Abstract
The injection of computer-simulated conductances through the dynamic clamp technique has allowed researchers to probe the intercellular and intracellular dynamics of cardiac and neuronal systems with great precision. By coupling computational models to biological systems, dynamic clamp has become a proven tool in electrophysiology with many applications, such as generating hybrid networks in neurons or simulating channelopathies in cardiomyocytes. While its applications are broad, the approach is straightforward: synthesizing traditional patch clamp, computational modeling, and closed-loop feedback control to simulate a cellular conductance. Here, we present two example applications: artificial blocking of the inward rectifier potassium current in a cardiomyocyte and coupling of a biological neuron to a virtual neuron through a virtual synapse. The design and implementation of the necessary software to administer these dynamic clamp experiments can be difficult. In this chapter, we provide an overview of designing and implementing a dynamic clamp experiment using the Real-Time eXperiment Interface (RTXI), an open- source software system tailored for real-time biological experiments. We present two ways to achieve this using RTXI’s modular format, through the creation of a custom user-made module and through existing modules found in RTXI’s online library.
Keywords: Dynamic clamp, RTXI, Cardiac electrophysiology, Neural electrophysiology, Neuronal networks, Artificial conductance block, Reciprocal neuronal coupling
1 Introduction
1.1 Dynamic Clamp
The dynamic clamp technique is an extension of traditional intra-cellular recording techniques that allows for the introduction of an artificial conductance, controlled by the experimentalist, into a biological cell [1–5]. Dynamic clamp uses closed-loop feedback control between a cell’s recorded membrane potential and an injected current calculated by a mathematical model (Fig. 1). This feedback loop, when iterated quickly and accurately enough, is sufficient to mimic one or more biological conductances in the targeted cell. Consequently, a versatile dynamic clamp system allows investigators to simultaneously control many electrophysiological parameters within a single experiment.
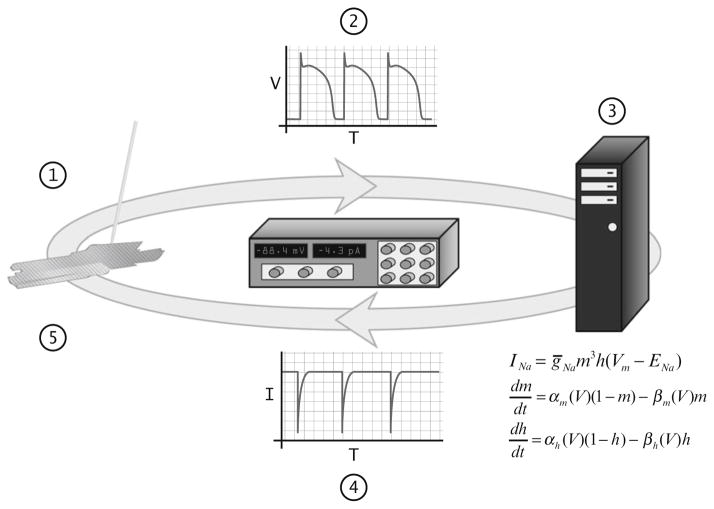
Fig. 1.
The fundamental steps of the dynamic clamp feedback loop when injecting an artificial conductance. When run at high enough rates (10–50 kHz), this feedback loop can sufficiently mimic a biological conductance. (Step 1 ) Membrane voltage from a cell is measured using the patch clamp technique and (Step 2) sent from the amplifier to the computer’s data acquisition system. (Step 3 ) Voltage data is used in a computational current model to calculate current, represented here by a sodium current. (Step 4) Calculated current data is sent to the patch clamp amplifier and (Step 5) injected into the cell
1.2 Real-Time eXperiment Interface
At the core of the dynamic clamp experimental setup is the software, which acts as the interface between the mathematical models and the patch-clamp hardware used to measure the parameters of the biological cell. Rapid sampling of membrane potential and fast computation of the conductance is required to adequately recapitulate a conductance [6]. Due to these requirements, real-time control is required to accomplish the dynamic clamp technique. For numerous technical reasons, real-time control is not possible on standard operating systems and software. Thus, a specialized operating system and software are required. RTXI is a fast and versatile software system adopted by many labs across many fields of electrophysiology to administer dynamic clamp experiments. The software provides real-time control using a real-time Linux operating system (Fig. 2). Free and open-source, the platform has been tailored to fit the needs of the biological research community. RTXI uses a modular architecture, which allows investigators to construct complex experimental protocols using a combination of built-in, community-shared, and self-made modules. More information on the software, installation, and use can be found on its Web site (www.rtxi.org).
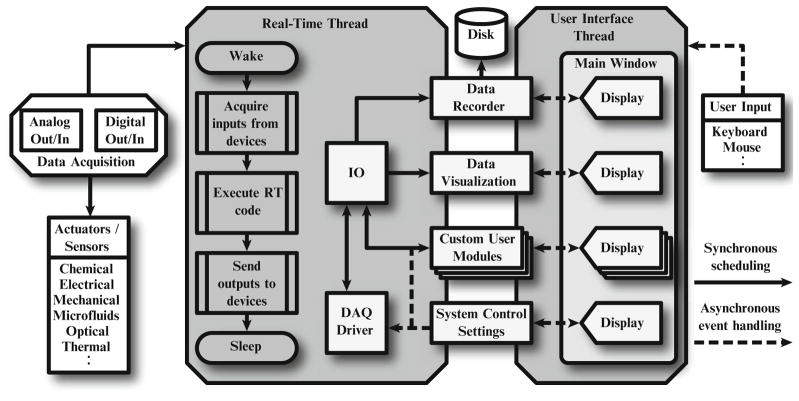
Fig. 2.
Diagram of the RTXI software architecture. RTXI uses a two thread architecture where modules communicate and share data in real-time through a signal and slot system. On every clock cycle, a prioritized real-time thread wakes and sequentially runs its tasks—data acquisition, execution of real-time instructions of loaded modules, and signal output. Once all tasks are complete, the thread sleeps until the following cycle. Data acquisition and output is performed using the open-source COMEDI (www.comedi.org) drivers, which allows RTXI to interface with many multifunction data acquisition cards. A separate user interface thread is responsible for all graphical components and user input. This approach allows for all real-time instructions to take priority over graphical components during computation
1.3 Simulating Artificial Conductance
In this chapter, we detail two different ways to build a dynamic clamp protocol using RTXI. In the first case, dynamic clamp will be used to artificially block a potassium conductance in a cardiac myocyte (Fig. 3a). By injecting a conductance with the opposite magnitude of a biologically present current, an artificial block can be achieved. This type of experiment is useful for testing the validity of a computational model in a biological setting, and when no available drugs block a current specifically. In this chapter, the described experimental protocol will use a custom module developed based on the equations describing the cardiac inward rectifier current (IK1). We will detail the steps required to convert the mathematical equations of a current model into a RTXI module capable of implementing this dynamic clamp experiment. In the second case, dynamic clamp will be used to construct a hybrid network by reciprocally coupling two neurons through alpha synapses (Fig. 3b). This hybrid network will consist of one real patch- clamped neuron, and a virtual Hodgkin–Huxley neuron model. Rather than manually constructing a module to accomplish this experiment, the protocol will be developed using already available modules: bundled with RTXI or available through its online library.
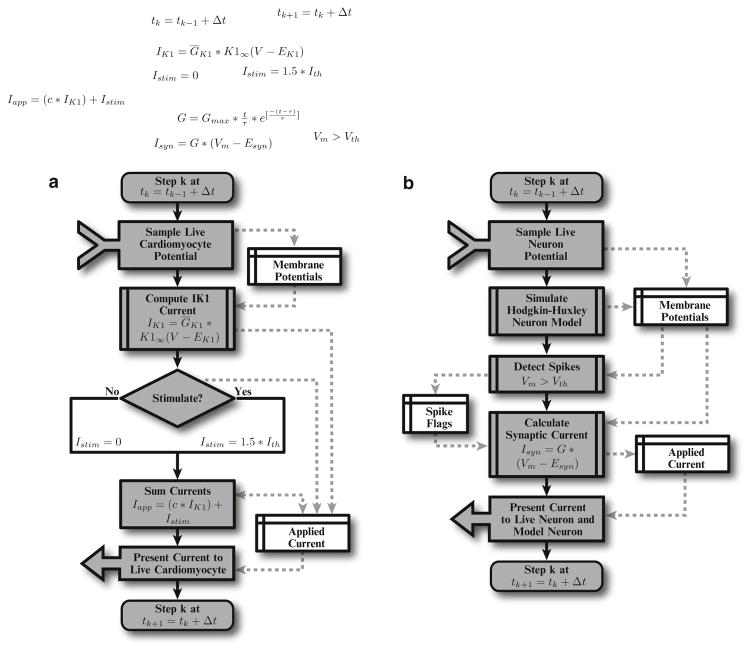
Fig. 3.
Flowcharts depicting dynamic clamp and RTXI module activity for a time step. The time step Δt is a parameter that would ideally be at most 1/20 the smallest time constant in the current model. (a) Overview of the dynamic clamp procedure to block the IK1 current in cardiomyocytes. Voltage is sampled from the myocyte and passed to a module that computes the IK1 current. Stimulation is provided at a set cycle length, typically for 1 ms at 1.5× the determined threshold to invoke action potentials. The stimulation and the IK1 current, scaled by the parameter c (for full block, c = −1), is then injected into the cardiomyocyte. (b) Overview of the dynamic clamp procedure to reciprocally couple a live neuron and a virtual neuron by virtual alpha synapses to construct a neural network. Voltage is sampled from a live neuron and a simulated Hodgkin–Huxley neuron model. When the membrane potential of either the model or virtual neuron passes a certain threshold (Vth) indicating a spike has occurred, the spike flag is updated. Using the spike information and membrane potential, the synaptic current is computed and injected into the live neuron and model neuron
This chapter focuses on the setup, customization, and use of the RTXI platform to implement a dynamic clamp experiment. As such, it is assumed the reader is familiar with the example applications, cardiomyocyte conductances and hybrid neuronal networks, and with the more general techniques of patch clamping.
2 Materials
In this list of materials, Subheadings 2.1–2.3 are collections of the absolute essential materials required to prepare and patch-clamp isolated cardiomyocytes or neurons in slice. Subheading 2.4 describes the required patch clamp solutions needed in both experimental settings, cardiac and neural. Subsequent sections deal with the materials specific to dynamic clamp. Subheading 2.5 lists the required components, hardware and software, for a digital dynamic clamp system run on a personal computer.
2.1 Cardiomyocyte Isolation Equipment
Anesthetic appropriate for animal model (e.g., sodium pentobarbital for guinea pigs).
Heparin 1,000 units/ml.
Langendorff perfusion system containing water bath, heat exchanger, peristaltic pump, bubble trap, and tubing.
Gas dispersion tube with fritted cylinder and medical grade oxygen.
Aortic cannula, appropriately sized for animal model.
Surgical scissors, forceps, two fine bent forceps, scalpel, bulldog clip, and surgical suture.
50 ml conical tubes and corresponding centrifuge.
250 μm cell strainer.
Shaking water bath.
Collagenase, Type II and Protease Type XIV.
Common Tyrode’s solution, calcium-free Tyrode’s solution, 1-Enzyme solution, 2-Enzyme solution, and wash solution (Table 1) made with endotoxin-free water.
Table 1.
Cardiomyocyte isolation solutions
| Solution | Common Tyrode’s | Calcium-free Tyrode’s | 1-Enzyme | 2-Enzyme | Wash |
|---|---|---|---|---|---|
| CaCl2 (mM) | 2 | 0.025 | 0.025 | 0.375 | |
| EGTA (μM) | 100 | ||||
| Collagenase (mg/ml) | 1 | 1 | |||
| Protease XIV (mg/ml) | 0.05 | ||||
| BSA (mg/ml) | 5 |
In addition, all solutions are buffered to a pH of 7.2 with NaOH and contain the following: 130 mM NaCl, 5.4 mM KCl, 3.5 mM MgCl2, 0.4 mM NaH2PO4, 5 mM HEPES, and 10 mM Dextrose EGTA ethyleneglycolbis-N,N,N′, N′-tetraacetic acid, BSA bovine serum albumin
2.2 Brain Tissue Slice Preparation Equipment
Air tight anesthesia chamber with isoflurane.
Appropriately sized animal guillotine.
Surgical scissors, Scoopula, scalpel.
Horizontal serial slicer (vibratome) with walled slice chamber.
Filter paper, fine artist’s paint brush.
Bubble chamber with tubing and oxygen.
Artificial cerebrospinal fluid (ACSF) (Table 2; see Note 1).
Table 2.
Patch clamp solutions
| Solution | Superfusion Tyrode’s | ACSF | IC cardiomyocyte | Artifi cial neuroplasm |
|---|---|---|---|---|
| NaCl (mM) | 137 | 126 | 2 | |
| KCl (mM) | 4.5 | 3 | 142 | |
| MgCl2 (mM) | 2 | 2 | ||
| MgSO4 (mM) | 1 | 2 | ||
| NaH2PO4 (mM) | 1.25 | |||
| NaHCO3 (mM) | 26 | |||
| K-Gluconate (mM) | 135 | |||
| Dextrose (mM) | 10 | 10 | ||
| CaCl 2 (mM) | 2 | 2 | 3.8 | |
| HEPES (mM) | 10 | 10 | 10 | |
| EGTA (mM) | 10 | 1.25 | ||
| Tris–ATP (mM) | 2 | |||
| Tris–GTP (mM) | 0.4 | |||
| Na2ATP (mM) | 5 | |||
| pH | 7.35 | 7.4 | 7.1 | 7.25 |
| Osmolarity (mOsm) | 300 | 300 | 290 | 290 |
EGTA ethyleneglycol-bis-N,N,N′, N′-tetraacetic acid
2.3 Patch Clamp Hardware
Inverted microscope or infrared differential interference contrast microscope depending on application (see Note 2).
Stage chamber with temperature controller and influx/efflux tubing for extracellular solutions.
One micromanipulator.
One current-clamp amplifier (Axon Instruments MultiClamp 700B; Molecular Devices, USA) with head stage (see Note 3).
Borosilicate glass electrodes drawn on a horizontal puller. Pipette specifications are dependent on application (see Note 4).
2.4 Patch Clamp Solutions
2.5 Dynamic Clamp System
Personal computer running a hard real-time operating system (Real-Time Application Interface (RTAI) patched Linux Kernel) (see Note 5).
A personal computer with the dynamic clamp software, RTXI, installed (see Note 6).
Multipurpose A/D-D/A device installed in the computer (PCI-6259; National Instruments, USA) and shielded connector block (BNC-2110; National Instruments, USA) (see Note 7).
Bayonet Neill–Concelman (BNC) cables.
3 Methods
Subheading 3.1 describes the setup of a typical dynamic clamp system running RTXI, which revolves around adequate communication between RTXI and the patch clamp amplifier. The simplest setup uses a personal computer, running RTXI, interfacing with a patch clamp amplifier using an analog voltage input and output channel through a data acquisition (DAQ) device (Fig. 4). Most patch clamp amplifiers possess the necessary analog output and analog external input to interface with RTXI. Depending on the capabilities of the amplifier, greater integration can be achieved, such as automatic detection of amplifier mode or compensation for changes in output gain (see Note 8). Also, RTXI modules designed specifically for controlling several of the most common models of patch clamp amplifiers are available (see Note 9).
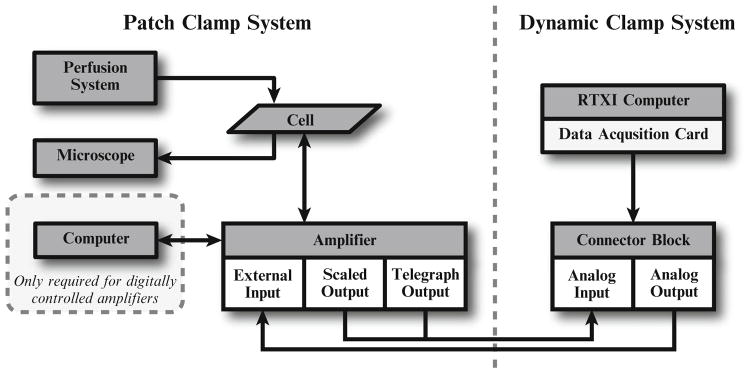
Fig. 4.
Diagram of a typical dynamic patch clamp setup. Arrows indicate flow and direction of information. The traditional patch clamp setup is shown on the left, including a computer required for digitally controlled amplifiers (see Note 7). The dynamic clamp system is shown on the right. The RTXI computer interfaces with the patch clamp amplifier through a connector block linked to a data acquisition card. BNC cables from the connector block to the amplifier connect the two devices
In subsequent sections, descriptions of two dynamic clamp experiments from the field of cardiac electrophysiology and neural electrophysiology are presented. These two experiments are accomplished using different approaches in development and execution of the experiment protocol. Subheading 3.2 describes the injection of an artificial potassium conductance, specifically IK1, into a cardiomyocyte. Focus is on development of a custom RTXI module written using the programming language C++. As shown in Fig. 3a, this module will sample membrane voltage, calculate a current based on equations describing the IK1 current, and inject this current into a patched cardiomyocyte. Subheading 3.3 describes the construction of a neuronal hybrid network with a real and virtual neuron through an alpha synapse (see Fig. 3b). Focus is on using already available RTXI modules, either built into RTXI or shared by the community. Each of these sections contains brief descriptions of cell/tissue preparation and patch clamping. More thorough descriptions can be found throughout this book.
3.1 Dynamic Clamp Setup
-
RTXI Installation:
RTXI requires a real-time Linux operating system (see Note 5). Typically, these operating systems are patched, compiled manually, and then installed on a personal computer. Afterwards, RTXI and other required software are installed on the real-time operating system. This process has been simplified through the use of a “Live USB.” The Live USB is a complete bootable real-time Linux operating system with RTXI pre-installed packaged onto a USB drive. Normally the Live USB is used to install the package onto a hard drive like any other operating system. However, it also can be run from the USB directly, resulting in a portable real-time software system. For more details on the installation of RTXI onto a personal computer, visit the installation Web page (www.rtxi.org/install).
-
Dynamic Clamp Hardware Setup:
The basic dynamic clamp setup is outlined in Fig. 4. Information is passed between RTXI and the amplifier through a data acquisition device. Several patch-clamp amplifier models are digitally controlled, in contrast to the traditional knob and button interface. The controlling software is usually only available on both Windows or Mac operating systems, and not Linux. In these situations, a second computer is required to control the amplifier.
Install the data acquisition card into the personal computer with RTXI installed, and attach the connector block.
Using a BNC cable, connect the labeled AO 0 (Analog Output 0) of the connector block to the external input of the amplifier.
Connect the labeled AI 0 (Analog Input 0) to the Output of the amplifier.
-
Interfacing RTXI and the Patch-Clamp Amplifier:
RTXI receives information from the amplifier in the form of a voltage signal read through a data acquisition device. Amplifiers apply various gains to these signals to achieve adequate signal to noise ratios. In order for RTXI to accurately read and send signals to the amplifier, these gains must be compensated for. For example, if a gain of 10 is applied to an amplifier output, the corresponding input must be reduced by a factor of 10 by RTXI. Unit mismatches are a frequent source of error. This section explains how to manually change the scaling values for the input/output signals in RTXI. As previously discussed, there are modules available that simplify or automate this process (see Note 9). In this and subsequent sections, various commands will need to be entered by the reader using the standard Linux terminal. These commands will be preceded by “$”.
-
Open RTXI with the terminal.
Open the terminal (System → Terminal in Ubuntu 9.10).
$ rtxi
-
Navigate to the system tab, and open the control panel.
System → Control Panel.
-
Activate the 0 channel for analog input and the 0 channel for analog output.
Set channel: Input|0.
Toggle the Active button.
Click the Apply button.
Repeat for channel: Output|0.
Adjust the scaling values accordingly (see Note 10).
-
Set thread rate of RTXI to 10 kHz (see Note 11).
Switch to Thread Tab.
Set Frequency: 10,000|Hz.
Click the Apply button.
-
3.2 Artificial Blocking of the Inward Rectifier K Current in Cardiomyocytes Through Dynamic Clamp
A dynamic clamp experiment to artificially block a current requires a computational model that adequately simulates the current in the target cell (Fig. 3a). In this example, the target cell is an isolated guinea pig ventricular myocyte. In this section, we use equations of the IK1 current from the Livsthitz Rudy 2009 guinea pig ventricular myocyte model [7]. From these equations, we describe a basic workflow to construct a RTXI module that can calculate the IK1 current in real-time (see Subheading 3.2.1). In Subheading 3.2.2 we then describe the isolation of ventricular myocytes from a guinea pig heart [8]. Finally, we implement the module in an IK1 conductance block experiment (see Subheading 3.2.3). This section can be used as a guideline to develop a similar dynamic clamp experiment to introduce, block, or scale any modeled current in other species.
3.2.1 Design and Construction a Custom RTXI Module
The ability for the user to create a custom module is the most powerful feature of RTXI, allowing the development of unique and complex experimental protocols. While some familiarity with C++ is required, by no means does the reader need to be an expert programmer. The custom module will be built using RTXI’s default_ gui_model library. This library was designed to create modules quickly and easily without in-depth understanding of software’s architecture. Starting with a template, this section explains the code necessary for each program part of the IK1 block RTXI module.
Open the terminal.
-
Create a directory for a new RTXI module.
$ mkdir IK1_Block
-
Copy the module program template files from your RTXI installation directory (example directory used).
$ sudo cp /home/rtxi/doc/my_plugin/* IK1_Block
-
Enter the module directory.
$ cd IK1_Block
-
Rename the template files
$ sudo mv my_plugin.cpp IK1_Block.cpp
$ sudo mv my_plugin.h IK1_Block.h
-
Open the template files in the preferred code editing program (see Note 12).
$ emacs IK1_Block.cpp IK1_Block.h
-
Add the necessary code to each part of the template file.
-
(a)
IK1_Block.h (Fig. 5).
Replace all instances of my_plugin with IK1_Block.
Declare all variables.
Save file.
-
(b)
IK1_Block.cpp
-
(a)
-
Edit the program Makefile
-
(c)
Replace all instances of my_plugin with IK1_Block.
-
(d)
Save file.
-
(c)
-
Compile and install the program.
-
(e)
$ sudo make install.
-
(e)
Fig. 5.
Header file for IK1 Block RTXI module. The C++ header file contains the class definitions and variable declarations required for the IK1 block module. These variables are model parameters, experimental parameters, or module control flags, and are kept in memory during the life of the module. This is in contrast to variables declared in the source code of module, which are only kept in memory for the duration of the function call
Fig. 6.
Source file for IK1 Block RTXI module—vars structure, constructor, and destructor. (Line 12) The vars structure defines the inputs, outputs, parameters, and states. An INPUT creates a slot for the module to acquire data from the DAQ device or another module. An OUTPUT is a signal that can be sent to the DAQ device or another module. Using RTXI’s signal and slot architecture, modules can share data through connections between inputs and outputs. A STATE is a numeric data-type that cannot be modified by the user. These are typically used to track values of intermediate or computed quantities. A PARAMETER is another numeric data- type, but can be modified on the fly by user input through the GUI. These module data-types are also what the Data Recorder module is capable of recording. (Line 35) The module constructor is the function first run upon module loading, and normally does not require any change. (Line 44 ) The module destructor is the function run upon module unloading. This is typically where any garbage collection takes place, such as release of any dynamically allocated memory through the use of the new operator
Fig. 7.
Source file for IK1 Block RTXI module—execute function. The execute function represents the real-time code that is run every time step during program execute. It is imperative that the execute code must be as efficient as possible. (Line 4) Voltage is first sampled from an input, typically from a DAQ device or a model, and used in the calculation of IK1 current. (Line 6) The pace parameter controls the modules periodic stimulus. When changed to 1, stimulation begins based on the BCL (basic cycle length), Stim Mag (stimulation magnitude), and Stim Length (stimulation length) parameters. (Line 15 ) The experiment parameter controls IK1 current injection. (Line 15) When the experiment variable is set to 1, voltage is used to calculate the IK1; (Inset) original IK1 equations are shown. (Line 23) This calculated current is then passed through an output, which is usually the analog output of a DAQ device connected to a patch clamp amplifier
Fig. 8.
Source file for IK1 Block RTXI module—update function. The update function is called during several module events. (Line 6 ) The INIT case is run during module initialization. (Line 18) The setParameter(“parameter name,” variable) and setState(“state name,” variable) functions are used to associate the data- types described in the vars structure with program variables. (Line 37) The modify case is called whenever the modify button is clicked. Parameters are set to corresponding values retrieved from the GUI using the getParameter(“parameter name”) function. Any other variables dependent on the parameters are also updated, such as bclConverted and stimlengthConverted. (Lines 46 and 47) These variables are unit-less integers calculated by dividing BCL or stimulation length by RTXI’s thread period. Through the use of integers, rounding errors that may occur during the timing of the stimulus are eliminated. (Lines 50 and 53) The un-pause and pause states are called during toggling of the pause button. (Line 56) The period case is run when the RTXI thread rate is changed through the control panel. All variables dependent on the RTXI thread rate are updated
3.2.2 IK1 Block: Experiment Setup in RTXI
-
Open RTXI and the newly built module.
$ rtxi
Modules → Load User Module → IK1_Block.so.
-
Load additional modules required for dynamic clamp experiment.
Utilities → Patch Clamp → Membrane Test.
System → Oscilloscope.
System → Data Recorder.
-
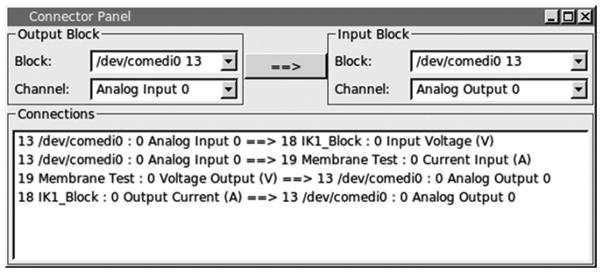
Connect the modules’ input/output (Fig. 9).
Open connector window (System → Connector).
Set the Output block: IK1_Block—Output Current.
Set the Input block: /dev/comedi0—Analog Output 0 (see Note 13).
Click Arrow button to complete connection.
-
Repeat for the following combinations:
/dev/comedi0—Analog Input 0|IK1_Block—Input Voltage.
Membrane_Test—Voltage Output|/dev/comedi0—Analog Output 0.
/dev/comedi0—Analog Input 0|Membrane_Test—Current Input.
-
Set oscilloscope to display cardiomyocyte membrane voltage (see Note 14).
Open oscilloscope properties (right-click on oscilloscope → Properties).
Set Channel: IK1_Block|Input|Input V.
Toggle the Active button.
Set Scale: 20 mV/V.
Click the Apply button.
Click Display tab.
Set Time Scale: 100 ms/div.
Set Screen Refresh: 50.
-
Setup Data Recorder module (see Note 15).
Set Channel: IK1_Block|Input|Input Voltage.
Click button (>) to add channel to record list.
Repeat for channel: IK1_Block|Output|Output Current.
-
Save workspace for future experiments.
File → Save Workspace.
Choose filename and click Save button.
Fig. 9.
RTXI connector settings for IK1 block dynamic clamp experiment. The connector panel showcases the signal and slot architecture of RTXI. The analog input 0 channel of a DAQ device (signal) is connected to the input of the IK1_Block module and Membrane Test module (slot). Conversely, the outputs of these two modules (signal) are connected to the analog output 0 channel (slot). The analog output channel is in turn connected to a patch clamp amplifier
3.2.3 Single Cardiomyocyte Isolation
Inject the guinea pig (400–500 g) with 500 units of heparin by intraperitoneal injection, allow at least 15 min for heparin to act.
Anesthetize the guinea pig by intraperitoneal injection of sodium pentobarbital.
Excise the heart by thoracotomy.
Once extracted, place the heart in oxygenated and 0 °C Common Tyrode’s solution. Immediately massage the heart to push blood out of the major chambers.
Using fine bent forceps and scissors, remove excess tissue around aorta: lungs, pericardium, and fat.
Using a pair of fine bent forceps and surgical suture, cannulate the aorta and begin retrograde perfusion with oxygenated Common Tyrode’s solution at 37 °C at 16 ml/min for 5 min at a pressure of at least 80 mm Hg. Heart should begin to beat vigorously and clear blood after 30 s.
Perfuse oxygenated calcium-free Tyrode’s solution for 4 min. Heart should cease to beat.
Perfuse oxygenated 2-enzyme solution for 1–4 min. Length of time is determined by appearance and feel of tissue. Perfusion is stopped once tissue becomes soft, bloated, and the color lightens.
While cannulated, cut only the ventricles into 5 ml of 1-enzyme solution using scissors and mince the tissue using a scalpel.
Once minced, transfer tissue and solution into a 50 ml conical tube, and add additional 10 ml of 1-enzyme solution.
Shake tissue containing solution for 30 s.
Filter the now cloudy solution with a 250 μm cell strainer while keeping tissue in the tube. Add 15 ml of 1-enzyme solution to the remaining tissue, and place in shaking water bath at 37 °C for 3 min.
Centrifuge the filtered solution for 1 min at 400 rpm and remove supernatant. Resuspend the cell pellet in 5 ml of wash solution.
Repeat the previous steps two more times, producing a total of three aliquots of cells in wash solution.
Centrifuge all aliquots for 1 min at 400 rpm, remove supernatant and resuspend in wash solution.
Inspect the cell aliquots. Dilute if necessary. Healthy cells should be quiescent with visible striations and crisp edges (see Note 16).
3.2.4 Dynamic Clamp: Blocking of the IK1 Current in Cardiomyocytes
-
Patch cardiomyocyte.
Add a drop from a cell aliquot into the bath. Fill the bath with extracellular solution, and wait 5 min for cells to adhere to glass.
Begin perfusion with extracellular solution warmed to 37 °C.
Fill a pulled patch pipette with appropriate intracellular solution, and attach it to amplifier headstage.
With the inverted microscope, select a target cardiomyocyte. Cell should not be touching any other cell or debris.
Set patch clamp amplifier to voltage clamp mode.
While measuring electrode resistance (see Note 17), apply light negative pressure to form a gigaohm seal.
Apply large and quick pulses of negative pressure to rupture the membrane to gain electrical access.
-
Measure membrane capacitance of the cell with Membrane Test module (see Note 18).
Click on the radio button corresponding to −80 mV.
Toggle down Pulse button.
Switch to the Membrane Properties tab.
Set pull-down menu: Continuous.
Toggle down Acquire to begin membrane parameter calculation.
Input measured Cm into the Cm parameter of IK1_Block module, and click the Modify button.
Switch to Resistance Tab in Membrane Test module.
Toggle up Pulse button.
Set patch clamp amplifier to current clamp mode.
-
Start stimulation of cardiomyocyte.
Toggle Pause button up to un-pause IK1_block module.
Change the Stim parameter of the dynamic clamp module to 1, and click the Modify button.
-
Find appropriate stimulation magnitude by slowly increasing the preset value.
Add 0.5 to the Stim Mag parameter, and click the Modify button.
Repeat these steps until an action potential is evoked to find threshold current.
To ensure reliable action potentials, increase threshold current by 50 %.
-
Stimulate the cell for 5 min at a basic cycle length (BCL) of 1,000 ms to allow cell to reach a steady state (see Note 19), and balance bridge to compensate for access resistance.
Change the BCL parameter to 1,000, and click the Modify button.
Balance bridge to compensate for access resistance in order to remove stimulus artifact (see Note 20).
-
Record baseline action potentials for 2 min with the Data Recorder module.
Click Choose File, input filename, and choose save location.
Click the Start Recording button.
After 2 min, click the Stop Recording button to end data collection.
-
Start IK1 block dynamic clamp.
Change the Scale parameter to -1 for full conductance block.
Change the Experiment parameter to 1 for conductance injection.
Click the Modify button.
-
Record dynamic clamped action potentials for 2 min with the Data Recorder module.
Click Choose File, input filename, and choose save location.
Click the Start Recording button.
After 2 min, click the Stop Recording button.
Observe depolarized resting membrane potential and shortened action potentials (see Fig. 10).
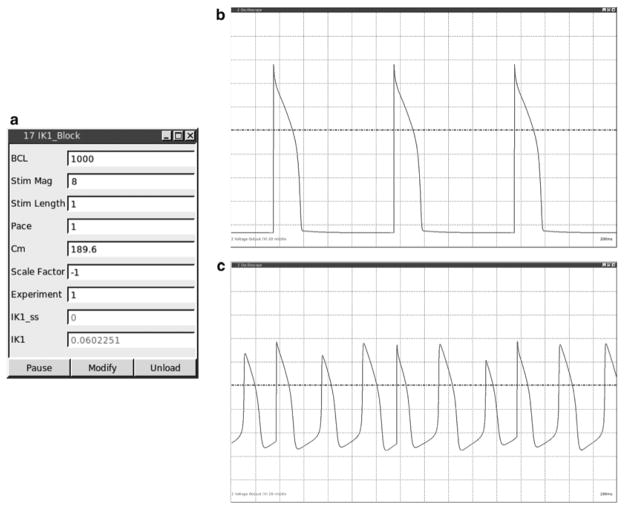
Fig. 10.
Results for IK1 conductance block in ventricular guinea pig cardiomyocyte through dynamic clamp. (a) Screenshot of the resulting GUI in RTXI of the constructed IK1_Block module during experiment. (b) Screenshot of membrane potential of the cardiomyocyte, showing evoked action potentials from a 1,000 BCL pacing protocol using the IK1_Block module using RTXI’s Oscilloscope module in real-time. (c) Subsequent screenshot after IK1 conductance block dynamic clamp is turned on. Plot now shows pacemaker-like activity of the cardiomyocyte after conductance block is initiated
3.3 Reciprocal Coupling Between a Virtual Neuron and Live Neuron Through Dynamic Clamp
Using the library of modules built into RTXI and freely available on the RTXI Web site (www.rtxi.org), numerous types of dynamic clamp experiments can be performed without any need of programming. This section describes using a Hodgkin–Huxley model neuron and alpha synapse model to construct a simple neuronal network (see Fig. 3b). This network can easily be expanded through additional virtual or real neurons. While no programming is required, the available module source code allows for alteration of the model equations.
3.3.1 Installation of a RTXI Module and Experiment Setup
-
Download the community built modules—Spike Detector and Reciprocal Coupling.
Visit the RTXI Web site (www.rtxi.org).
On the Web site, navigate to Modules → Neural Electrophysiology.
Download the Spike Detector and Reciprocal Coupling modules.
-
Navigate to the download directory of the modules, unpack the contents, and install the module.
$ cd ~/Downloads/ (see Note 21).
$ tar –xjvf SpikeDetect.tar.gz
$ cd SpikeDetect
$ sudo make install
Repeat for Reciprocal Coupling module.
3.3.2 Reciprocal Coupling: RTXI Experiment Setup
-
Open RTXI and load required modules for dynamic clamp experiment.
$ rtxi
Modules → Load User Module → spikedetect.so (referred to as Spike Detector 1) (see Note 22).
Repeat to load a second Spike Detector module (referred to as Spike Detector 2).
Modules → Load User Module → coupling.so.
Utilities → Model HH Neuron.
Utilities → Patch Clamp → Membrane Test.
System → Oscilloscope (referred to as Oscilloscope 1).
Repeat to load a second Oscilloscope (referred to as Oscilloscope 2).
System → Data Recorder.
-
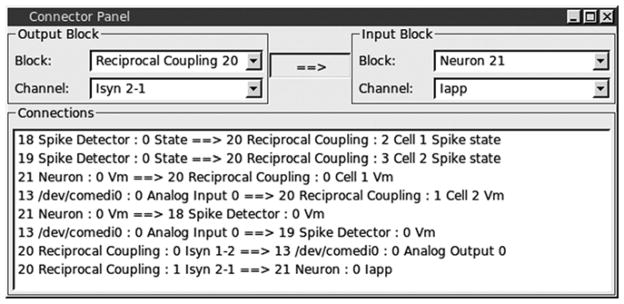
Connect the modules’ input/output (see Fig. 11).
Open connector window (System → Connector).
Set Output block: Spike Detector 1—State.
Set the Input block: Coupling—Cell 1 Spike State.
Click Arrow button to complete connection.
-
Repeat for the following combinations:
Spike Detector 2—State|Coupling—Cell 2 Spike State.
Neuron—Vm|Coupling—Cell 1 Vm.
/dev/comedi0|Coupling—Cell 2 Vm.
Neuron—Vm|Spike Detector 1—Vm.
/dev/comedi0—Analog Input 0|Spike Detector 2—Vm.
Coupling—Isyn 1-2|/dev/comedi0—Analog Output 0.
Coupling—Isyn 2-1|Neuron—Iapp.
-
Set oscilloscope to display virtual neuron membrane potential on Oscilloscope 1 and live neuron membrane potential on Oscilloscope 2.
Open Oscilloscope 1 properties (Right click on Oscilloscope 1 → Properties).
Set Channel: Neuron|Output|Vm.
Toggle the Active button.
Set Scale: 20 mV/V.
Click the Apply button.
Click Display tab.
Set Time Scale: 100 ms/div.
Set Screen Refresh: 50.
Repeat for Oscilloscope 2 for Channel: /dev/ comedi0|Output|Analog Input 0.
-
Setup Data Recorder module.
Set Channel to: Reciprocal Coupling|Input|Cell 1 Vm.
Click button (>) to add channel to record list.
-
Repeat for the following under Reciprocal Coupling:
Input|Cell 2 Vm.
Input|Cell 1 Spike State.
Input|Cell 2 Spike State.
Output|Isyn 1-2.
Output|Isyn 2-1.
State|Phase Diff.
State|Time.
-
Save workspace for future experiments.
File → Save Workspace.
Choose filename and click Save button.
Fig. 11.
RTXI connector settings for reciprocal coupling of a live and virtual neuron dynamic clamp experiment. Inputs and outputs are not only used to connect modules to the DAQ device, but also to other modules. One spike detector retrieves input through a DAQ device, while another retrieves its input from the Neuron module. The outputs of both modules are then passed to the Reciprocal Coupling module
3.3.3 Brain Tissue Slice Preparation
Place rat into sealed chamber with isoflurane vapor until it is anesthetized.
Decapitate animal with guillotine. Open skull with scissors.
With Scoopula, remove brain and place in 0 °C ACSF.
With scalpel, isolate chunk(s) of tissue containing the brain region(s) of interest.
Glue tissue to the bottom of a walled slice chamber with cyanoacrylate glue. Fill chamber with 0 °C ACSF.
Attach chamber to vibratome. Cut 300–400 μm thick slices. Place tissue into chamber containing ACSF bubbled with 95 % O2 and 5 % CO2.
Allow tissue to recover for at least 1 h before transferring to microscope stage chamber.
3.3.4 Dynamic Clamp: Reciprocal Neuronal Coupling
-
Patch neuron.
With a microscope using dual interference contrast microscopy, select a target neuron in slice tissue.
With micromanipulator, place electrode tip against the cell body.
Set patch clamp amplifier to voltage clamp mode.
While measuring electrode resistance, apply light negative pressure to form a gigaohm seal between electrode and neuron.
Apply negative pressure to pop through the membrane.
Balance bridge and compensate for capacitance (see Note 23).
Observe live neuronal spiking on Oscilloscope 2.
Set patch clamp amplifier to current clamp mode.
Un-pause Spike Detector 1 and Spike Detector 2.
-
Start Neuron simulation.
Toggle Pause button up to un-pause Neuron module.
Set Iapp_offset: 9.
Click the Modify button.
Observe virtual neuronal spiking on Oscilloscope 1.
-
Setup Reciprocal Coupling module.
Set Gmax 2-1: 9e4 (see Note 24)
Check the Automate Coupling Delay checkbox.
Click the Modify button.
Toggle Pause button up.
-
Start data recording with the Data Recorder module for 2 min.
Click Choose File, choose save directory, and input an appropriate filename.
Click the Start Recording button.
-
Begin reciprocal coupling.
Toggle Couple Neurons button down.
-
Stop data recording after 5 min.
Click the Stop Recording button of the Data Recorder module.
Observe neuronal synchrony (see Fig. 12).
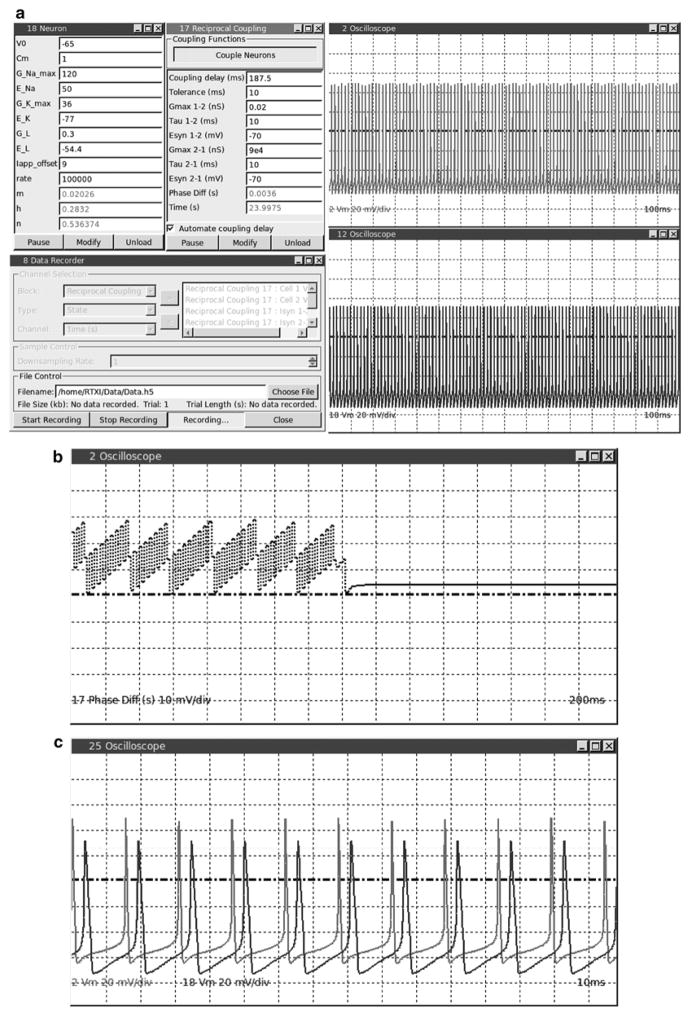
Fig. 12.
Results for reciprocal coupling between a live and virtual neuron through dynamic clamp. (a) Screenshot of the RTXI workspace during the experiment. Top and bottom oscilloscopes display the membrane potential of the real and virtual neuron, respectively. (b) A plot using the Oscilloscope module of the calculated phase difference performed by the Reciprocal Coupling module. Reciprocal coupling dynamic clamp was switched on in the middle of the plot, resulting in a constant phase difference c) Screenshot of Oscilloscope module while plotting the membrane potential of both neurons, live and virtual, using the trigger function. Note the synchrony of the spikes due to the artificial coupling
4 Notes
The artificial cerebrospinal fluid and the artificial neuroplasm listed here are merely examples that we find sufficient for our studies with entorhinal cortical stellate neurons. In addition, you may wish to add synaptic blockers to chemically isolate the desired hybrid network from all the other neurons in living tissue.
Inverted microscopes are usually preferred in patch clamp rigs because they provide good visualization of cells, and adequate access of the micromanipulator and patch pipettes above the cell bath. Thus, inverted microscopes are recommended for patching cardiomyocytes. However, these microscopes are unable to adequately visualize neurons in slices due to the thickness of the tissue. Differential interference contrast microscopes are preferred for their ability to visualize live, unstained neurons within the slices.
Almost all patch clamp amplifier models can be used in the dynamic clamp setup described in this chapter. At the minimum, the amplifier must have an external analog output and analog input to interface with the dynamic clamp computer. It is also important to note the maximum current the amplifier can apply, especially for cardiomyocyte recordings. Cardiomyocytes may need upwards of 14 nA of a 1 ms current pulse to evoke an action potential. The amplifier should also be able to bridge balance in order to adequately compensate for access resistance. This is especially important in dynamic current clamp, where the level of current is being changed on the microsecond time scale.
Pipette shape and tip size are largely dependent on the cell type. For cardiomyocytes, we recommend a pipette tip resistance of 2–3 MΩ, and to approach the cell as close to perpendicular as possible due to the relative flag surface of the membrane. For neurons, we recommend a pipette tip resistance of 6–7 MΩ, and to approach the cell from a 45° angle.
In order to adequately mimic a biological conductance, real-time control is required to execute the task of calculation and injection of the current. The dynamic clamp software must operate on the same time scale as the currents it is trying to mimic. Hard real-time operating systems allows for execution of these tasks at a guaranteed timing. With a standard operating system, multitasking allows a single CPU to switch from multiple tasks to give the appearance all programs are being executed at the same time. This makes the operating system inherently unpredictable, and makes real-time processing unreliable. The software packages, RTAI (www.rtai.org) or Xenomai (www.xenomai.org), are available to modify the Linux operating system into a hard real-time operating system. These free and open-source extensions are used by RTXI to provide real-time control.
Any modern computer system compatible with the Linux operating system can be used in the dynamic clamp setup. This chapter assumes RTXI was installed through its Live USB installation, which uses the Ubuntu distribution of the Linux operating system. It is important to note that modifications to the CPU clock rate can disrupt real-time control. Thus, it is important to prevent the adjustment of the CPU clock rate during experimentation. This can be done by disabling any features, such as “Turbo Mode,” through the computer’s BIOS.
RTXI uses the open-source library of DAQ drivers, Comedi (www.comedi.org), to interface with DAQ devices. A list of supported hardware can be found on the Comedi Web site.
Many patch clamp amplifiers have telegraphs, which output analog voltage signals that represent the amplifier’s current configuration, i.e., the output gain. Others also allow control of a setting through an analog voltage input. For example, the Axon Multiclamp 700B mode can be changed through a mode external input. These additional amplifier input and outputs can be used by RTXI for better communication with the amplifier.
An RTXI module can be designed to take advantage of the various telegraphs of an amplifier, and automatically compensate for any configuration changes. For example, an amplifier telegraph could signal an RTXI module when the output gain was changed. The module could then modify settings within RTXI, signal-scaling values in this case, and compensate accordingly. Several amplifier control modules are bundled with the RTXI installation for common models of patch clamp amplifiers: Axon Multiclamp 700 Series, Axon Axopatch 1D, and the Axon 200 Series. The capabilities of these modules are limited to the amplifier’s features, but all are capable of compensating for amplifier gains through input from the amplifier itself or the user.
Gains are applied to amplifier inputs and outputs to improve signal to noise ratio. These gains are described in the amplifier’s manual, and must be compensated for by RTXI in order to accurately measure a signal. For example, if output gain is set to 10×, RTXI must divide that signal by a factor of 10. This is done by scaling factors that can be set through RTXI’s control panel (System → Control Panel). Each channel can have a unique scaling factor set to it. As previously mentioned, RTXI modules for common amplifier models that automatically compensate for these gains are available for download (see Note 8).
The RTXI thread rate controls the real-time period of the system, and can be set in the Thread tab of the Control Panel (System → Control Panel). The real-time period controls how often a module’s execute section is run, as well as the sampling rate of active inputs and outputs of the DAQ device. In a dynamic current clamp experiment, thread rate is usually set to the speed necessary to adequately recapitulate the current being injected. However, it is limited by several factors. First, the computation time required to run the model equations. Second, the time required to sample active inputs and outputs from the DAQ device. The sum of these must consistently be below the real-time period. Measurement of the computation time and real-time period can be done through the Real-Time Benchmarks module (System → Real-Time Benchmarks).
While any code editor can be used, commands listed use Emacs as an example. Emacs can be installed on an Ubuntu operating system through the following command: “sudo apt-get install emacs”.
Based on the naming scheme of the Comedi (www.comedi.org) driver, “/dev/comedi0” refers to the first detected DAQ device, i.e., “/dev/comedi1” and “/dev/comedi2” would be the subsequently detected devices. While it is assumed only one DAQ device is being used, RTXI is capable of interfacing with many DAQ devices simultaneously.
RTXI’s digital oscilloscope module is capable of visualizing any of RTXI’s module data-types: INPUT, OUTPUT, STATE, and PARAMETER. The settings window can be accessed by right-clicking a loaded Oscilloscope module to bring up its context menu, and selecting Properties. Multiple channels can be displayed in a single instance.
The Data Recorder (System → Data Recorder) is capable of recording any module data-types: INPUT, OUTPUT, STATE, and PARAMETER. Rate of recording is equivalent as RTXI’s thread rate, but can be down-sampled. Files are recorded in the HDF5 (www.hdfgroup.org/HDF5/) format and imported directly into MATLAB using the included scripts located in the following directory: /[rtxi install directory]/ hdf/. HDF5 Data files can also be converted into ASCII format through the use of the rtxi_hdf_reader program with the following command: rtxi_hdf_reader –r [sample start]:[sample rate]:[sample end] [file name]>[output file], i.e., rtxi_hdf_ reader –r 1:1:end test.h5 > test.dat.
Health of isolated cardiomyocytes can be negatively affected at many steps of the isolation protocol. Typical issues include the following: inadequate placement of the cannula (past aortic valve will result in inadequate perfusion), use of water containing endotoxins, unclean perfusion equipment, too short or too long perfusion of the enzyme solution, or too low or too high collagenase concentration in enzyme solution. The length of time and amount of collagenase to use is determined through trial and error, and the times and concentrations listed should be used as starting guidelines.
Pipette tip resistance can be measured in a number of ways: with the amplifier’s built-in resistance measurement, through the amplifier’s included software, or with RTXI’s Membrane Test module (Utilities → Membrane Test).
Capacitance can be measured with the amplifier’s included software, or with RTXI’s Membrane Test module (Utilities → Membrane Test). Accurate measurement of cell capacitance is vital to scale the calculated model current. The Membrane Test module calculates cell capacitance through measurement of the capacitive transient evoked through a standard square wave voltage pulse. Measurement should be performed at resting membrane potential to eliminate the effect of the fast sodium current.
Determining if a steady state for a cardiomyocyte has been reached can be done by plotting action potential duration (APD) versus time. Over the course of several hundred beats, APD will gradually decrease until it reaches a stable value. It is of note that beat to beat variability will still exist once the cell has stabilized. If APD continues to decrease without stabilizing, the health of the cell should be in question.
Stimulus artifact during current injection results from the voltage drop caused by the access resistance of the electrode. Through the use of bridge balance compensation, this voltage drop can be compensated for and eliminated. While the stimulus artifact can be ignored and removed during data analysis in traditional patch clamp experiments, the artifact can have significant effects during dynamic clamp. During artificial conductance injection, the stimulus artifact will be included in the current calculation and result in an inaccurate solution.
The “~” represents the current user’s home directory. “~/ Downloads” is the typical directory where the installed Internet browser will download files.
The reciprocal coupling setup uses two instances of the Spike Detector module, one each for the real neuron and virtual neuron. RTXI assigns an ID number to each module listed on the top left of the module’s window. Use this ID number to differentiate between multiple instances of an identical module.
Depending on amplifier model, bridge balance and capacitance compensation will need to be performed during current-clamp (Axon Axopatch 200 Series) mode or voltage-clamp mode (Axon MultiClamp 700 Series).
Both the real and virtual neurons receive synaptic input with a true conductance (nS) from the Reciprocal Coupling module. The Hodgkin–Huxley neuron model bundled with RTXI, however, uses parameters with units that are in conductance densities (mS/cm2). Thus, while the Gsyn 2 > 1 is listed as 9e4 nS (90 μS), the value interpreted by the Hodgkin–Huxley neuron model is 90 μS/cm2, to match the units of the Hodgkin–Huxley model.
Acknowledgments
This work was supported by NIH R01 RR020115 (to D.J.C., R.J.B., and J.A.W.). We thank R.J. Lin for invaluable programming assistance, and F.R. Fernandez for helpful comments regarding this chapter.
References
- 1.Robinson HP, Kawai N. Injection of digitally synthesized synaptic conductance transients to measure the integrative properties of neurons. J Neurosci Meth. 1993;49:157–165. doi: 10.1016/0165-0270(93)90119-c. [DOI] [PubMed] [Google Scholar]
- 2.Sharp AA, O'Neil MB, Marder E. Dynamic clamp: computer-generated conductances in real neurons. J Neurophysiol. 1993;69:992–995. doi: 10.1152/jn.1993.69.3.992. [DOI] [PubMed] [Google Scholar]
- 3.Dorval AD, Christini DJ, White JA. Real-Time Linux dynamic clamp: a fast and flexible way to construct virtual ion channels in living cells. Ann Biomed Eng. 2001;29:897–907. doi: 10.1114/1.1408929. [DOI] [PubMed] [Google Scholar]
- 4.Lin RJ, Bettencourt J, White JA, et al. Real-time experiment interface for biological control applications. Conf Proc IEEE Eng Med Biol Soc. 2010;1:4160–4163. doi: 10.1109/IEMBS.2010.5627397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Economo MN, Fernandez FR, White JA. Dynamic clamp: alteration of response properties and creation of virtual realities in neurophysiology. J Neurosci. 2010;30:2407–2413. doi: 10.1523/JNEUROSCI.5954-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettencourt JC, Lillis KP, Stupin LR, White JA. Effects of imperfect dynamic clamp: computational and experimental results. J Neurosci Meth. 2008;169:282–289. doi: 10.1016/j.jneumeth.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livshitz L, Rudy Y. Uniqueness and stability of action potential models during rest, pacing, and conduction using problem-solving environment. Biophysical J. 2009;97:1265–1276. doi: 10.1016/j.bpj.2009.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan X, Laurita KR, Pruvot EJ, Rosenbaum DS. Molecular correlates of repolarization alternans in cardiac myocytes. J Mol Cell Cardiol. 2005;12:196–206. doi: 10.1016/j.yjmcc.2005.06.004. [DOI] [PubMed] [Google Scholar]