miR-155 expression is induced in adipocytes and adipose tissue submitted to inflammatory conditions in obesity context in murine and human models and participate to a pro-inflammatory loop by targeting PPARg.
Abstract
Context:
Obesity alters adipose tissue's metabolic and endocrine functions and causes a chronic local and systemic low-grade inflammatory state to develop, generating obesity-associated complications. In the last decade, many entities contributing to and regulating this inflammatory state have been identified, among which are microRNAs.
Objective:
This study aimed to identify microRNA regulated in inflamed adipocytes and adipose tissue, and its effect on adipocyte biology.
Design and Results:
We screened the expression profile of TNFα-treated adipocytes (a major pro-inflammatory protein expressed in obese adipose tissue), and identified miR-155 as the most responsive microRNA. The involvement of TNFα on the basal miR-155 expression was confirmed in the adipose tissue of Tnfa−/− mice where miR-155 was significantly reduced. Also, mice overexpressing p65 or invalidated for p65 in adipose tissue respectively increased and decreased miR-155 expression, in line with the involvement of the nuclear factor κB (NF-κB) pathway in miR-155 induction. miR-155 expression was higher in obese subjects' adipose tissue than in that of normal-weight subjects, and correlated with TNFα expression and body mass index. Gain and loss of function of miR-155 showed its effect on adipocyte function, probably via its ability to target PPARγ mRNA 3′UTR. Interestingly, miR-155 overexpression also resulted in an increased inflammatory state in adipocytes.
Conclusion:
Altogether, these data are evidence of a proinflammatory loop mediated by NF-κB and miR-155 that could participate in the amplification of inflammatory status in adipocytes.
Adipose tissue is a major contributor of the chronic inflammatory response observed in obesity. Obesity is characterized by an increased production of cytokines such as IL6, IL1β, TNFα, or various chemokines, including monocyte chemotactic protein 1 and other mediators of the immune response (1). These proteins can act in an autocrine, paracrine, or endocrine fashion to control various metabolic functions, and the modulation of their expression is mainly driven by JNK and I-κ-B kinase signaling directly (1) but may also be influenced by microRNAs (miRNAs) (2).
miRNAs are short, functional RNAs transcribed from noncoding regions of the genome (3). They act as negative regulators that suppress gene functions through translational repression by targeting 3′ UTR in mRNAs of protein-coding genes, inducing instability of mRNAs, or directly by inhibiting protein production (4). It is now well established that miRNA expression is tissue- and cell-type-specific, and plays essential roles in many biological processes including proliferation, apoptosis, development, and differentiation (5). Various miRNAs have been associated with physiopathological disorders related to obesity such as oxidative stress, impaired adipogenesis, insulin signaling, apoptosis, angiogenesis, and inflammation (2, 6). Specifically, the role of miRNAs in inflamed adipose tissues has been highlighted in several studies. This is notably the case for miR-132, which controls the expression of IL8 and CCL2 (7). In addition, several miRNAs, including miR-126 and miR-193b, affect the secretion of CCL2 from human adipocytes (8). miR-145 also increases the expression and release of TNFα by adipocytes (9), whereas TNFα treatment of adipocytes results in an induction of miR-146b (10) and miR-130 (11). Recently, using high-throughput methodology, it was reported that 18 miRNAs were modulated in response to inflammatory conditions (lipopolysaccharide or macrophage-conditioned medium) (12).
In the present study we set out to identify miRNAs regulated in human adipocytes under inflammatory conditions. In TNFα-treated adipocytes, miR-155 was up-regulated in adipose biopsies of both obese subjects and aP2-p65 mice, and down-regulated in Tnfa-knockout mice. The effect of miR-155 on adipocytes was studied, and revealed that this miRNA could affect adipocyte function negatively and promote inflammation, probably by targeting peroxisome proliferator–activated receptor γ (PPARγ).
Materials and Methods
Cell culture
Adipocyte cells were grown at 37°C in a 5% CO2 humidified atmosphere. The 3T3-L1 preadipocytes (ATCC) were seeded in 3.5-cm diameter dishes at a density of 15 × 104 cells per well, and grown in DMEM supplemented with 10% fetal bovine serum (FBS), at 37°C, as previously reported (13). To induce differentiation, 2-day postconfluent 3T3-L1 preadipocytes (day 0) were stimulated for 72 hours with 0.5mM isobutylmethylxanthine, 0.25 μmol/L dexamethasone, and 1 μg/mL insulin in DMEM supplemented with 10% FBS. The cultures were then treated with DMEM supplemented with 10% FBS and 1 μg/mL insulin.
The human preadipocytes (three independent cultures) were obtained from Promocell, and cultured according to the manufacturer's instructions for 12 days.
Adipocytes of human or murine origin were incubated with different doses of TNFα (5, 10, 15, and 20 ng/mL) or exposed to 15 ng/mL of TNFα at different times (0, 3, 6, 12, and 24 h).
Raw 264.7 macrophages (ECACC) were grown in DMEM supplemented with 10% FBS and 2% HEPES as previously reported (14, 15).
Primary adipocytes from F-p65-Ko mice (16) were differentiated from ear fibroblasts and treated with TNFα (20 ng/mL) for 24 hours.
Cell culture incubations
To identify signaling pathways involved in miR-155 regulation, 3T3-L1 cells were treated with specific inhibitors of mitogen-activated protein (MAP) kinases (JNK, p38) and nuclear factor κB (NF-κB) signaling (JNK inhibitor II [10μM], SB 202190 [20μM], and BAY 117082 [10μM], respectively) for 1 hour (all obtained from Calbiochem, Merck Millipore) and then stimulated or not with TNFα (15 ng/mL) for 24 hours. All the treatments were performed on day 8. To explore the combined effect of rosiglitazone and miR-155, 3T3-L1 adipocytes were incubated with or without Rosiglitazone (0.1μM, 24 h) followed by a 24-hour transfection with miR-155 mimic.
miR-155 mimic or antagomir transfections
Single-stranded oligonucleotides reproducing the biological effect of miR-155 (miR-155 mimic, 50nM; QIAGEN) or directed against miR-155 (miR-155 antagomir; QIAGEN) were transfected in 3T3-L1 adipocytes with Hiperfect transfection reagent (QIAGEN) according to the manufacturer's instructions. Measurements were performed 24 hours after transfection.
Microarrays
The miRNA expression microarrays were run on mature human adipocytes (three independent cultures) incubated with different doses of TNFα (5, 10, and 15 ng/mL) for 24 hours. Gene expression microarrays were run on 3T3-L1 adipocytes (three independent cultures) transfected with miScript miR-155 mimic as described previously. RNA quality control was performed on an Agilent 2100 Bioanalyzer, according to the manufacturer's instructions. RNA was hybridized to the Agilent Human miRNA array or Whole Mouse Genome array. All labeling, hybridization, washing, and scanning was performed as described in the manufacturer's protocol and as previously reported (17–19). The arrays were scanned using an Agilent Scanner. Data were extracted using Agilent Feature Extraction v10.5.1.1 software and analyzed with Agilent GeneSpring GX v11.0.2 software. Pathway analyses were performed with MetaCore and GSEA software (Gene Set Enrichment Analysis) as previously described (14, 20, 21). A false discovery rate q-value < 0.25 for normalized enrichment score was considered significant. A heatmap was obtained with PermutMatrix.
Cloning of miR-155 promoter
miR-155 promoter region (939 base pairs [bp]) was cloned into expression vector pGL3 basic (Promega) using the following oligonucleotides: 5′-TGCCCTGTCAAAATATATTCACTG-3′ for forward primer and 5′-GGATAAGGGATATGCAACTTAAAC-3′ for reverse primer. The product was transfected into 3T3-L1 cells. Each transfection reaction contained 240 ng of miR-155 promoter fragment in pGL3-basic vector (Promega) and 5 ng of pGL4.73 (Promega) containing Renilla luciferase for normalization. Twenty-four hours after transfection, cells were treated with specific inhibitors of MAP kinases (JNK, p38) and NF-κB signaling (JNK inhibitor II [10μM], SB 202190 [20μM] and BAY 117082 [10μM], respectively) for 1 hour (all obtained from Calbiochem, Merck Millipore) and then stimulated or not with TNFα (15 ng/mL) for 24 hours. Both firefly and Renilla luciferase were measured with Dual-Glo Luciferase Assay System (Promega), and firefly activity was normalized with Renilla activity. Experiments were performed at least three times (n = 6 per condition).
Human white adipose tissue biopsies
Eleven lean (body mass index [BMI], 22.5 ± 0.5 kg/m2) and fourteen obese (BMI, 31.7 ± 0.9 kg/m2) male subjects were recruited for this study. The lean and obese volunteers were age 44 ± 7 years and 44 ± 5 years, respectively. Subcutaneous adipose tissue biopsies were performed between 0630 and 0730 hours after an overnight fast. Biopsies were obtained by needle aspiration in the periumbilical area under local anesthesia. Adipose tissue samples were rinsed in physiologic serum, immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction. The experimental protocol was performed in accordance with the guidelines in the Declaration of Helsinki and was approved by the Ethical Committee of the Auvergne Region (agreement No. AU 800, March 2010). Participants gave their written informed consent to participation in this study.
Adipose tissue miR-155 quantification in mice
In the present study, epididymal RNA was used to quantify miR-155 expression levels in various animal models. aP2-p65 mice were generated on the C57BL/6J background as described elsewhere (21, 22). All of the mice were housed in the animal facility at the Pennington Biomedical Research Center with a 12:12-hour light-dark cycle and constant temperature (22–24°C). The male mice were fed chow diet (MF 5001, 11% calories in fat) and the epididymal fat tissue was collected at 20 weeks. The mice were housed at four per cage with free access to water and diet. F-p65-KO mice fed a high-fat diet at 8 weeks (D12331, 36% wt/wt or 58% calories in fat; Research Diets) (16). Tnfa−/− mice were fed a high-fat, high-carbohydrate, semiliquid diet ad libitum for 12 weeks (23). The high-fat diet (from UPAE) contained 17.4% protein (casein), 45% fat (lard for the most part), and 37.6% carbohydrates (25.6% cornstarch and 12% sucrose), expressed as percentage of total energy.
Cotransfection of PPARγ 3′UTR mRNA and miR-155
PPARγ 3′UTR cloned in psiCHECK2 vector (a generous gift from Dr Michael Karbiener, University of Graz, Austria) (50 ng) was cotransfected with miScript miR-155 mimic (50nM) in Cos-1 cells using jetPEI (Polyplus transfection) and Hiperfect Reagent (QIAGEN), respectively, according to the manufacturer's instruction. Renilla and firefly luciferase activities were measured 24 hours after cotransfection with Dual-Glo Luciferase Assay System (Promega). Empty psiCHECK2 vector was used for background measurement. miR-130a was used as a positive control for the experiment. Renilla activity was under the control of PPARγ mRNA 3′UTR, and firefly luciferase activity was used for normalization.
RNA isolation and qPCR
Total cellular RNA was extracted using TRIzol reagent according to the manufacturer's instructions. miR-155 in supernatant was extracted of 150 μL of untreated or TNFα treated (15 ng/mL, 24 h) 3T3-L1 adipocyte supernatants using the miRNeasy Micro Kit according to the manufacturer's instructions (QIAGEN). To quantify miR-155, cDNAs were first synthesized from 1 μg of total RNA in 20 μL using 5X miScript Hiflex Buffer, 10X nucleics mix and miScript reverse transcriptase according to the manufacturer's instructions (QIAGEN). Real-time quantitative RT-PCR analyses were performed using the Mx3005P Real-Time PCR System (Stratagene). Reactions were performed in a 12.5-μL volume containing 6.25 μL of 2X QuantiTect SYBR Green PCR Master Mix (QIAGEN), 1.25 μL of 10X miScript Universal Primer (QIAGEN), 1.25 μL of 10X miScript Primer Assay (Hs_miR-155_2 miScript Primer Assay, Mm_miR-155_1 miScript Primer Assay, Hs_RNU6B miScript Primer Assay (QIAGEN), Hs_SNORD68 miScript Primer Assay (QIAGEN), and 2.5 μL of RNase-free water. For each condition, the expression was quantified in duplicate, and the RNU6B or SNORD68 were used as the endogenous controls in the comparative cycle threshold (CT) method.
To quantify mRNA expression levels, cDNAs were synthesized from 1 μg of total RNA using random primers and Moloney murine leukemia virus reverse transcriptase. Real-time quantitative RT-PCR analyses were performed using the Mx3005P Real-Time PCR System (Stratagene) as previously described (17, 24). For each condition, the expression was quantified in duplicate, and the ribosomal protein 18S rRNA was used as the endogenous control in the comparative cycle threshold (CT) method. The sequences of the primers used for qPCR determination of gene expression are given in Supplemental Table 1.
Oil Red O staining
3T3-L1 adipocytes were washed with PBS and fixed with 10% formalin for 30 minutes. After two washes with distilled water, cells were stained for at least 1 hour at room temperature in freshly diluted Oil Red O solution (six parts Oil Red O [Sigma-Aldrich] in stock solution and four parts H2O; Oil Red O stock solution is 0.5% Oil Red O in isopropanol). Oil Red O solution was removed and the cells were washed with distilled water. Images were then acquired under the microscope. The dye was extracted by adding 100% isopropanol for 10 minutes, and the absorbance was measured at 492 nm.
Macrophage-migration assays
Boyden chamber assays were performed using cell-culture inserts with a 3-μm membrane pore size (Transwell Millipore). The 3T3-L1 adipocytes were incubated with TNFα or transfected with miR-155 mimic for 24 hours. The 3T3-L1-conditioned media was transferred to plates containing inserts. RAW 264.7 macrophages were seeded onto these inserts at a density of 900 cells/cm2. After migration for 4 hours at 37°C, the macrophages in the lower compartment were counted as previously reported (14, 15).
Statistical analysis
Data are expressed as means ± SEM. Significant differences between the control and treated groups were determined using ANOVA followed by a PLSD Fischer post-hoc test using Statview software; P < .05 was considered statistically significant. Significant differences between two groups were determined using a t test; P < .05 was considered statistically significant (*, P < .05; **, P < .01, ***, P < .001). Correlation analyses were performed using Spearman coefficient correlation (ρ) using SPSS v17 (IBM).
Results
miR-155 expression is induced by TNFα
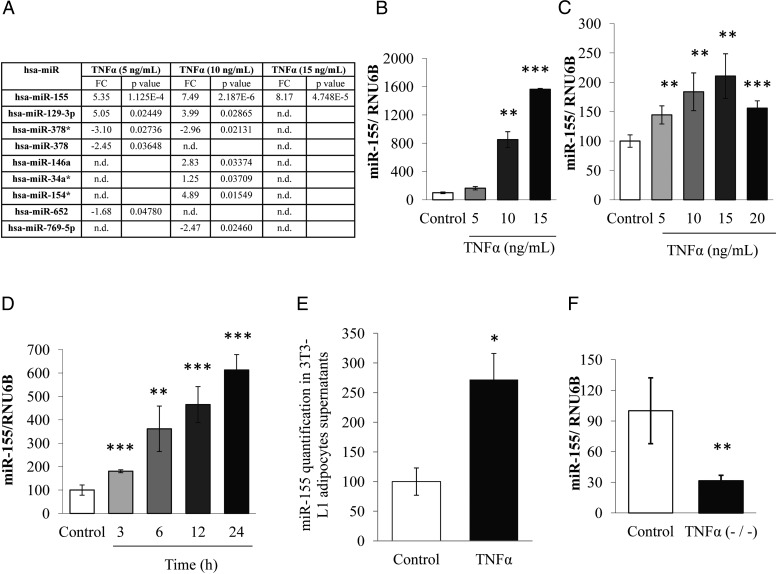
To identify miRNA expression modulation after exposure to TNFα in primocultures of human adipocytes, we made use of miRNA microarray technology. Microarray analysis showed that miR-155 was the only miRNA that was substantially up-regulated by TNFα at different doses (5, 10, and 15 ng/mL) for 24 hours. In addition, among the set of the nine miRNAs positively or negatively regulated, miR-155 seemed to be the most strongly induced in adipocytes for the three concentrations of TNFα (Figure 1A). These results were confirmed by qPCR in human adipocytes (Figure 1B), where TNFα dose-dependently induced miR-155 expression, as well as in 3T3-L1 adipocytes. For this purpose, cells were incubated with different doses of TNFα (5, 10, 15, and 20 ng/mL) for 24 hours. qPCR analysis showed that TNFα increased miR-155 levels significantly and dose dependently in 3T3-L1 adipocytes. The maximum effect was obtained at 15 ng/mL (2.1-fold increase vs control) (Figure 1C). Similarly, the kinetics of miR-155 expression was studied in the presence of TNFα (15 ng/mL). The maximum effect was obtained after 24 hours of 3T3-L1 incubation (1.8, 3.6, 4.7, and 6.1-fold increase after 3, 6, 12, and 24 hours, respectively) (Figure 1D). The effect of TNFα was also evaluated on the secretion of miR-155 in cultured adipocyte supernatant. qPCR analysis showed that TNFα increased miR-155 secretion by 2.72-fold in comparison with control condition (Figure 1E). To confirm the role of TNFα on miR-155 expression, we used mice invalidated for Tnfa gene. miR-155 expression was measured in epididymal adipose tissue of these mice. As shown in Figure 1E, the levels of miR-155 were significantly decreased 3.3-fold in Tnfa-knockout mice compared with wild-type mice.
Figure 1.
miR-155 is regulated by TNFα in human and 3T3–L1 adipocytes. A, The expression of miRNA in TNFα (5, 10, and 15 ng/mL for 24 h) incubated human adipocytes was quantified by microarrays and represented in a Table, as described in Materials and Methods. B, miR-155 expression in human adipocytes and the effect of 24 hours' incubation with TNFα was confirmed by qPCR. C, The effect of TNFα on miR-155 expression was studied in 3T3–L1 adipocytes. Cells were differentiated for 10 d and then incubated with different doses of TNFα (5, 10, 15, and 20 ng/mL) for 24 h or (D) exposed to 15 ng/mL of TNFα at different times (3, 6, 12, and 24 h). E, miR-155 was quantified in 3T3–L1 adipocyte supernatant under TNFα condition (15 ng/mL, 24 h). F, The expression of miR-155 was quantified by qPCR in epididymal adipose tissue of TNFα knockout mice (TNFα−/−). RNU6 or SNORD68 were used as the endogenous controls. The data are expressed as relative expression ratios of at least three independent experiments. The values are presented as means ± SEM. n.d., not detected. *, P < .05; **, P < .01; ***, P < .001.
NF-κB is involved in miR-155 regulation by TNFα
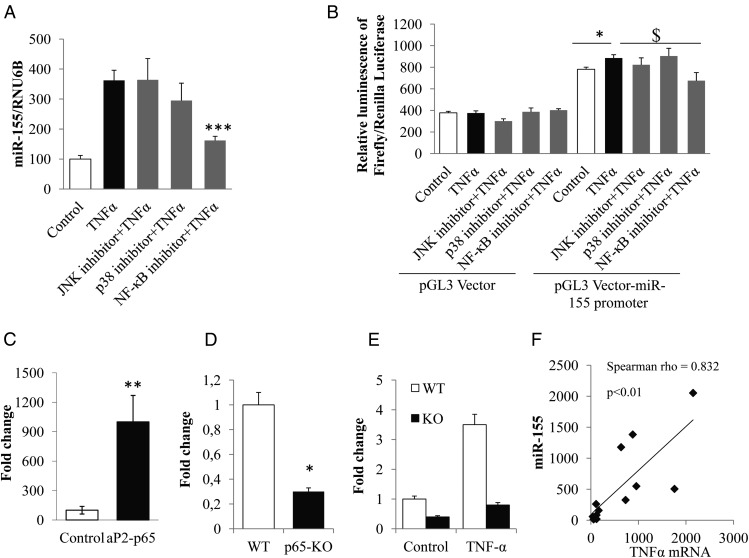
To identify signaling pathways involved in miR-155 regulation by TNFα, 3T3-L1 cells were incubated with specific inhibitors of MAP kinases (JNK, p38) and NF-κB signaling, and then stimulated or not with TNFα. The results show that NF-κB inhibition resulted in a significant decrease in miR-155 expression (2.2-fold compared with TNFα condition) (Figure 2A) whereas no effect of JNK and p38 inhibitors were observed, suggesting a specific role of NF-κB in this regulation. To confirm the transcriptional origin of the regulation of miR-155 under TNFα effect, we cloned the 939-bp genomic sequence upstream of the transcription start site of the mouse miR-155 gene in pGL3-basic vector. pGL3 empty vector or pGL3-miR-155 were transfected in 3T3-L1 cells were incubated with JNK, p38, and NF-κB inhibitors for 1 hour and then stimulated or not with TNFα for 24 hours. Compared with empty vector, pGL3-miR-155 promoter activity was induced by TNFα (2.36-fold increase vs control, Figure 2B), suggesting that the observed regulation has a transcriptional origin. Moreover, the results show that NF-κB inhibition resulted in a significant decrease in the miR-155 promoter activity (1.31-fold compared with pGL3-miR-155 promoter under TNFα stimuli) (Figure 2B). MatInspector software analysis of the cloned region of the miR-155 promoter revealed that putative NF-κB binding site (aaGGGActtgtctcc sequence located between −292 and −306 bp from the transcription start site; matrix similarity score = 0.835) could be responsible of this transcriptional regulation. To confirm the role of NF-κB in miR-155 regulation in vivo, we used a transgenic model overexpressing NF-κB p65 in adipose tissue [aP2-p65 mice (22)], together with mice invalidated for p65 in adipose tissue [F-p65-KO (16)]. miR-155 expression, quantified by qPCR in epididymal adipose tissue of these models was significantly increased in aP2-p65 mice compared with controls (Figure 2C), whereas in F-p65-KO, miR-155 expression was significantly decreased in adipose tissue (Figure 2D), and in primary adipocytes in basal condition (control) and in the TNFα-stimulated condition (Figure 2E). There was also a direct, positive correlation between miR-155 expression and mRNA levels coding for TNFα (Spearman ρ = 0.83, P < .01) in aP2-p65 mice (Figure 2F).
Figure 2.
NF-κB is implicated in miR-155 regulation by TNFα. A, 3T3–L1 cells were treated with specific inhibitors of MAP kinases (JNK, p38) or NF-κB signaling (JNK inhibitor II [10 μM], SB 202190 [20 μM], and BAY 117082 [10 μM], respectively) for 1 h and then stimulated with TNFα (15 ng/mL) for 24 h, and miR-155 expression was measured by qPCR. RNU6B was used as the endogenous control. The data are expressed as relative expression ratios. The values are presented as means ± SEM. ***, P < .001 for NF-κB inhibitor + TNFα condition compared with TNFα condition. B, The 939-pb genomic sequence upstream of the mouse miR-155 was cloned into the expression vector pGL3 basic. The product was transfected into 3T3–L1 cells and treated with specific inhibitors of MAP kinases (JNK, p38) or NF-κB signaling (JNK inhibitor II [10 μM], SB 202190 [20 μM], and BAY 117082 [10 μM], respectively) for 1 h and then stimulated with TNFα (15 ng/mL) for 24 h. The luminescence results were presented as the means ± SEM. *, P < .05 for pGL3 Vector-miR-155 promoter + TNFα compared with pGL3 Vector-miR-155 promoter; $, P < .05 for pGL3 Vector-miR-155 promoter + NF-κB inhibitor + TNFα compared with pGL3 Vector-miR-155 promoter + TNFα. C, The expression of miR-155was quantified by qPCR in epididymal adipose tissue of aP2–p65 transgenic mice. RNU6B was used as the endogenous control. The expression is expressed in fold change for mean ± SE. *, P < .05. D, The expression of miR-155 was quantified by qPCR in epididymal adipose tissue of p65-KO mice. RNU6B was used as the endogenous control. The expression is expressed in fold change for mean ± SE. *, P < .05. E, the effect of TNFα on miR-155 expression in primary adipocytes was studied by qPCR. The adipocytes were differentiated from ear fibroblasts and treated with TNFα (20 ng/mL) for 24 h. RNU6B was used as the endogenous control. The expression is expressed in fold change for mean ± SE. *, P < .05. F, The correlation between miR-155 expression and TNFα mRNA in epididymal adipose tissue of aP2–p65 transgenic mice was established using the Spearman correlation coefficient (ρ).
Obesity-associated inflammation induces adipose miR-155 expression in humans
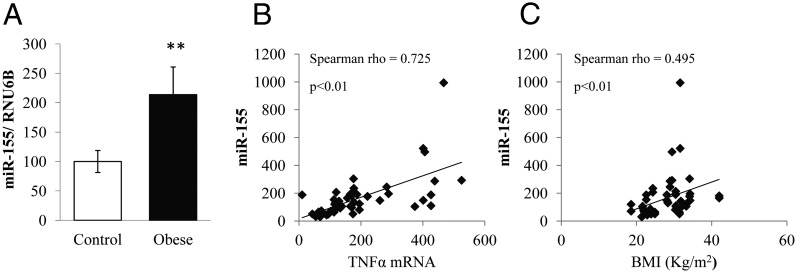
The miR-155 expression was assessed in sc adipose tissue biopsies of lean and obese subjects. The qPCR analysis showed that miR-155 was significantly overexpressed in adipose tissue of obese subjects compared with lean controls (Figure 3A).
Figure 3.
Inflammation associated with obesity induces adipose miR-155 expression in humans. A, The expression of miR-155 was quantified by qPCR in adipose tissue biopsies of obese or normal-weight subjects (control). RNU6B was used as the endogenous control. The expression is expressed in fold change for mean ± SE. **, P < .01 compared with controls. B, The correlation between miR-155 expression and TNFα mRNA or BMI in adipose tissue of obese patients (C) was established using the Spearman correlation coefficient (ρ).
Furthermore, there was a positive, significant correlation between miR-155 and mRNA coding for TNFα (Spearman ρ = 0.73; P < .01) on the one hand and between miR-155 and BMI on the other (Spearman ρ = 0.50; P < .01) (Figure 3, B and C).
miR-155 affects adipocyte biology
To study the effect of a modulation of miR-155 expression on adipocyte biology, we performed transfections with oligonucleotides mimicking miR-155 (miR-155 mimic) or oligonucleotides inhibiting miR-155 (miR-155 antagomir). In a validation experiment, we studied the effects of these compounds on miR-155 expression in basal conditions. As expected, miR-155 was strongly induced in 3T3-L1 subjected to miR-155 mimic transfection (113-fold increase vs control) (Supplemental Figure 1A), whereas in the presence of miR-155 antagomir, we observed a decrease in miR-155 expression (1.4-fold vs control) (Supplemental Figure 1B). Interestingly, miR-155 antagomir reduced expression of miR-155 under TNFα treatment (by 3.48-fold) (Supplemental Figure 1C).
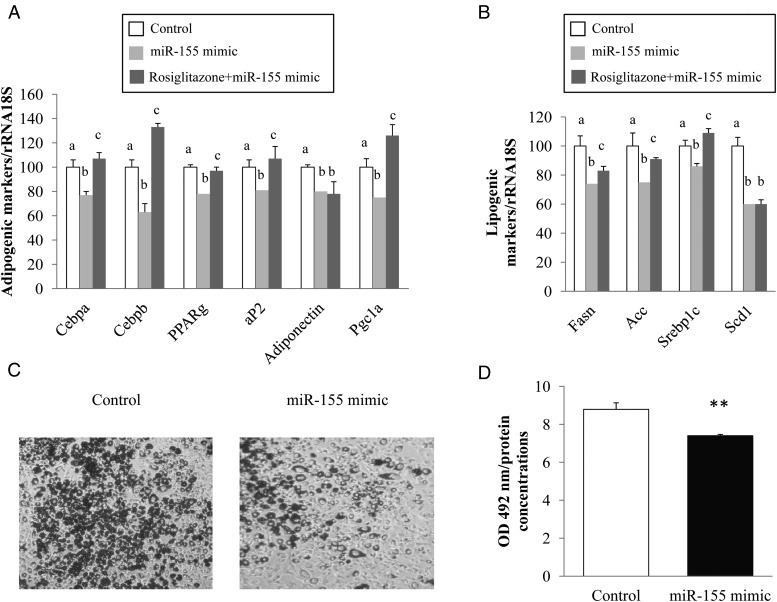
We went on to determine the effect of miR-155 mimic transfection on several key genes of the adipocyte phenotype. For this purpose, the mRNA levels of CCAAT enhancer binding protein-α (Cebpa), CCAAT enhancer binding protein-β (Cebpb), peroxisome proliferator-activated receptor gamma (Pparg), adipocyte fatty acid binding protein (aP2), adiponectin and PPARγ-coactivator 1α (Pgc-1a) were quantified by RT-qPCR. Interestingly, the expression of all these genes decreased in adipocytes transfected with miR-155 mimic compared with controls (1.3-, 1.6-, 1.3-, 1.23-, 1.25- and 1.33-fold, respectively) (Figure 4A). In line with this finding, intracellular lipid content, quantified by Oil Red O staining, was significantly reduced by miR-155 mimic compared with controls (Figure 4, C and D). We also studied the effect of miR-155 mimic on lipogenesis. The mRNA levels of fatty acid synthase (Fasn), acetyl coenzyme A carboxylase (Acc), sterol responsive element binding protein 1c (Srebp1c) and stearoyl coenzyme A desaturase 1 (Scd1) were quantified by RT-qPCR. The expression of these genes decreased in adipocytes transfected with miR-155 mimic compared with controls (1.35-, 1.33-, 1.16- and 1.67-fold, respectively) (Figure 4B). We also investigated the preventive effect of rosiglitazone, a well-known thiazolidinedione on miR-155 mimic, 3T3-L1 adipocytes were incubated with rosiglitazone (0.1μM, 24 h) followed by a 24-hour transfection with miR-155 mimic. PCR analysis showed that rosiglitazone significantly blunted the down-regulation of Cebpa, Cebpb, Pparg, aP2, Pgc1a, Fasn, Acc, and Srebp1c compared with miR-155 mimic condition (1.4, 2.1, 1.2, 1.3, 1.6, 1.2, 1.2, and 1.3-fold, respectively). This effect was not significant for adiponectin and Scd1 (Figure 4, A and B).
Figure 4.
Effect of miR-155 mimic and rosiglitazone on expression of adipogenic and lipogenic markers and lipid accumulation in 3T3–L1 adipocytes. 3T3–L1 adipocytes were incubated with or without rosiglitazone (0.1 μM, 24 h) followed by a 24-h transfection with miR-155 mimic. A, The mRNA levels of Cebpa, Cebpb, Pparg, aP2, adiponectin, and Pgc1a were quantified through qPCR. B, The mRNA levels of Fasn, Acc, Srebp1c, and Scd1 were quantified through qPCR. C, 3T3–L1 adipocytes were transfected with miR-155 mimic. Twenty-four hours after transfection, intracellular lipid was stained with Oil Red O. D, the stained Oil Red O was extracted with isopropanol. The absorbance of the extracted Oil Red O was spectrophotometrically determined at 492 nm after solubilization in isopropanol, and normalized to protein concentrations. The data are expressed as relative expression ratios of at least three independent experiments. The values are presented as the means ± SEM. **, P < .01 compared with control. Bars not sharing the same letters are significantly different P < .05.
Conversely, we also studied the effect of miR-155 antagomir transfection on the same adipocyte genes in the inflammatory conditions induced by TNFα. Cebpa, Pparg, aP2, adiponectin, Fasn, Acc, Srebp1c, and Scd1 were quantified by RT-qPCR. The expression of all these genes decreased in adipocytes treated with TNFα compared with controls (6.3, 2.8, 2.9, 4.2, 3.8, 2.5, 1.4, and 8.3-fold, respectively) (Supplemental Figure 2, A and B). Interestingly, miR-155 antagomir significantly limited the decrease in Pparg, adiponectin, Fasn, Acc, Srebp1c, and Scd1 expression induced by TNFα, but this decrease did not reach statistical significance in the case of Cebpa and aP2.
Taken together, these results suggest that miR-155 can reproduce the effects of TNFα, and conversely, miR-155 inhibition by an antagomir partially limits the effects of the TNFα on the expression of adipogenic and lipogenic markers.
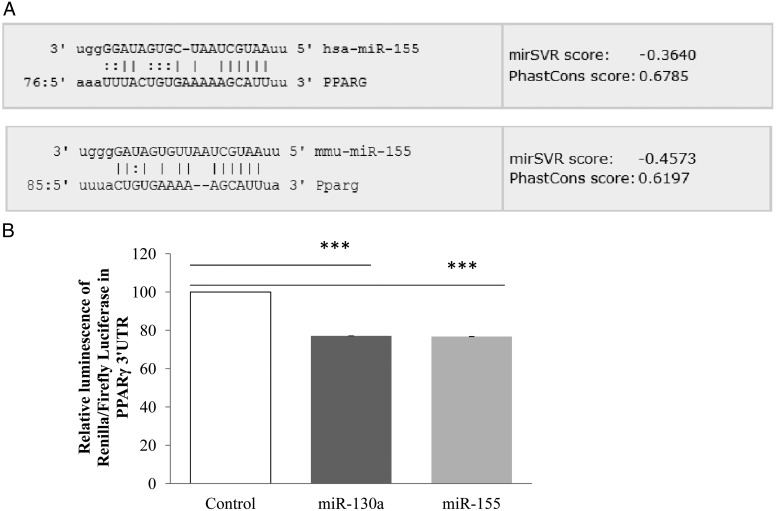
miR-155 directly targets PPARγ
Because most of the genes tested, except for CEBPβ [a direct target of miR-155 (25)] are under the control of PPARγ, we speculated that PPARγ might be a direct target of miR-155. We thus screened the PPARγ 3′UTR region (of human and mouse origin), using microrna.org software and we identified PPARγ 3′UTR as a putative miR-155 target (Figure 5A). To confirm the effect of miR-155 on PPARγ, we transfected PPARγ 3′UTR with miR-155 mimic. miR-130a was used as a positive control for the experiment, given that it has already been reported to target PPARγ (26). The ratio of Renilla to firefly luciferase activity was significantly decreased in both miR-155 and miR-130a (1.3-fold vs control) (Figure 5B), validating our experimental conditions, and strongly suggesting that PPARγ may be considered as a miR-155 target.
Figure 5.
miR-155 directly targets PPARγ. A, PPARγ 3′UTR region (of human and mouse origin) was analyzed in silico by microrna.org software. B, PPARγ 3′UTR cloned in psiCHECK2 vector was transfected with miR-155 mimic in 3T3–L1 adipocytes for 24 h. miR-130a was used as a positive control for experiments. Renilla activity was under the control of PPARγ 3′UTR, and firefly activity was used for normalization. The data are expressed as relative expression ratios of at least three independent experiments. The values are presented as means ± SEM. ***, P < .001 compared with control.
miR-155 induces inflammatory response, chemokine expression, and macrophage migration in 3T3-L1 adipocytes
To study the global effect of an overexpression of miR-155, microarray experiments were performed with RNA extracted from 3T3-L1 adipocytes transfected with miR-155 mimic. The data were analyzed using MetaCore software. This analysis revealed that immune response linked to toll-like receptor signaling, NF-κB activation, chemotaxis, and chemokines were overrepresented compared with controls (P < .05; 22 pathway maps are shown in Supplemental Table 2). Several maps linked to chemokines and chemokine receptors were strongly affected after miR-155 transfection, namely “Chemotaxis_CCL19- and CCl21-mediated chemotaxis,” “Cell adhesion_Chemokines and adhesion,” “Signal transduction_Soluble CXCL16 signaling,” “Immune response_CXCR4 signaling via second messenger,” “Immune response_CCR5 signaling in macrophages and T lymphocytes,” “Chemotaxis_CCR1 signaling,” “Chemotaxis_CXCR4 signaling pathway,” and “Immune response_CCR3 signaling in eosinophils” (Supplemental Table 2). MetaCore analysis according to process networks confirmed that “Cell adhesion_Leucocyte chemotaxis” was significantly overrepresented (P < .05), and among genes up-regulated in this network, we identified several chemokines (Ccl5, Ccl17, Ccl13, Ccl19, Cx3cl1, GRO-2, Cxcl13) (data not shown). Interestingly, we also observed that Il-10 expression was down-regulated in our set of data. The regulation of some of these genes, randomly chosen (Ccl5, Cxcl1 and Il-10) was confirmed by qPCR (Supplemental Figure 3). Finally, a GSEA analysis also confirmed that several pathways or gene families related to inflammation (response to virus, I κb kinase NF-κb cascade, positive regulation of I kappab kinase NF-κb cascade, defense response) were strongly overrepresented in miR-155-transfected cells (Supplemental Table 3).
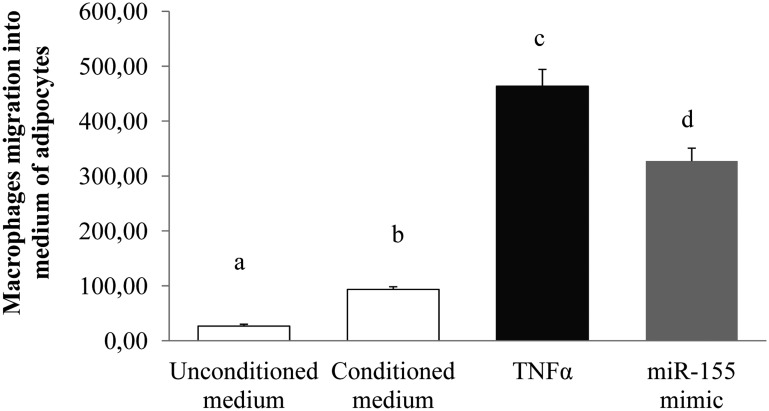
To examine the functional consequences of chemokine expression modulations under miR-155 mimic effect, macrophage migration experiments were performed using unconditioned medium as control or adipocyte-conditioned media. In presence of miR-155 mimic, 3T3-L1-conditioned medium induced a significant migration of macrophages (3.5-fold), similarly with those obtained in TNFα adipocytes conditioned medium condition (Figure 6).
Figure 6.
miR-155 induces macrophages migration into 3T3–L1 adipocytes. 3T3–L1 adipocytes were incubated with TNFα or transfected with miR-155 mimic for 24 h. RAW 264.7 macrophages migration in 3T3–L1-conditioned media (4 h, 37°C) was subsequently performed. The values are presented as means ± SEM. Bars not sharing the same letters are significantly different, P < .05.
Discussion
In the present study, using miRNA expression microarrays, we found that miR-155 was up-regulated by TNFα in human adipocyte primocultures. miR-155 is processed from an exon of noncoding RNA transcribed from the B-cell integration cluster gene located on chromosome 21 (27). This miR is considered as a multifunctional miRNA (28), given that it has been associated with the regulation of different immune-related processes, such as hematopoiesis (29), innate immunity (30), B-cell and T-cell differentiation (31), and cancer (32).
The up-regulation of miR-155 by TNFα was confirmed by qPCR in the human and murine mature 3T3-L1 adipocytes and in 3T3-L1 adipocytes supernatants. Moreover, the contribution of TNFα in the regulation of miR-155 was confirmed in vivo by using Tnfa−/− mice (23) that displayed a reduced miR-155 expression in adipose tissue, suggesting that miR-155 expression level is controlled by the TNFα level in adipose tissue. The up-regulation of miR-155 was also observed in biopsies of obese subjects where the induction of miR-155 was correlated with TNFα expression and to BMI. It is noteworthy that obesity-associated accumulation of fat mass (reflected by the increase in BMI) is strongly linked with an increased expression of proinflammatory markers such as TNFα (33). These results are fully consistent with the literature describing the ability of TNFα to induce the expression of miR-155 in various cell types such as retinal pigment epithelial cells (34), mesanglial cells (35), synovial fibroblasts (36), preadipocytes (37), and adipocytes (12). Altogether, these data strongly suggest that the induction of miR-155 mediated by TNFα in adipocytes and in adipose tissue could explain at least in part the observed induction of miR-155 in inflamed adipose tissue of obese subjects.
From a molecular point of view, we found that the miR-155 promoter was activated by TNFα. This finding suggests that the TNFα-mediated regulation of miR-155 is at least in part due to an increased transcription of miR-155, although a stabilization of pri or premiR-155 cannot be excluded. In addition, this transcriptional induction of miR-155 seems to be mediated at least in part by NF-κB, given that the use of specific NF-κB inhibitor blunted the TNFα-mediated induction of miR-155. Conversely, we identified in silico putative response elements of NF-κB: although the functionality of this response element is at present not established, these data are consistent with several studies describing the role of NF-κB. The NF-κB signaling pathway has been described as a major regulator of the expression of miR-155 in different cellular models, such as macrophages (30), monocytes (38), fibroblasts (39) or trophoblast cells (40). In addition, Liu et al (37) have shown that TNFα up-regulates miR-155 expression into 3T3-L1 preadipocytes through the NF-κB binding site located 1.01 kb upstream of the miR-155 transcription start site, which seems to be different from the putative binding site we identified in silico. To confirm the involvement of NF-κB signaling in the induction of miR-155, we used a aP2-p65 transgenic mouse model that displays a constitutive overexpression of p65 in adipose tissue (22). In line with this finding, we observed a strong up-regulation of miR-155, closely correlated with Tnfa expression, in epidydimal adipose tissue of these mice. Conversely, inactivation of p65 in adipose tissue (F-p65-KO mice), resulted in a down-regulated miR-155. Taken together, these results confirm that NF-κB activation can mediate miR-155 expression.
To gain further insight into the effect of the overexpression of miR-155 on adipocyte function we used a gain or loss of function approach, through the transfection of oligonucleotides mimicking or inhibiting miR-155. Using this strategy, we found that an overexpression of miR-155 could decrease expression levels of adipogenic (Pparg, Cebpa, aP2 and adiponectin) and lipogenic markers (Fasn, Srebp1c, Acc and Scd1) in mature adipocytes, which could be likened to a dedifferentiation process, as highlighted by the decrease in intracellular lipid droplets in differentiated adipocytes incubated with miR-155 mimic. It is noteworthy that rosiglitazone, a well-known thiazolidinedione partly blunted the effect of miR-155 mimic.
Conversely, antagomir partly blunted the effect of TNFα on these adipogenic and lipogenic markers, suggesting that the effects of TNFα on them are partly mediated by miR-155. Although the ability of miR-155 to block adipogenesis has already been reported in 3T3-L1 preadipocytes (37) and in brown preadipocytes (41), its ability to affect the expression of adipogenic markers in mature adipocytes is reported here for the first time. This effect could be due to the modulation of PPARγ mediated by miR-155. We observed that PPARγ expression decreased by miR-155 overexpression, and the TNFα-mediated down-regulation of PPARγ was partly blunted by antagomir. Direct regulation of Pparg mRNA expression by miR-155 is strongly suggested by the identification and demonstration of the functionality of a recognition element for miR-155 in the 3′UTR of PPARγ (identified in silico). To confirm the effect of miR-155 on PPARγ, we transfected PPARγ 3′UTR with miR-155 mimic. Results suggest that PPARγ may be considered as a miR-155 target, but we cannot exclude an indirect effect of mir-155 on PPARγ via CEBPβ. This transcription factor is also targeted by miR-155 (25, 37, 42) and is down-regulated in our conditions. In addition, the ability of CEBPβ to regulate PPARγ has been demonstrated in preadipocytes (43). Whether such regulation also occurs in mature adipocytes is at present not clear, and will require further experiments.
This regulation of PPARγ by TNFα through miR-155 could represent a new mechanism of regulation in addition to others already reported (44), involving a direct down-regulation of CEBPδ and a consequent down-regulation of PPARγ, a loss of DNA-binding activity of PPARγ and an activation of corepressors by TNFα.
Using a microarray approach, we demonstrated that miR-155 overexpression in adipocytes was associated with an increase in inflammatory tone. These new findings are in line with the literature, where miR-155 expression has been associated with inflammatory diseases such as rheumatoid arthritis (36), vascular inflammation (45), and inflammation in the nervous system (46). In this study, we showed that overexpression of miR-155 in adipocytes induced a global inflammatory response as highlighted by Metacore and GSEA analysis, and more specifically increased the expression of a large set of chemokines together with a reduced Il-10 expression, which displays an anti-inflammatory effect (47). Interestingly, chemokines defined as “cytokines with selective chemoattractant properties,” coordinate leukocyte movement to sites of inflammation or injury (48). In obesity, chemokines are secreted notably by inflamed adipocytes, and are involved in leukocyte recruitment in adipose tissue, thus strongly participating in the establishment of the proinflammatory status (49). In agreement we demonstrated that overexpression of miR-155 in adipocytes increased macrophage migration mediated by the conditioned medium of adipocytes. From a molecular point of view, the regulation of the expression of chemokines under miR-155 effect could be mediated at least in part via the miR-155-mediated down-regulation of PPARγ. It has been reported that knock-down of adipocyte PPARγ resulted in up-regulation of chemokine expression (50), and so we could speculate that by targeting PPARγ expression, miR-155 mediated up-regulation of chemokines and overall inflammatory process, given that the anti-inflammatory effect of PPARγ is well established (51).
Altogether, these data demonstrate that miR-155 is induced by inflammation and that miR-155, in turn, participates in the overall effect of TNFα, and more globally to the amplification of the inflammatory phenotype. The control of miR-155 expression in adipose tissue could thus offer new ways to attenuate inflammatory state during obesity.
Acknowledgments
We thank Laurence Louis (UMR 910 INSERM, Marseille) for technical assistance with the microarray experiments.
Author contributions: J.-F.L. and J.Y. designed the experiments; E.K., J.A., M.E.A., B.R., E.G., L.W., J.S., and S.W. carried out experiments and analyzed data; E.K., F.T., P.B., and J.F.L jointly drafted the paper; J.-F.L. takes the full responsibility for the work as a whole.
This work was partially funded through a grant from the Fondation Coeur et Artères to J.-F.L., and National Institutes of Health Grants DK068036 and R56 DK068036-10A1 to Y.J.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- Acc
- acetyl coenzyme A carboxylase
- aP2
- adipocyte fatty acid binding protein
- BMI
- body mass index
- bp
- base pair
- Cebpa
- CCAAT enhancer binding protein-α
- Cebpb
- CCAAT enhancer binding protein-β
- CT
- comparative cycle threshold
- Fasn
- fatty acid synthase
- FBS
- fetal bovine serum
- MAP
- mitogen-activated protein
- NF-κB
- nuclear factor κB
- Pgc-1a
- and PPARγ-coactivator 1α
- PPARγ
- peroxisome proliferator-activated receptor γ
- Scd1
- stearoyl coenzyme A desaturase 1
- Srebp1c
- sterol responsive element binding protein 1c.
References
- 1. Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–445. [DOI] [PubMed] [Google Scholar]
- 2. Arner P, Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol. 2015;11:276–288. [DOI] [PubMed] [Google Scholar]
- 3. Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. [DOI] [PubMed] [Google Scholar]
- 4. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. [DOI] [PubMed] [Google Scholar]
- 5. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- 6. Ge Q, Brichard S, Yi X, Li Q. microRNAs as a new mechanism regulating adipose tissue inflammation in obesity and as a novel therapeutic strategy in the metabolic syndrome. J Immunol Res. 2014;2014:987285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Strum JC, Johnson JH, Ward J, et al. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol. 2009;23:1876–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arner E, Mejhert N, Kulyté A, et al. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes. 2012;61:1986–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lorente-Cebrian S, Mejhert N, Kulyte A, et al. MicroRNAs regulate human adipocyte lipolysis: Effects of miR-145 are linked to TNF-alpha. PLoS One. 2014;9:e86800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shi C, Zhu L, Chen X, et al. IL-6 and TNF-α induced obesity-related inflammatory response through transcriptional regulation of miR-146b. J Interferon Cytokine Res. 2014;34:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim C, Lee H, Cho YM, Kwon OJ, Kim W, Lee EK. TNFalpha-induced miR-130 resulted in adipocyte dysfunction during obesity-related inflammation. FEBS Lett. 2013;587(23):3853–3858. [PubMed] [Google Scholar]
- 12. Ortega FJ, Moreno M, Mercader JM, et al. Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants. Clin Epigenetics. 2015;7:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Landrier JF, Gouranton E, El Yazidi C, et al. Adiponectin expression is induced by vitamin E via a peroxisome proliferator-activated receptor gamma-dependent mechanism. Endocrinology. 2009;150:5318–5325. [DOI] [PubMed] [Google Scholar]
- 14. Karkeni E, Marcotorchino J, Tourniaire F, et al. Vitamin d limits chemokine expression in adipocytes and macrophage migration in vitro and in male mice. Endocrinology. 2015;156:1782–1793. [DOI] [PubMed] [Google Scholar]
- 15. Marcotorchino J, Romier B, Gouranton E, et al. Lycopene attenuates LPS-induced TNF-α secretion in macrophages and inflammatory markers in adipocytes exposed to macrophage-conditioned media. Mol Nutr Food Res. 2012;56:725–732. [DOI] [PubMed] [Google Scholar]
- 16. Gao Z, Zhang J, Henagan TM, et al. P65 inactivation in adipocytes and macrophages attenuates adipose inflammatory response in lean but not in obese mice. Am J Physiol Endocrinol Metab. 2015;308:E496–E505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Landrier JF, Gouranton E, Reboul E, et al. Vitamin E decreases endogenous cholesterol synthesis and apo-AI-mediated cholesterol secretion in Caco-2 cells. J Nutr Biochem. 2010;21:1207–1213. [DOI] [PubMed] [Google Scholar]
- 18. Gouranton E, Aydemir G, Reynaud E, et al. Apo-10′-lycopenoic acid impacts adipose tissue biology via the retinoic acid receptors. Biochim Biophys Acta. 2011;1811:1105–1114. [DOI] [PubMed] [Google Scholar]
- 19. Milenkovic D, Deval C, Gouranton E, et al. A Modulation of miRNA expression by dietary polyphenols in apoE deficient mice: A new mechanism of the action of polyphenols. PLoS One. 2012;7:e29837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Romier B, Tourniaire F, Marcotorchino J, et al. Bioeffects of a combination of trace elements on adipocyte biology. Metallomics. 2013;5:524–531. [DOI] [PubMed] [Google Scholar]
- 21. Tourniaire F, Romier-Crouzet B, Lee JH, et al. Chemokine expression in inflamed adipose tissue is mainly mediated by NF-κB. PLoS One. 2013;8:e66515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tang T, Zhang J, Yin J, et al. Uncoupling of inflammation and insulin resistance by NF-kappaB in transgenic mice through elevated energy expenditure. J Biol Chem. 2010;285:4637–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Salles J, Tardif N, Landrier JF, et al. TNFα gene knockout differentially affects lipid deposition in liver and skeletal muscle of high-fat-diet mice. J Nutr Biochem. 2012;23:1685–1693. [DOI] [PubMed] [Google Scholar]
- 24. Landrier JF, Malezet-Desmoulins C, Reboul E, Marie Lorec A, Josephe Amiot M, Borel P. Comparison of different vehicles to study the effect of tocopherols on gene expression in intestinal cells. Free Radic Res. 2008;42:523–530. [DOI] [PubMed] [Google Scholar]
- 25. Worm J, Stenvang J, Petri A, et al. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Res. 2009;37:5784–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee EK, Lee MJ, Abdelmohsen K, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2011;31:626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. [DOI] [PubMed] [Google Scholar]
- 28. Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: A typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497–505. [DOI] [PubMed] [Google Scholar]
- 29. Masaki S, Ohtsuka R, Abe Y, Muta K, Umemura T. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem Biophys Res Commun. 2007;364:509–514. [DOI] [PubMed] [Google Scholar]
- 30. O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kluiver J, Poppema S, de Jong D, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. [DOI] [PubMed] [Google Scholar]
- 32. Gironella M, Seux M, Xie MJ, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–16175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: Direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. [DOI] [PubMed] [Google Scholar]
- 34. Kutty RK, Nagineni CN, Samuel W, Vijayasarathy C, Hooks JJ, Redmond TM. Inflammatory cytokines regulate microRNA-155 expression in human retinal pigment epithelial cells by activating JAK/STAT pathway. Biochem Biophys Res Commun. 2010;402:390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Imaizumi T, Tanaka H, Tajima A, et al. IFN-γ and TNF-α synergistically induce microRNA-155 which regulates TAB2/IP-10 expression in human mesangial cells. Am J Nephrol. 2010;32:462–468. [DOI] [PubMed] [Google Scholar]
- 36. Stanczyk J, Pedrioli DM, Brentano F, et al. Altered expression of microRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. [DOI] [PubMed] [Google Scholar]
- 37. Liu S, Yang Y, Wu J. TNFα-induced up-regulation of miR-155 inhibits adipogenesis by down-regulating early adipogenic transcription factors. Biochem Biophys Res Commun. 2011;414:618–624. [DOI] [PubMed] [Google Scholar]
- 38. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sheedy FJ, O'Neill LA. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann Rheum Dis. 2008;67 Suppl 3:iii50–iii55. [DOI] [PubMed] [Google Scholar]
- 40. Dai Y, Diao Z, Sun H, Li R, Qiu Z, Hu Y. MicroRNA-155 is involved in the remodelling of human-trophoblast-derived HTR-8/SVneo cells induced by lipopolysaccharides. Hum Reprod. 2011;26:1882–1891. [DOI] [PubMed] [Google Scholar]
- 41. Chen Y, Siegel F, Kipschull S, et al. A miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun. 2013;4:1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. He M, Xu Z, Ding T, Kuang DM, Zheng L. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPbeta. Cell Mol Immunol. 2009;6:343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lefterova MI, Zhang Y, Steger DJ, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ye J. Regulation of PPARγ function by TNF-α. Biochem Biophys Res Commun. 2008;374:405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Staszel T, Zapala B, Polus A, et al. Role of microRNAs in endothelial cell pathophysiology. Pol Arch Med Wewn. 2011;121:361–366. [PubMed] [Google Scholar]
- 46. Cardoso AL, Guedes JR, Pereira de Almeida L, Pedroso de Lima MC. miR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology. 2012;135:73–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Isomaki P, Luukkainen R, Saario R, Toivanen P, Punnonen J. Interleukin-10 functions as an antiinflammatory cytokine in rheumatoid synovium. Arthritis Rheum. 1996;39:386–395. [DOI] [PubMed] [Google Scholar]
- 48. Viola A, Luster AD. Chemokines and their receptors: Drug targets in immunity and inflammation. Annu Rev Pharmacol Toxicol. 2008;48:171–197. [DOI] [PubMed] [Google Scholar]
- 49. Lolmède K, Duffaut C, Zakaroff-Girard A, Bouloumie A. Immune cells in adipose tissue: Key players in metabolic disorders. Diabetes Metab. 2011;37:283–290. [DOI] [PubMed] [Google Scholar]
- 50. Nguyen MT, Chen A, Lu WJ, et al. Regulation of chemokine and chemokine receptor expression by PPARγ in adipocytes and macrophages. PLoS One. 2012;7:e34976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martin H. Role of PPAR-γ in inflammation. Prospects for therapeutic intervention by food components. Mutat Res. 2010;690:57–63. [DOI] [PubMed] [Google Scholar]