Abstract
Background
Whereas several longitudinal metabolomics studies have been conducted in individuals with normal eGFR at baseline, disease progression among individuals with established CKD has not been rigorously examined.
Methods
We performed a nested case-control study of rapid CKD progression in the Chronic Renal Insufficiency Cohort (CRIC) Study, profiling baseline plasma from 200 individuals each with eGFR slope < −3 ml/min/1.73m2/yr (cases) or between −1 and +1 ml/min/1.73m2/yr (controls), matched on baseline eGFR and proteinuria. To directly assess how the kidney modulates circulating metabolites, we profiled plasma from the aorta and renal vein of 25 hospital-based individuals.
Results
At baseline, cases and controls had a mean eGFR of 41.7 ± 13.3 and 45.0 ± 14.5 mL/min/1.73m2, respectively. Ten plasma metabolites were nominally associated with CKD progression in logistic regression models adjusted for age, sex, race/ethnicity, hypertension, systolic and diastolic blood pressure, diabetes, eGFR, and proteinuria; no metabolite achieved the Bonferroni-adjusted significance threshold (p<0.0003). In cross-sectional analysis, all six of the metabolites that were higher in cases than controls were significantly associated with eGFR at baseline. By contrast, threonine, methionine, and arginine were lower in cases than controls and had no association with baseline eGFR. Further, in the hospital-based cohort that underwent renal arteriovenous sampling, these three metabolites were net released from the kidney. Combining these metabolites into a panel of markers further strengthened their association with CKD progression.
Conclusion
Our results motivate interest in arginine, methionine, and threonine as potential indicators of renal metabolic function and markers of renal prognosis.
Keywords: biomarkers, CKD, CKD progression, metabolism, metabolomics
INTRODUCTION
Metabolomics, or metabolite profiling, refers to the systematic analysis of metabolites (i.e., sugars, amino acids, organic acids, nucleotides, etc) in a biologic specimen [1–3]. Because of long-standing interest in small molecules as potential uremic toxins, initial applications of metabolomics in nephrology research examined plasma or dialysate from individuals with ESRD [4–6]. At the other end of the spectrum of renal health, recent large scale profiling in population-based cohorts including the Framingham Heart Study (FHS), the Cooperative Health Research in the Region of Augsburg (KORA) Study, the TwinsUK Registry, and the Atherosclerosis Risk in Communities Study (ARIC) have identified cross-sectional associations between metabolite levels and eGFR within the normal range [7–10]. Because longitudinal follow-up is available in these cohorts, these data sets have also permitted assessment of the association of these baseline metabolite measures with the future onset of CKD.
Whereas studies of incident CKD have compared individuals who do or do not cross an eGFR threshold of 60 ml/min/1.73 m2, identifying markers of progression among individuals with established disease is arguably of greater interest. The relative paucity of high-quality cohorts enriched for advanced renal disease, however, presents a challenge. Here, we conducted a nested case-control study of disease progression in the Chronic Renal Insufficiency Cohort (CRIC) Study, a longitudinal, multi-center cohort study of individuals with CKD. Because of the possibility of confounding due to baseline difference in kidney function, we tested the cross-sectional association between the markers identified in the case-control analysis with baseline eGFR. In addition, in order to provide a physiologic understanding of how the kidneys modulate these potential metabolite markers, we analyzed metabolite levels in the arterial and renal venous circulation from individuals undergoing invasive catheterization.
MATERIALS AND METHODS
CRIC
The CRIC Study was initiated in 2001 by the NIDDK to investigate the natural history of kidney disease as well as the risk factors for disease progression and cardiovascular complications. Between 2003–2008, 3939 individuals with mild to moderate kidney disease were recruited at 13 sites across the US [11,12]. By design, approximately 50% of study participants had diabetes at study entry when they were between the age of 21 and 74 years, with an eGFR of 20–70 ml/min/1.73 m2. The institutional review board of the Perelman School of Medicine at the University of Pennsylvania approved the study. The study adhered to the Declaration of Helsinki and all participants provided written informed consent.
Study Design
We used a nested case-control design, comparing a subset of the CRIC Study population demonstrating rapid progression of their renal disease (cases, n=200) to a subset whose renal dysfunction was stable over time (controls, n=200). Cases were randomly chosen from individuals with an eGFR slope < −3 ml/min/1.73 m2/yr. Controls were chosen from individuals with an eGFR slope between −1 and +1 ml/min/1.73 m2/yr. For each case, a control was selected that was in the same eGFR (<30, 30-<40, 40-<50, and ≥50 ml/min/1.73 m2) and proteinuria (≤0.1, 0.1-<0.5, 0.5-<1.5, and ≥1.5 g/24h) category at study entry. eGFR was calculated from serum creatinine and cystatin C using a CRIC Study equation [13] and eGFR slope was derived using all available eGFR values using linear regression. Proteinuria was measured from 24h urine collections. Cardiovascular disease (CVD) and smoking status were self-reported whereas diabetes was defined as a fasting glucose ≥126 mg/dL, a random glucose ≥200 mg/dL, or use of insulin or antidiabetic medications. Systolic blood pressure (SBP) and diastolic blood pressure (DBP) was measured in triplicate following a standardized protocol with the participant seated using a Tycos Classic hand cuff and aneroid sphygmomanometer (Welch Allyn, Skaneateles Falls, NY, USA). The baseline values were the mean of all three SBP and DBP measurements, separately. Hypertension was defined as SBP ≥140 mmHg and/or DBP ≥90 mmHg and/or self-reported antihypertensive medication use. Creatinine, glucose, and uric acid were measured using coupled enzymatic reactions with spectrophotometric quantification on the Ortho Vitros 950 (Ortho Clinical Diagnostics, Raritan, NJ, USA). Metabolomics assays were performed on fasting plasma samples obtained at study entry and stored at −80°C.
Arterial and Renal Venous Plasma Sampling
Details on this protocol have been published [7]. In brief, we recruited consecutive patients referred to the Massachusetts General Hospital Cardiac Catheterization Laboratory for right and left heart catheterization. During the procedure, plasma was sampled from catheters placed in the ostium of a renal vein and the abdominal aorta. All patients were fasting and all samples were obtained prior to coronary artery catheterization (and administration of iodinated contrast dye). eGFR for study participants undergoing venous sampling was calculated using the MDRD equation [14]. The study protocol was approved by the institutional review board of the Massachusetts General Hospital, adhered to the Declaration of Helsinki, and all participants provided written informed consent.
Metabolite Profiling
Two liquid chromatography-mass spectrometry (LC-MS) methods were used to measure polar plasma metabolites. Amino acids, amino acid metabolites, acylcarnitines, dipeptides, nucleotides, and other cationic polar metabolites were measured in 10 μL of plasma using hydrophilic interaction liquid chromatography coupled with positive ion mode analysis on a Q Exactive Plus Orbitrap MS (Thermo Scientific, Waltham, MA, USA). Sugars, sugar phosphates, organic acids, bile acids, nucleotides and other anionic polar metabolites were measured in 30 μL of plasma using hydrophilic interaction liquid chromatography and multiple reaction monitoring in the negative ion mode on a 5500 QTRAP MS (AB Sciex, Framingham, MA, USA). Details on plasma extraction, isotope internal standards, and chromatographic conditions for both methods have been published [4,15]. Metabolite identifications were confirmed using synthetic mixtures of reference compounds as well as characterized pooled plasma reference samples. All samples for a given study design (i.e. CRIC or renal arterio-venous sampling) were assayed in a single LC-MS run.
Statistical Analyses
We compared demographic and laboratory parameters between cases and controls using unpaired t-tests or Chi-squared tests, as appropriate. We compared select metabolites measured by LC-MS with spectrophotometric measurements using Pearson correlation coefficients. We compared medication levels between individuals with or without CVD and cotinine levels between non-smokers and smokers using the Wilcoxon rank-sum test. In the primary CRIC analysis, we fit logistic regression models to assess the associations between log transformed metabolite levels and rapid progression of CKD. The primary model adjusted for age, sex, race/ethnicity, hypertension, SBP, DBP, diabetes, eGFR and proteinuria; eGFR, and proteinuria were included as continuous variables. With 160 endogenous metabolites measured in this cohort, the Bonferroni adjusted significance threshold was 0.0003. We considered a p-value <0.05 as nominally significant. In addition, we examined the cross-sectional association of log transformed metabolite levels with baseline eGFR in linear regression models adjusted for age, sex, race/ethnicity, hypertension, SBP, DPB, diabetes, and proteinuria, again with a Bonferroni adjusted significance threshold of p<0.0003. For the study of renal arteriovenous metabolite gradients, we compared arterial and venous metabolite levels using paired t-tests. Based on these initial analyses highlighting threonine, methionine, and arginine as potential markers of renal metabolic function independent of baseline GFR, data for these three metabolites were log transformed, standardized, and then summed to create a composite score. We hypothesized that this panel of metabolites provides a more complete assessment of renal metabolism and hence would have a stronger association with case status than any of the three metabolites alone. This panel of metabolites was examined in the primary model, as well as models further adjusting for CVD and smoking, and for CVD, smoking, hemoglobin, and FGF-23 levels. All analyses were performed using SAS software, version 9.4 (SAS Institute, Cary, NC, USA).
RESULTS
CRIC Study
Baseline characteristics of the CRIC study sample are shown in Table 1. Rapid progressors (cases) were older (60.1 versus 57.8 years), more likely to be black (55% versus 35%), and had a higher prevalence of hypertension (95% versus 87%), diabetes (55% versus 44%), active smoking (20% versus 12%), and CVD (47% versus 36%) than individuals with stable kidney function (controls). Cases also had lower hemoglobin (12.7 versus 13.3 mg/dL) and higher FGF-23 (205.7 versus 159.4 RU/mL) levels, but there was no significant difference in bicarbonate, phosphate, cholesterol, insulin, or BMI. Although the groups were matched on the basis of eGFR and proteinuria category, the mean eGFR at baseline was lower among cases compared to controls (41.7 ± 13.3 versus 45.0 ± 14.5 mL/min per 1.73 m2). There was no difference in proteinuria. The mean eGFR slope was −4.1 ± 1.0 and −0.1 ± 0.6 mL/min/1.73 m2 per year among cases and controls, respectively. The mean follow-up was 2.84 +/− 0.44 years, with no difference between cases and controls.
Table 1.
Baseline characteristics of the CRIC Study sample
| Rapid progressors (cases) | Stable CKD (controls) | p-value | |
|---|---|---|---|
| Clinical characteristics | |||
| Age (years) | 60.1 ± 10.2 | 57.8 ± 11.3 | 0.04 |
| Male sex (%) | 57 | 64 | |
| Race/ethnicity | white 31% | white 51% | |
| black 55% | black 35% | <0.001 | |
| other 14% | other 14% | ||
| Hypertension (%) | 95 | 87 | 0.004 |
| Systolic blood pressure (mmHg) | 134.5 ± 21.4 | 121.9 ± 20.0 | <0.001 |
| Diastolic blood pressure (mmHg) | 72.1 ± 13.8 | 70.1 ± 12.7 | 0.14 |
| Body mass index (kg/m2) | 32.5 ± 7.8 | 32.1 ± 7.1 | 0.53 |
| Diabetes (%) | 55 | 44 | 0.03 |
| Cardiovascular disease (%) | 47 | 36 | 0.04 |
| Smokers (%) | 20 | 12 | 0.03 |
| Laboratory measures | |||
| Serum creatinine (mg/dL) | 1.86 ± 0.6 | 1.82 ± 0.6 | 0.58 |
| Baseline eGFR (mL/min/1.73 m2)† | 41.7 ± 13.3 | 45.0 ± 14.5 | 0.02 |
| Follow-up eGFR (mL/min/1.73 m2) | 24.7 ± 10.5 | 45.0 ± 11.2 | <0.001 |
| 24h urine protein (g) | 0.9 ± 1.6 | 0.8 ± 1.3 | 0.27 |
| Hemoglobin (mg/dL) | 12.7 ± 1.7 | 13.3 ± 1.9 | 0.002 |
| Bicarbonate (mmol/L) | 24.3 ± 3.4 | 24.7 ± 2.9 | 0.12 |
| Phosphate (mg/dL) | 3.7 ± 0.7 | 3.6 ± 0.6 | 0.05 |
| FGF-23 (RU/mL) | 205.7 ± 249.1 | 159.4 ± 137.0 | 0.02 |
| Insulin (μIU/mL) | 25.7 ± 25.6 | 26.8 ± 33.4 | 0.73 |
| Total cholesterol (mg/dL) | 177.2 ± 44.3 | 176.1 ± 38.4 | 0.78 |
| HDL cholesterol (mg/dL) | 47.7 ± 13.0 | 48.6 ± 15.7 | 0.55 |
| LDL cholesterol (mg/dL) | 95.2 ± 34.4 | 94.7 ± 29.8 | 0.89 |
| Triglycerides (mg/dL) | 158.2 ± 112.7 | 154.0 ± 96.6 | 0.66 |
baseline eGFR using the CKD-EPI equation was 43.9 ± 13.9 for cases and 44.6 ± 13.4 mL/min/1.73 m2 for controls
Conversion factor for units: serum creatinine in mg/dL to mmol/L, x88.4
Metabolite Profiling in CRIC
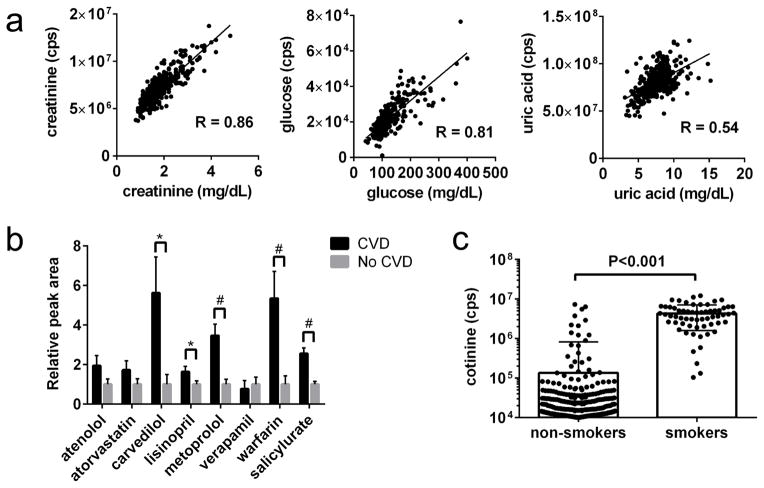
A total of 160 polar metabolites were measured in baseline plasma obtained from cases and controls. First, we sought to examine this metabolite data in relation to several cross-sectional phenotypes captured in CRIC (Figure 1). Figure 1A depicts the correlation for three metabolites assayed by our LC-MS methods that had already been measured in CRIC using alternative (spectrophotometric) methods. The Pearson correlation coefficients were 0.86 for creatinine, 0.81 for glucose, and 0.54 for uric acid. Figure 1B shows data on select medications (or medication metabolite) monitored by our platform. Individuals with self-reported CVD had higher plasma levels of metoprolol, carvedilol, lisinopril, warfarin, and salicylurate (a metabolite of aspirin) than individuals without CVD. Finally, Figure 1C shows plasma levels of the nicotine metabolite cotinine in non-smokers versus smokers, demonstrating markedly elevated mean levels in the latter group, albeit with notable overlap in the distributions.
Figure 1. Examination of select metabolomics data in relation to CRIC phenotyping.
(a) Correlation between LC-MS (y-axis) and spectrophotometric measures (x-axis) of creatinine, glucose, and uric acid. (b) Mean medication levels in individuals with (n=166) or without (n=234) self-reported CVD at baseline. Bars represent SEM. *p < 0.05, #p < 0.001. (c) Distribution of plasma cotinine levels in self-reported non-smokers and smokers; note y-axis is log scale.
Metabolite Associations with Rapid CKD Progression
Endogenous metabolites with a nominal association (p < 0.05) with CKD progression in logistic regression, adjusted for age, sex, race/ethnicity, hypertension, SBP, DBP, diabetes, baseline eGFR and proteinuria, are shown in Table 2 (full results are shown in Supplementary data, Table S1). No metabolite met the Bonferroni-adjusted threshold for significance. Of the ten metabolites nominally associated with CKD progression, six (uric acid, glucuronate, 4-hydroxymandelate, 3-methyladipate/pimelate, cytosine, and homogentisate) were higher in cases than controls, whereas four (threonine, methionine, phenylalanine, and arginine) were lower in cases than controls. Because many circulating small molecules (including metabolites) accumulate with falling GFR, we also examined the cross-sectional association between metabolite levels and baseline eGFR in linear regression models adjusted for age, sex, race/ethnicity, hypertension, SBP, DBP, diabetes, and proteinuria. Notably, all six of the metabolites that were higher in cases than controls had a significant (p<0.0003) association with baseline eGFR, with an inverse correlation between metabolite level and eGFR (Table 2). These findings raise the possibility that the association between these metabolites and subsequent GFR decline is confounded by baseline differences in renal function not fully captured by one-time GFR estimation. By contrast, threonine, methionine and arginine – three metabolites that were lower in cases than controls – had no association with eGFR at baseline (Table 2).
Table 2.
Metabolites associated with rapid CKD progression in CRIC at p < 0.05 and their baseline association with eGFR
| CKD Progression (longitudinal) | Association with baseline eGFR (cross-sectional) | |||
|---|---|---|---|---|
|
| ||||
| Metabolite | OR per SD (95% CI)* | p-value† | β (95% CI)** | p-value |
| Lower in cases than controls | ||||
| threonine | 0.35 (0.16, 0.78) | 0.01 | 0.35 (−0.93, 1.62) | 0.59 |
| methionine | 0.35 (0.13, 0.93) | 0.04 | 0.52 (−0.73, 1.78) | 0.41 |
| phenylalanine | 0.23 (0.05, 0.95) | 0.04 | −2.06 (−3.34, −0.77) | 0.002 |
| arginine | 0.46 (0.21, 0.98) | 0.05 | 0.49 (−0.79, 1.78) | 0.45 |
| Higher in cases than controls | ||||
| uric acid | 9.72 (2.17, 43.45) | 0.003 | −4.17 (−5.41, −2.93) | <0.0001 |
| glucuronate | 1.84 (1.19, 2.87) | 0.007 | −3.00 (−4.29, −1.70) | <0.0001 |
| 4-hydroxymandelate | 5.00 (1.45, 17.29) | 0.01 | −4.21 (−5.46, −2.97) | <0.0001 |
| 3-methyladipate/pimelate | 1.95 (1.15, 3.3) | 0.01 | −4.06 (−5.27, −2.86) | <0.0001 |
| cytosine | 1.61 (1.03, 2.53) | 0.04 | −4.47 (−5.70, −3.24) | <0.0001 |
| homogentisate | 3.32 (1.03, 10.72) | 0.05 | −4.31 (−5.56, −3.07) | <0.0001 |
Abbreviations: OR = odds ratio; SD = standard deviation; CI = confidence interval; β = beta coefficient in linear regression model
Model adjusted for age, sex, race/ethnicity, hypertension, SBP, DBP, diabetes, eGFR, and proteinuria
Model adjusted for age, sex, race/ethnicity, hypertension, SBP, DBP, diabetes, and proteinuria
No metabolite reached Bonferroni adjusted significance threshold of 0.0003
Renal Arterio-Venous Metabolite Gradients
We have previously reported metabolomics data on a pilot study of nine individuals who underwent aortic and renal venous sampling, thus providing an initial view of how metabolites are modulated at the organ level [7]. Here, we present data from the complete set of 25 individuals enrolled in the protocol. Clinical characteristics of this hospital-based cohort are shown in Supplementary data, Table S2. Two individuals had ESRD and were on hemodialysis. The remaining 23 individuals had a mean eGFR of 68.1 ± 19.2 mL/min/1.73 m2. Whereas creatinine levels did not change from aorta to renal vein in the two individuals with ESRD, they fell in all the others. Figure 2 shows the ratio of renal venous to arterial creatinine in each individual. As expected, the decrement in creatinine was lower in individuals with lower eGFR (and negligible in the two individuals with ESRD). The mean venous to arterial ratio (V/A) for creatinine was 0.81. Table 3 shows the mean V/A for the metabolites with nominal associations with CKD progression in CRIC (full results are shown in Supplementary data, Table S3). Of these ten metabolites, seven had a V/A ratio ≤1. By contrast, three metabolites had a mean V/A >1, indicating net renal release of these metabolites. Notably, in CRIC all three of the metabolites – threonine, methionine, and arginine – were lower at baseline among individuals with rapid CKD progression compared to individuals with stable CKD and had no association with baseline eGFR.
Figure 2. Renal venous to arterial creatinine ratio in 25 individuals.
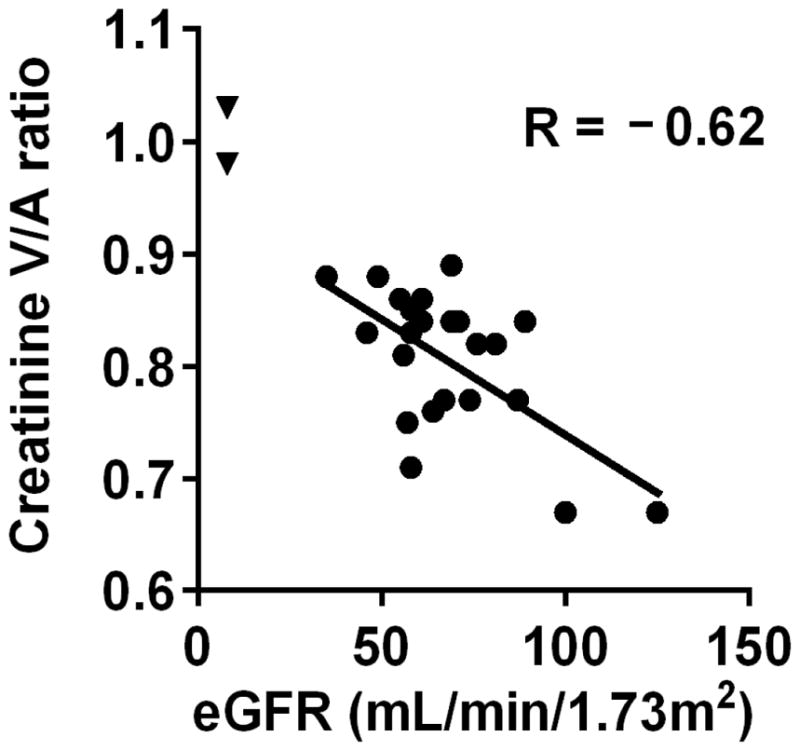
Each data point represents one individual, with eGFR (estimated by MDRD) plotted on the x-axis and creatinine V/A ratio plotted on the y-axis. The two triangles represent the 2 study subjects with ESRD.
Table 3.
Renal arteriovenous gradients for metabolites nominally associated with rapid CKD progression in CRIC
| Metabolite | # of individuals with V/A <1 | # of individuals with V/A ≥1 | Mean V/A Ratio | p-value |
|---|---|---|---|---|
| homogentisate* | 22 | 0 | 0.37 | <0.001 |
| 4-hydroxymandelate* | 22 | 0 | 0.37 | <0.001 |
| 3-methyladipate-pimelate | 23 | 0 | 0.60 | <0.001 |
| cytosine | 21 | 2 | 0.80 | <0.001 |
| glucuronate | 23 | 0 | 0.85 | <0.001 |
| uric acid | 20 | 3 | 0.98 | 0.02 |
| phenylalanine | 10 | 13 | 1.00 | 0.9 |
| threonine | 3 | 20 | 1.07 | <0.001 |
| methionine | 2 | 21 | 1.10 | <0.001 |
| arginine | 0 | 23 | 1.16 | <0.001 |
| creatinine | 23 | 0 | 0.81 | <0.001 |
Abbreviation: V/A = venous to arterial ratio
no data on these metabolites for one study participant
A Panel of Markers of Renal Metabolic Function
Next, we created a panel of markers comprised of the three metabolites with V/A ratio >1 highlighted by our invasive catheterization studies above. The association between this panel of markers and eGFR decline in CRIC is shown in Table 4. In the logistic regression model adjusted for age, sex, race/ethnicity, hypertension, SBP, DBP, diabetes, eGFR and proteinuria (Model 1), the panel of markers had an OR of 0.87 (0.79–0.96) per SD (P = 0.004). Individuals in the second tertile had an OR of 0.81 (0.48–1.37) and individuals in the third tertile had an OR of 0.44 (0.26–0.76) compared to individuals in the first tertile of the marker panel (P = 0.003 for trend). Results were unchanged when the model was further adjusted for CVD and smoking status at baseline (Model 2) or CVD, smoking status, hemoglobin, and FGF-23 levels (Model 3).
Table 4.
Panel of Metabolite Markers of Renal Metabolic Function and Rapid CKD Progression in CRIC
| Adjustment | Arginine + Methionine + Threonine*
|
|||||
|---|---|---|---|---|---|---|
| OR per SD (95% CI) | p-value | OR 1st tertile | OR 2nd tertile (95% CI) | OR 3rd tertile (95% CI) | p-value for trend | |
| Model 1 | 0.87 (0.79, 0.96) | 0.004 | 1.0 | 0.81 (0.48, 1.37) | 0.44 (0.26, 0.76) | 0.003 |
| Model 2 | 0.87 (0.79, 0.96) | 0.004 | 1.0 | 0.78 (0.46, 1.33) | 0.44 (0.26, 0.76) | 0.003 |
| Model 3 | 0.86 (0.78, 0.95) | 0.003 | 1.0 | 0.73 (0.43, 1.25) | 0.41 (0.23, 0.72) | 0.002 |
Abbreviations: OR = odds ratio; SD = standard deviation; CI = confidence interval
Metabolites were natural log transformed, standardized, and then summed to create a composite score
Model 1: Adjusted for eGFR, age, sex, race/ethnicity, systolic blood pressure, diastolic blood pressure, hypertension, diabetes, and 24 hour urine protein
Model 2: Adjusted for all variables in Model 1 + cardiovascular disease and smoking status
Model 3: Adjusted for all variables in Model 2 + FGF-23 and hemoglobin levels
DISCUSSION
Current blood markers of CKD, i.e. serum creatinine and blood urea nitrogen, are unable to identify which individuals will progress and do not provide insight on underlying causal pathways. Increasing interest in CKD biomarker research has been directed towards metabolomics-based discovery because of the broad impact kidney function has on circulating metabolites and because metabolites may themselves play functional roles in CKD pathogenesis. In a nested case-control analysis in the CRIC Study, we found several metabolite alterations nominally associated with subsequent disease progression. Although these metabolite associations were not significant after adjusting for multiple testing, our data on renal arteriovenous gradients coupled with an analysis of cross-sectional metabolite associations with eGFR nominate select markers of renal metabolic function for future investigation.
In order to illustrate the value of metabolite profiling in the context of a well characterized clinical cohort, we examined metabolite measures in relation to several phenotypes ascertained in CRIC. For metabolites that had been assayed previously, the correlation with our LC-MS measures ranged from excellent (creatinine) to modest (uric acid). Given the multiplex nature of the LC-MS methods, it is possible that the individualized measures provide more precise results. However, we do note that our LC-MS data were generated in a single sample run, thus circumventing any issues with batch effect or inter-laboratory variability that may have affected the spectrophotometric data. In this regard, it is interesting that the association between uric acid and CKD progression is stronger for data generated by LC-MS (OR 9.72 [2.17–43.45], P = 0.003) than enzymatic assay (OR 2.13 [0.80–5.69], P = 0.13, not shown). In other studies, uric acid has been found to have a variable association with incident CKD and CKD progression, and whether it is a modifiable risk factors remains an area of controversy [16–20]. In addition to overlap with previously examined endogenous metabolites, our platform provides an objective read-out of medication and nicotine input; in the case of smoking our data indicate that some self-reported non-smokers are in fact subject to considerable nicotine exposure, either directly or as a result of second-hand smoke. By integrating both endogenous and exogenous (i.e. pharmaceutical or environmental) inputs, metabolomics has the potential to more finely subtype study participants and risk factors in epidemiologic studies.
Our primary analysis, a case-control examination of metabolites and CKD progression in CRIC, builds on and extends a growing body of literature. Several large studies have examined baseline metabolite levels in >1000 subjects each from FHS, KORA, TwinsUK, and ARIC in relation to the subsequent development of CKD, i.e. crossing below an eGFR of 60 mL/min/1.73m2 [7–10]. Niewczas et al. examined 80 individuals with diabetic nephropathy at the Joslin Diabetes Center, 40 of whom progressed to ESRD [21]. Acknowledging that incomplete overlap of platform coverage significantly limits comparison across studies, no dominant signal has emerged. In general, however, each of these prior studies have identified robust statistical associations between select metabolites and eGFR decline even after adjusting for multiple testing. Two major differences in our study warrant mention. First, unlike the prior studies we matched cases and controls on baseline eGFR and proteinuria. Second, we examined individuals starting at a much lower mean eGFR. The studies cited above had mean eGFRs of 93.6 (FHS), 81.0 (KORA), 93.8 (Twins UK), 105.3 (ARIC), and 81.0 mL/min/1.73 m2 (Joslin), whereas individuals in our study started with a mean eGFR of 43.3 mL/min/1.73 m2. It is possible that as kidney function declines, nonspecific accumulation dilutes the risk signal attributable to select metabolite alterations, particularly metabolite elevations.
In addition to filtering and secreting many plasma metabolites, the kidneys are also responsible for the net release of some metabolites back into circulation. For example, prior studies have firmly established the kidney as an important source of circulating arginine via the intra-renal transamination of citrulline to arginine [22,23]. Our data in CRIC raise the possibility that impairment of this metabolic function is associated with subsequent CKD progression. Interestingly, we found that two additional amino acids nominally associated with CKD progression in CRIC were both depleted in cases relative to controls and were net released across the kidney. None of these three metabolites were correlated with baseline eGFR, and a panel of markers comprised of all three metabolites had a stronger association with case status than each individual analyte. Thus, we raise the hypothesis that arginine, methionine, and threonine are markers of renal metabolic function and that at advanced stages of renal dysfunction their plasma levels provide insight on renal prognosis. Consistent with this paradigm, we note that other read-outs of known renal anabolic functions, i.e. hemoglobin (from erythropoietin) and 1,25-hydroxyvitamin D levels are associated with renal disease progression [24–27]. In addition to reporting on renal metabolic capacity, we do not rule out the possibility that select metabolite depletions could play a causal role in disease progression; for example, as the substrate for nitric oxide synthase, limited arginine bioavailability could have deleterious vascular effects pertinent to CKD progression.
Our study has several strengths. First, it is the largest metabolomics study to date of disease progression in established CKD, utilizing high quality samples from a well-phenotyped, racially diverse cohort. Second, our LC-MS based platform has an established record in biomarker studies of ESRD, diabetes, and cancer as well as genome-wide association study, generating robust associations that in many cases have been confirmed by other investigators [4,15,28–31]. Third, to our knowledge the summary of renal arteriovenous metabolite gradients we have generated is a unique data set and is provided in full in the Supplement. We believe this will be a valuable resource for the interpretation of metabolomics biomarker studies, in nephrology and beyond.
Our study also has several limitations. Most important is the lack of a replication cohort. Clearly, follow-up studies are required to substantiate the metabolite signals identified in our analysis, including the panel of markers of renal metabolic function. Although the construction of this panel was informed by physiologic data, we acknowledge that combining markers significantly increases the potential number of comparisons, necessitating rigorous validation. Our focus on plasma is an additional limitation. Recently, Sharma et al. found decreased levels of several metabolites linked to mitochondrial metabolism in the urine of individuals with diabetic kidney disease [32]. We focused initially on plasma to build on our prior plasma based studies of incident CKD and ESRD and in order to integrate our results with our data on arteriovenous gradients. Future studies should clearly test urine metabolites as markers of CKD progression as well. Future studies should also investigate a wider breadth of metabolites. Whereas we utilized a targeted approach to measure a subset of the metabolome, methods capable of measuring thousands of metabolite peaks (many of unknown identity) are being increasingly deployed in biomarker research.
In conclusion, metabolite profiling in a nested case-control study in the CRIC Study revealed modest metabolite associations with CKD progression. The major difference with prior renal metabolomics studies demonstrating stronger statistical associations with eGFR decline is the considerably lower starting eGFR in our study, which likely results in the nonspecific accumulation of many solutes. In conjunction with an examination of renal arteriovenous gradients our data nevertheless generate interest in metabolite depletion as a potential indicator of renal health. Future efforts will be directed at replicating these observations, as well as expanding the scope of metabolites measured, in a larger sampling of the CRIC Study. These efforts will also investigate whether metabolite profiling can supplement or even clarify other variables captured in CRIC including other biomarkers, medication adherence and comorbidities, and differences in dietary habits and the microbiome.
Supplementary Material
Acknowledgments
This study was supported by NIH grants U01DK060990, K08-DK-090142, and the Extramural Grant Program of Satellite Healthcare, a not-for-profit renal care provider.
Footnotes
Conflict of Interest Statement: the authors report no conflicts of interest regarding this publication
References
- 1.Lindon JC, Holmes E, Bollard ME, Stanley EG, Nicholson JK. Metabonomics technologies and their applications in physiological monitoring, drug safety assessment and disease diagnosis. Biomarkers. 2004;9:1–31. doi: 10.1080/13547500410001668379. [DOI] [PubMed] [Google Scholar]
- 2.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. Hmdb: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhee EP. Metabolomics and renal disease. Curr Opin Nephrol Hypertens. 2015;24:371–379. doi: 10.1097/MNH.0000000000000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee EP, Souza A, Farrell L, Pollak MR, Lewis GD, Steele DJ, Thadhani R, Clish CB, Greka A, Gerszten RE. Metabolite profiling identifies markers of uremia. J Am Soc Nephrol. 2010;21:1041–1051. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato E, Kohno M, Yamamoto M, Fujisawa T, Fujiwara K, Tanaka N. Metabolomic analysis of human plasma from haemodialysis patients. Eur J Clin Invest. 2011;41:241–255. doi: 10.1111/j.1365-2362.2010.02398.x. [DOI] [PubMed] [Google Scholar]
- 6.Godfrey AR, Williams CM, Dudley E, Newton RP, Willshaw P, Mikhail A, Bastin L, Brenton AG. Investigation of uremic analytes in hemodialysate and their structural elucidation from accurate mass maps generated by a multi-dimensional liquid chromatography/mass spectrometry approach. Rapid Commun Mass Spectrom. 2009;23:3194–3204. doi: 10.1002/rcm.4235. [DOI] [PubMed] [Google Scholar]
- 7.Rhee EP, Clish CB, Ghorbani A, Larson MG, Elmariah S, McCabe E, Yang Q, Cheng S, Pierce K, Deik A, Souza AL, Farrell L, Domos C, Yeh RW, Palacios I, Rosenfield K, Vasan RS, Florez JC, Wang TJ, Fox CS, Gerszten RE. A combined epidemiologic and metabolomic approach improves ckd prediction. J Am Soc Nephrol. 2013;24:1330–1338. doi: 10.1681/ASN.2012101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu B, Zheng Y, Nettleton JA, Alexander D, Coresh J, Boerwinkle E. Serum metabolomic profiling and incident ckd among african americans. Clin J Am Soc Nephrol. 2014;9:1410–1417. doi: 10.2215/CJN.11971113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goek ON, Prehn C, Sekula P, Romisch-Margl W, Doring A, Gieger C, Heier M, Koenig W, Wang-Sattler R, Illig T, Suhre K, Adamski J, Kottgen A, Meisinger C. Metabolites associate with kidney function decline and incident chronic kidney disease in the general population. Nephrol Dial Transplant. 2013;28:2131–2138. doi: 10.1093/ndt/gft217. [DOI] [PubMed] [Google Scholar]
- 10.Sekula P, Goek ON, Quaye L, Barrios C, Levey AS, Romisch-Margl W, Menni C, Yet I, Gieger C, Inker LA, Adamski J, Gronwald W, Illig T, Dettmer K, Krumsiek J, Oefner PJ, Valdes AM, Meisinger C, Coresh J, Spector TD, Mohney RP, Suhre K, Kastenmuller G, Kottgen A. A metabolome-wide association study of kidney function and disease in the general population. J Am Soc Nephrol. 2015 Oct 8; doi: 10.1681/ASN.2014111099. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman HI, Appel LJ, Chertow GM, Cifelli D, Cizman B, Daugirdas J, Fink JC, Franklin-Becker ED, Go AS, Hamm LL, He J, Hostetter T, Hsu CY, Jamerson K, Joffe M, Kusek JW, Landis JR, Lash JP, Miller ER, Mohler ER, 3rd, Muntner P, Ojo AO, Rahman M, Townsend RR, Wright JT. The chronic renal insufficiency cohort (cric) study: Design and methods. J Am Soc Nephrol. 2003;14:S148–153. doi: 10.1097/01.asn.0000070149.78399.ce. [DOI] [PubMed] [Google Scholar]
- 12.Lash JP, Go AS, Appel LJ, He J, Ojo A, Rahman M, Townsend RR, Xie D, Cifelli D, Cohan J, Fink JC, Fischer MJ, Gadegbeku C, Hamm LL, Kusek JW, Landis JR, Narva A, Robinson N, Teal V, Feldman HI. Chronic renal insufficiency cohort (cric) study: Baseline characteristics and associations with kidney function. Clin J Am Soc Nephrol. 2009;4:1302–1311. doi: 10.2215/CJN.00070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson AH, Yang W, Hsu CY, Joffe MM, Leonard MB, Xie D, Chen J, Greene T, Jaar BG, Kao P, Kusek JW, Landis JR, Lash JP, Townsend RR, Weir MR, Feldman HI. Estimating gfr among participants in the chronic renal insufficiency cohort (cric) study. Am J Kidney Dis. 2012;60:250–261. doi: 10.1053/j.ajkd.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. Modification of diet in renal disease study group. Ann Intern Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 15.Rhee EP, Ho JE, Chen MH, Shen D, Cheng S, Larson MG, Ghorbani A, Shi X, Helenius IT, O’Donnell CJ, Souza AL, Deik A, Pierce KA, Bullock K, Walford GA, Vasan RS, Florez JC, Clish C, Yeh JR, Wang TJ, Gerszten RE. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 2013;18:130–143. doi: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiner DE, Tighiouart H, Elsayed EF, Griffith JL, Salem DN, Levey AS. Uric acid and incident kidney disease in the community. J Am Soc Nephrol. 2008;19:1204–1211. doi: 10.1681/ASN.2007101075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obermayr RP, Temml C, Gutjahr G, Knechtelsdorfer M, Oberbauer R, Klauser-Braun R. Elevated uric acid increases the risk for kidney disease. J Am Soc Nephrol. 2008;19:2407–2413. doi: 10.1681/ASN.2008010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chonchol M, Shlipak MG, Katz R, Sarnak MJ, Newman AB, Siscovick DS, Kestenbaum B, Carney JK, Fried LF. Relationship of uric acid with progression of kidney disease. Am J Kidney Dis. 2007;50:239–247. doi: 10.1053/j.ajkd.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Madero M, Sarnak MJ, Wang X, Greene T, Beck GJ, Kusek JW, Collins AJ, Levey AS, Menon V. Uric acid and long-term outcomes in CKD. Am J Kidney Dis. 2009;53:796–803. doi: 10.1053/j.ajkd.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jalal DI, Chonchol M, Chen W, Targher G. Uric acid as a target of therapy in CKD. Am J Kidney Dis. 2013;61:134–146. doi: 10.1053/j.ajkd.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niewczas MA, Sirich TL, Mathew AV, Skupien J, Mohney RP, Warram JH, Smiles A, Huang X, Walker W, Byun J, Karoly ED, Kensicki EM, Berry GT, Bonventre JV, Pennathur S, Meyer TW, Krolewski AS. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: Metabolomic study. Kidney Int. 2014;85:1214–1224. doi: 10.1038/ki.2013.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owen EE, Robinson RR. Amino acid extraction and ammonia metabolism by the human kidney during the prolonged administration of ammonium chloride. J Clin Invest. 1963;42:263–276. doi: 10.1172/JCI104713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tizianello A, De Ferrari G, Garibotto G, Gurreri G, Robaudo C. Renal metabolism of amino acids and ammonia in subjects with normal renal function and in patients with chronic renal insufficiency. J Clin Invest. 1980;65:1162–1173. doi: 10.1172/JCI109771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossert J, Froissart M. Role of anemia in progression of chronic kidney disease. Semin Nephrol. 2006;26:283–289. doi: 10.1016/j.semnephrol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Iseki K, Kohagura K. Anemia as a risk factor for chronic kidney disease. Kidney Int Suppl. 2007:S4–9. doi: 10.1038/sj.ki.5002481. [DOI] [PubMed] [Google Scholar]
- 26.Kendrick J, Cheung AK, Kaufman JS, Greene T, Roberts WL, Smits G, Chonchol M. Associations of plasma 25-hydroxyvitamin d and 1,25-dihydroxyvitamin d concentrations with death and progression to maintenance dialysis in patients with advanced kidney disease. Am J Kidney Dis. 2012;60:567–575. doi: 10.1053/j.ajkd.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izumaru K, Ninomiya T, Nagata M, Usui T, Yoshida D, Yonemoto K, Fukuhara M, Tsuruya K, Kitazono T, Kiyohara Y. Serum 1,25-dihydroxyvitamin d and the development of kidney dysfunction in a japanese community. Circ J. 2014;78:732–737. doi: 10.1253/circj.cj-13-0422. [DOI] [PubMed] [Google Scholar]
- 28.Rhee EP, Feldman HI. Metabolite markers of incident ckd risk. Clin J Am Soc Nephrol. 2014;9:1344–1346. doi: 10.2215/CJN.05910614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O’Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayers JR, Wu C, Clish CB, Kraft P, Torrence ME, Fiske BP, Yuan C, Bao Y, Townsend MK, Tworoger SS, Davidson SM, Papagiannakopoulos T, Yang A, Dayton TL, Ogino S, Stampfer MJ, Giovannucci EL, Qian ZR, Rubinson DA, Ma J, Sesso HD, Gaziano JM, Cochrane BB, Liu S, Wactawski-Wende J, Manson JE, Pollak MN, Kimmelman AC, Souza A, Pierce K, Wang TJ, Gerszten RE, Fuchs CS, Vander Heiden MG, Wolpin BM. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med. 2014;20:1193–1198. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalim S, Clish CB, Wenger J, Elmariah S, Yeh RW, Deferio JJ, Pierce K, Deik A, Gerszten RE, Thadhani R, Rhee EP. A plasma long-chain acylcarnitine predicts cardiovascular mortality in incident dialysis patients. J Am Heart Assoc. 2013;2:e000542. doi: 10.1161/JAHA.113.000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma K, Karl B, Mathew AV, Gangoiti JA, Wassel CL, Saito R, Pu M, Sharma S, You YH, Wang L, Diamond-Stanic M, Lindenmeyer MT, Forsblom C, Wu W, Ix JH, Ideker T, Kopp JB, Nigam SK, Cohen CD, Groop PH, Barshop BA, Natarajan L, Nyhan WL, Naviaux RK. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol. 2013;24:1901–1912. doi: 10.1681/ASN.2013020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.