R. (2005a). A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science 307, 1625–1630. [DOI] [PubMed] [Google Scholar]
- Revankar C.M., Cimino D.F., Sklar L.A., Arterburn J.B., Prossnitz E.R., (2005b). A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science Signalling 307, 1625. [DOI] [PubMed] [Google Scholar]
- Revankar C. M., Cimino D. F., Sklar L. A., Arterburn J. B., Prossnitz E. R. (2005c). A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science 307, 1625–1630. [DOI] [PubMed] [Google Scholar]
- Routledge E. J., White R., Parker M. G., Sumpter J. P. (2000). Differential effects of xenoestrogens on coactivator recruitment by estrogen receptor (ER) α and ERβ. Journal of Biological Chemistry 275, 35986–35993. [DOI] [PubMed] [Google Scholar]
- Schafer T. E., Lapp C. A., Hanes C. M., Lewis J. B., Wataha J. C., Schuster G. S. (1999). Estrogenicity of bisphenol A and bisphenol A dimethacrylate in vitro. Journal of Biomedical Materials Research 45, 192–197. [DOI] [PubMed] [Google Scholar]
- Soderblom E. J., Philipp M., Thompson J. W., Caron M. G., Moseley M. A. (2011). Quantitative label-free phosphoproteomics strategy for multifaceted experimental designs. Analytical Chemistry 83, 3758.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhir P. R., Hsu C. L., Wang M. J., Wang Y. T., Chen Y. J., Sung T. Y., Hsu W. L., Yang U. C., Chen J. Y. (2011). Phosphoproteomics identifies oncogenic Ras signaling targets and their involvement in lung adenocarcinomas. PloS One 6, e20199.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P., Dong J. (2006). Binding and activation of the seven-transmembrane estrogen receptor GPR30 by environmental estrogens: A potential novel mechanism of endocrine disruption. The Journal of Steroid Biochemistry and Molecular Biology 102, 175–179. [DOI] [PubMed] [Google Scholar]
- Thomas P., Pang Y., Filardo E. J., Dong J. (2005). Identity of an estrogen membrane receptor coupled to a G protein in human breast cancer cells. Endocrinology 146, 624–632. [DOI] [PubMed] [Google Scholar]
- Wang C., Lv X., Jiang C., Davis J. S. (2012). The putative G-protein coupled estrogen receptor agonist G-1 suppresses proliferation of ovarian and breast cancer cells in a GPER-independent manner. American Journal of Translational Research 4, 390.. [PMC free article] [PubMed] [Google Scholar]
- Wei W., Chen Z., Zhang K., Yang X., Wu Y., Chen X., Huang H., Liu H., Cai S., Du J. (2014). The activation of G protein-coupled receptor 30 (GPR30) inhibits proliferation of estrogen receptor-negative breast cancer cells in vitro and in vivo. Cell Death and Disease 5, e1428.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weißenborn C., Ignatov T., Poehlmann A., Wege A. K., Costa S. D., Zenclussen A. C., Ignatov A. (2014). GPER functions as a tumor suppressor in MCF-7 and SK-BR-3 breast cancer cells. Journal of Cancer Research and Clinical Oncology 140, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Sun Y., Mei X., Zhang C., Pan W., Shi W. (2014). 17β‐oestradiol enhances global DNA hypomethylation in CD4‐positive T cells from female patients with lupus, through overexpression of oestrogen receptor‐α‐mediated downregulation of DNMT1. Clinical and Experimental Dermatology 39, 525–532. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.