Abstract
We previously identified a locus linked to total cholesterol (TC) concentration in Pima Indians on chromosome 19p. To characterize this locus, we genotyped >2000 SNPs in 1838 Pimas and assessed association with log(TC). We observed evidence for association with log(TC) with rs2278426 (3.5% decrease/copy of the T allele; P=5.045 x 10−6) in the ANGPTL8 (angiopoietin-like 8) gene. We replicated this association in 2413 participants of the San Antonio Mexican American Family Study (SAMAFS: 2.0% decrease per copy of the T allele; P=0.005842). In a meta-analysis of the combined data, we found the strongest estimated effect with rs2278426 (P=2.563 x 10−7). The variant T allele at rs2278426 predicts an Arg59Trp substitution and has previously been associated with LDL-C and HDL-C. In Pimas and SAMAFS participants, the T allele of rs2278426 was associated with reduced HDL-C levels (P=0.000741 and 0.00002, respectively), and the combined estimated effect for the two cohorts was −3.8% (P=8.526 x 10−8). ANGPTL8 transcript and protein levels increased in response to both glucose and insulin. The variant allele was associated with increased levels of cleaved ANGPTL3. We conclude that individuals with the variant allele may have lower TC and HDL-C levels due to increased activation of ANGPTL3 by ANGPTL8.
Keywords: Cholesterol, Diabetes, Dyslipidemias, Genetics, HDL, Lipasin, Betatrophin, RIFL, C19ORF80, LOC55908
Introduction
Coronary heart disease (CHD) is the leading cause of death in the United States and contributes to an economic burden totaling over $142 billion (1). Major risk factors for CHD include hypertension (2–4) and abnormal levels of circulating lipids (5). Diet, physical activity levels, and behaviors such as tobacco use and alcohol consumption are among the most common, modifiable environmental factors affecting lipid levels (6–8). Genetic factors are also well-recognized determinants of dyslipidemia, and variants with significant effects on lipid levels have been identified for a number of monogenic familial disorders. For example, mutations in the low density lipoprotein receptor (LDLR) gene underlie familial hypercholesterolemia, which is marked by excessively high low-density lipoprotein cholesterol (LDL-C) levels (9), and defects in the ATP binding cassette A1 (ABCA1) gene lead to Tangier disease, which is characterized by reduced high-density lipoprotein cholesterol (HDL-C) levels (10–12). Although the underlying genetic bases for numerous other monogenic lipid-related disorders have been identified, these variants account for only a small percentage of CHD in the United States. In contrast, genetic determinants of lipid abnormalities leading to complex disorders like hypercholesterolemia and hypertriglyceridemia, which contribute substantially to the risk of CHD development in the general population, are only recently being identified.
Over ninety genome-wide linkage scans have been performed to identify loci affecting lipid levels and lipid-related traits, including total cholesterol (TC), HDL-C, LDL-C, and triglycerides (13). Although linkage for lipid traits has been observed on almost every chromosome, 19p13.3-q13.32 is the most frequently and consistently reported region of linkage for TC and/or LDL-C in the literature. Linkage to this region has been implicated for lipid traits in at least fifteen independent studies (14–25), and in a meta-analysis of published genome scans for quantitative lipid traits conducted in families ascertained for T2DM, we (26) observed strong and consistent support for linkage of TC on 19p13.3-p12 (6.57–38.05cM; P=0.00026), 19p12-q13.13 (38.05–69.53cM; P=0.00001), and 19q13.13-q13.43 (69.53–101.1cM; P=0.00033), and of LDL-C on 19p13.3-p12 (P=0.00041). Results from multiple genome-wide association studies (GWAS) have also identified loci underlying lipid traits on chromosome 19 (27–32). Combined, findings from both linkage and association studies support the presence of one or more loci that underlie variability in lipid traits on chromosome 19.
We have previously identified a locus linked to fasting serum TC concentration in Pima Indians on chromosome 19p (21). As part of our efforts to characterize this locus and identify variants with effects on lipid traits in this population, we sought to refine the region of linkage and identify alleles associated with TC concentration in Pima Indians. Here we show that genetic markers in the genes encoding dedicator of cytokinesis 6 (DOCK6) and angiopoietin-like 8 (ANGPTL8) are associated with TC and HDL-C concentration in Pima Indians and Mexican Americans. ANGPTL8, is known by a variety of alternative names (i.e., betatrophin, lipasin, TD26, LOC55909, C19orf80, and RIFL), and has recently emerged as a novel regulator of lipid metabolism (33–37) that is strongly expressed in adipocytes and regulated by fasting and insulin (35, 37). Previous studies involving cell culture and mouse models suggest an important role for ANGPTL8 in both the regulation of lipid metabolism (34–36) and the maintenance of glucose homeostasis (38). We provide evidence showing that ANGPTL8 expression is regulated by insulin and glucose in cultured human liver cells in a dose- and time-dependent manner. We also demonstrate that the rs2278426 variant in ANGPTL8, which causes an Arg59Trp substitution, affects levels of the activated form of ANGPTL3, a relationship that is modulated by insulin. These results confirm a role for ANGPTL8 in lipoprotein metabolism and provide novel support for functional consequences of the rs2278426 variant.
Materials and Methods
Study populations
From 1965 to 2007, the Pima Indians from the Gila River Indian Community participated in a longitudinal survey. Approximately every 2 years, each resident ≥5 years of age was invited to receive a standardized medical examination (39). In addition to a 75 g glucose tolerance test, total fasting serum cholesterol and HDL-C concentrations were measured enzymatically (40), and measurements used in the genome scan were those taken from the last available examination for each individual (21). Assay accuracy was monitored and verified by the CDC Laboratory Program Office, the College of American Pathologists Surveys Program, or the American Association of Bioanalysts. The interassay coefficient of variation was 1.97% and 4.83% for TC and HDL-C measurements, respectively.
Characteristics of the Pima Indian study sample are shown in Table 1. The sample included all individuals who participated in the genome-wide linkage study (n=912) and a random sample of participants in the longitudinal Pima study, excluding first-degree relatives (n=926). Inclusion criteria for the study sample were 1) age ≥20 years, 2) total cholesterol levels available at examination, and 3) known diabetes status. Individuals taking lipid medications were excluded from the study. Approvals were obtained from the Institutional Review Boards of the National Institute of Diabetes and Digestive and Kidney Diseases, and the Translational Genomics Research Institute. NIDDK gave regular reports of study findings to the Council of the Gila River Indian Community. All subjects provided written informed consent.
Table 1.
Characteristics of American Indian study sample (N=1838)
| Mean ± SD (or %) | Median | Range | |
|---|---|---|---|
| Age (yrs) | 45.5 ±12.8 | 44.5 | 20.2–88.0 |
| Male | 41.8 | - | - |
| Diabetic | 58.3 | - | - |
| BMI (kg/m2) | 35.8±8.6 | 34.4 | 11.5–72.5 |
| TC (mg/dl) | 178.2 ±41.9 | 174.0 | 36.0–387.0 |
| HDL-C (mg/dl) | 46.0 ±13.8 | 44.0 | 10.0–128.0 |
| TG (mg/dl) | 152.7 ±106.9 | 128.0 | 16.0–1240.0 |
| Calc-LDL-C (mg/dl) | 101.7 ±32.8 | 100.0 | 18.8–260.6 |
Inclusion criteria for the study sample were 1) age ≥20 years, 2) total cholesterol levels available at examination, and 3) known diabetes status.
We validated the most strongly associated DOCK6 and ANGPTL8 markers in a Mexican American cohort using family data obtained from two studies: the San Antonio Family Heart Study (SAFHS) and the San Antonio Family Diabetes/Gallbladder Study (SAFDGS). Details of the SAFHS (41, 42) and SAFDGS (43) have been published elsewhere. Together, the SAFHS and SAFDGS comprise the San Antonio Mexican American Family Study (SAMAFS), which represents approximately 2,500 individuals from about 80 Mexican American complex pedigrees. The clinical characteristics of the individuals that are part of this replication study are reported in Table 2. Genome-wide SNP genotypic data (~1 million SNPs acquired using the Illumina platform) were available for SAMAFS individuals (44). For validation studies, we performed association analysis between selected DOCK6 and ANGPTL8 markers that were most strongly associated with fasting serum TC in the Pima population and TC concentrations in SAMAFS data. TC concentrations used in these analyses were taken from measures obtained at the most recent clinical examination (Pimas) or based on information at the last study examination (SAMAFS).
Table 2.
Characteristics of SAMAFS cohort (N=2413)
| Mean ± SD or % | |
|---|---|
| Age | 47.4 ± 17.2 |
| Male | 40.9 |
| T2D | 26.8 |
| BMI (kg/m2) | 31.1 ± 7.3 |
| TC (mg/dl) | 187.0 ± 39.9 |
| HDL-C (mg/dl) | 47.9 ± 13.7 |
| TG (mg/dl) | 147.9 ± 109.1 |
Selection of markers for genotyping
We genotyped markers in Pima samples in two stages; the purpose of the first stage was to refine the linkage interval, while that of the second was to fine-map the resulting reduced linkage interval by association methods. For the first stage, we selected a panel of 346 SNPs from the HapMap database spanning the linkage interval, and genotyped them in the Pima Indian families who participated in the original genome scan. Markers common to all four populations available in the HapMap database (i.e., Caucasian, African, Chinese, and Japanese) were selected based upon physical position using a minor allele frequency ≥0.10 and r2=0.80. Mean inter-marker distance was ~132 kb. For the second stage, we selected 1536 markers spanning the refined linkage interval using the same criterion as in the first stage, along with the Tagger program and genotype data obtained from the CEU population. This marker coverage provided an average density of 1 SNP/8.9 kb. Despite this density, several gaps in marker distribution were present, primarily due to genotyping failure; we therefore filled these regions with an additional 450 SNPs. In addition, a genome-wide association study performed in Pima Indians under the auspices of an unrelated project (45) identified approximately 300 SNPs spanning the region of interest that were not tagged by markers selected in CEU with an r2≥0.80 and thus, we assessed these markers in our study sample. In total, therefore, we genotyped an additional 2179 markers in the two study samples derived from the Pima Indian population comprising this investigation.
In addition to the chromosome 19 markers described above, we selected markers that were previously associated with total cholesterol levels or related traits in other GWA studies, including rs6511720, rs2228671, rs688, rs2228603, rs10401969, rs16996148, rs17545624, rs4803750, rs2075650, rs4420638, and rs16979595 (27, 28, 30–32) for genotyping.
SNP genotyping
We utilized the Illumina GoldenGate assay in conjunction with the Universal BeadChip technology and BeadArray Reader for genotyping according to the manufacturer’s protocol. Data from these images were analyzed with the Genome Studio v1.9.4 program (Illumina). Of the genotyped markers, 105 were monomorphic and 51 had a minor allele frequency <0.001, and these were excluded from further analyses. The observed genotype frequency for each SNP was assessed for deviation from that expected under Hardy-Weinberg equilibrium using a chi-square test. Control measures (i.e., encrypted samples and determination of Mendelian incompatibility) were employed to assess data quality. Genotypes with errors were recoded as missing before proceeding with analysis. After assessing each of the remaining SNPs for deviation from that expected under Hardy-Weinberg equilibrium, reproduction frequency rates among quality control samples, and error rates, we removed an additional 165 markers from analysis. In total, 1858 SNPs were carried forward for statistical analysis.
Statistical analysis
In the Pima studies, we analyzed log values of TC and HDL-C using measurements from the last exam at which diabetes status was known and age was at least 20 years. P-values were adjusted for age, sex, diabetes status, and exam date. Association between SNP genotype and logarithmically transformed TC or HDL-C concentration was tested using linear regression. We accounted for family membership, as some of the study sample participants were family members, using generalized estimating equation procedures. For the SAMAFS data, we analyzed the association with the TC or HDL-C levels using a classical additive measured genotype approach, allowing for non-independence among pedigree members, as implemented in SOLAR (http://solar.txbiomedgenetics.org). The TC or HDL-C values were adjusted for covariate effects of age, age x sex, age2, clinic exam year, diabetes status, and potential population stratification influences using the first three principal components (PCs). Results were combined across the Pima and SAMAFS studies by fixed effects meta-analysis using weights derived from the inverse of the variance of the effect estimates.
Cell culture and cell treatments
We purchased Hep G2 cells from ATCC (Manassas, VA) and cultured them in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% fetal bovine serum (FBS) according to the manufacturer’s protocol. Approximately 1.3 x104 cells/cm2 were seeded in 75 cm2 cell culture flasks (VWR International; Radnor, PA) containing 10 mL cell culture medium and placed at 37°C in a Hera Cell 5% CO2 incubator (ThermoFisher Scientific; Waltham, MA). Culture medium was replaced the first day after seeding and then every 48 hours until treatment.
For the insulin dose-response experiments, we seeded approximately 1.0 x 106 Hep G2 cells in 60 mm tissue culture plates (Corning Life Sciences; Lowell, MA) containing 10 mL EMEM supplemented with 10% FBS overnight in a 37° incubator. Insulin was diluted to 100 mM in sterile water per the manufacturer’s instructions (Sigma Aldrich; St. Louis, MO). Cells were serum-starved overnight and then treated for 24 hours with serum-free media supplemented with 10, 25, 50, 75, 100, or 150 nM insulin. For the time-course experiments, we incubated Hep G2 cells for 2, 4, 8, 12, 16, 24, 32, and 48 hours in media containing 100 nM insulin. For the glucose treatment, we seeded cells as described above and serum-starved cells overnight prior to initiating treatments with normal glucose (NG: 5.6 mM), high glucose (HG: 30 mM) or an osmotic control (OC: 5.6 mM glucose + 19.4 mM 3-0-methyl-D-glucopyranose) for 24 hours. Cells were grown to approximately 80% confluence at the time of treatment. All experiments were performed in triplicate.
RNA extraction and quantitative real-time PCR (qPCR) analysis
RNA was extracted using the RNeasy kit (Qiagen; Germantown, MD) following the manufacturer’s instructions and quantified using the NanoDrop 1000 spectrophotometer (Thermo Scientific; Wilmington, DE). First-strand cDNA was synthesized from 500 ng total RNA per reaction using the TaqMan RNA-to-Ct 1-Step kit (Life Technologies; Grand Island, NY) according to the manufacturer’s protocol, followed by qPCR analysis in conjunction with the ABI Prism 7900 HT Sequence Detector apparatus (Life Technologies). Cycle threshold value was generated using SDS software version 2.3 (Life Technologies). The −ΔΔCt method was used to determine fold-change of gene expression between samples. Data were normalized using beta-actin (ACTB) and then analyzed using ExpressionSuite Software 1.0.3 (Life Technologies). A two-tailed t-test was used to determine p-values.
Quantitative determination of ANGPTL8 protein concentration in cell culture media
We collected cell culture media into 15 ml tubes (Becton Dickinson; Franklin Lakes, NJ) and centrifuged at 4°C at 5000 x g for 5 minutes to remove particulates. The supernatant was concentrated using Amicon Ultracel 10K centrifugal filters (EMD Millipore; Billerica, MA) and then frozen at −80°C. We determined protein concentrations using the bicinchoninic acid assay (BCA) per the established protocol (ThermoFisher Scientific). We used a commercial sandwich enzyme-linked immunosorbent assay (ELISA) kit (EIAab; Wuhan, China) to assess the concentration of ANGPTL8 protein in the cell culture supernatant.
ANGPTL8/ANGPTL3 co-expression studies
Vectors containing ANGPTL3, ANGPTL8 wildtype (C allele), and ANGPTL8 variant (T allele) were designed using VectorBuilder and commercially synthesized (Cyagen Biosciences; Santa Clara, CA). Vector designs are available upon request. HEK293 cells were plated on 60 mm dishes and grown in Dulbecco’s Modified Eagle’s Medium (DMEM) with 5 mM glucose and 10% FBS until 90% confluent, then cells were serum-starved for 24 hours prior to transfection. Cells were washed twice with PBS, cultured with serum-free DMEM plus 5 mM glucose and treated with vector and Lipofectamine 2000 (Life Technologies) for six hours. Following transfection, the media was treated with 30 mM glucose or 100 nM insulin, incubated for an additional eight hours, and then collected and concentrated with Amicon Ultra 10K Centrifugal Filters (Millipore). Cells were lysed in RIPA buffer (Pierce). The conditioned media and cell lysates were quantitated using the BCA method, electrophoresed on precast 10% Bis-Tris Gels (Life Technologies), and transferred to Hybond P PVDF membranes (GE Healthcare; Buckinghamshire, UK). The membranes were blocked in 5% skim milk overnight at 4°C, and then incubated with primary antibody (1:500) against human ANGPTL3 specific to the C-terminal fibrinogen-like domain (Cayman Chemical; Ann Arbor, MI). Lysates were incubated in primary antibody (1:1000) against human ANGPTL8 (Abcam; Cambridge, MA). Following an overnight incubation, blots were washed three times for five minutes each in Tris-buffered Saline with Tween 20 (TBST), and then incubated with a 1:1000 dilution of rabbit anti-mouse antibody (Santa Cruz Biotechnology; Dallas, TX) for one hour. Blots were washed repeatedly in TBST and then developed using ECL Prime Western detection reagent (GE Healthcare) and Amersham Hyperfilm ECL (GE Healthcare).
Results
Association analysis of genotyped markers with TC concentration in Pima Indians
To narrow the 30 cM 1-LOD support interval of linkage for fasting serum TC concentration, we first genotyped 346 SNPs in Pima Indian families who participated in the original genome scan (21). Markers common to the Caucasian (CEU), African (YRI), Chinese (CHB), and Japanese (JPT) populations available in the HapMap database (http://www.hapmap.org) were selected based upon physical position using a minor allele frequency ≥0.10 and r2=0.80. Mean inter-marker distance was ~132 kb. Variance components linkage analysis, as implemented in the program Merlin (46), was performed. We observed resolution of the interval, with the peak of linkage at marker rs9807915 (Supplemental Fig. 1). The region of linkage was substantially narrowed to a 1-LOD support interval (rs1054623-rs10405035) of ~15.7 cM, which corresponds to a physical distance of ~6.8 Mb (chr 19:8,492,086-15,308,898).
To determine the extent to which loci previously identified in various GWAS (27, 28, 30–32) contributed to TC concentration in Pima Indians, we also genotyped 11 markers (rs6511720, rs2228671, rs688, rs2228603, rs10401969, rs16996148, rs17545624, rs4803750, rs2075650, rs4420638, and rs16979595) showing statistically significant evidence for association in previously published studies. Of the genotyped markers, only rs10401969, located in the SURP And G Patch Domain Containing 1 (SUGP1) gene, was nominally associated with fasting TC concentration (P=0.0437). None of the remaining markers showed statistically significant evidence for association with this trait in Pima Indians.
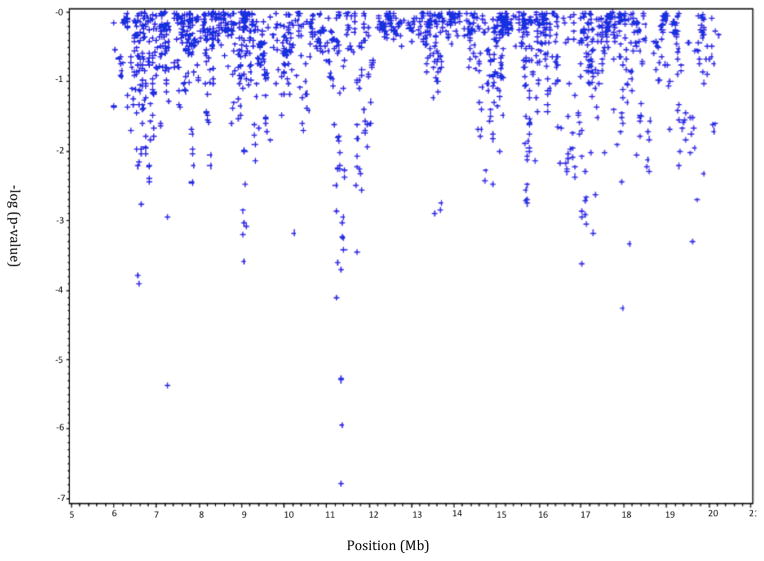
We next genotyped over 1800 markers spanning the refined linkage interval and assessed the evidence for association between genetic variants and log(TC) concentration. The results from the tests of general association for TC concentration for all genotyped SNPs are shown in Fig. 1. The strongest evidence for association with TC concentration was found with marker rs4804576, located in the gene encoding dedicator of cytokinesis 6 (DOCK6), which showed an decrease of 4.0% in TC concentration per copy of the A allele (P=2.0 x 10−7). We also observed evidence for association with marker rs2278426, located in the angiopoietin-like 8 gene (ANGPTL8), which showed a 3.5% decrease in TC concentration per copy of the T allele (P=5.0 x 10−6) and rs2116876, in DOCK6, which showed a 3.8% decrease per copy of the A allele; P=1.1 x 10−6. A summary of the association findings is shown in Table 3. The rs2278426 marker was not associated with diabetes (odds ratio=0.94 per copy of the T allele, P=0.44), nor was it associated with 2-hour post-load glucose concentration in nondiabetic individuals (P=0.38).
Figure 1. General test of association of all markers genotyped on Chr 19 with total cholesterol levels in Pima Indians.
Data were analyzed under an additive model and adjusted for age, sex, diabetes status, and exam date. Only SNPs with a MAF ≥0.01 were analyzed (N=1838).
Table 3.
Evidence for association between DOCK6/ANGPTL8 SNPs and TC concentration in American Indians
| SNP | Locus | 1a | 2 | Freqb | 11c | 12c | 22c | TC11d | TC12d | TC22d | Effecte | P-valuef |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs6511727 | DOCK6 | C | A | 0.71 | 861 | 699 | 139 | 175.3 | 181.0 | 179.3 | −2.4% | 0.0061933 |
| rs11878417 | DOCK6 | A | G | 0.72 | 870 | 645 | 134 | 175.3 | 181.8 | 178.8 | −2.5% | 0.0054833 |
| rs4804576 | DOCK6 | A | C | 0.48 | 385 | 788 | 465 | 170.1 | 179.0 | 184.6 | −4.0% | 0.0000002 |
| rs2278426 | ANGPTL8 | T | C | 0.50 | 430 | 856 | 425 | 170.7 | 178.7 | 183.6 | −3.5% | 0.0000050 |
| rs2116876 | DOCK6 | A | G | 0.50 | 412 | 861 | 417 | 170.9 | 178.1 | 184.6 | −3.8% | 0.0000011 |
| rs7248924 | DOCK6 | G | A | 0.54 | 498 | 761 | 367 | 173.5 | 179.9 | 181.4 | −2.6% | 0.0005884 |
| rs322132 | DOCK6 | A | G | 0.55 | 509 | 785 | 349 | 174.0 | 179.6 | 181.8 | −2.5% | 0.0009245 |
| rs12608933 | DOCK6 | A | T | 0.55 | 512 | 849 | 342 | 174.0 | 178.5 | 182.2 | −2.6% | 0.0005602 |
Numerical designations refer to each allele of marker;
Frequency of the “1” allele;
Number of individuals with each genotype;
Mean TC concentrations relative to each genotype;
Effect expressed as a multiplier (in %) per copy of the “1” allele;
P-value based on an additive model
Validation of markers in DOCK6 and ANGPTL8 with TC concentration in Mexican Americans
We evaluated those markers showing the strongest evidence for association with TC concentration in the DOCK6 and ANGPTL8 genes in an independent population of Mexican Americans from the SAMAFS. Genotype data for markers (rs6611727, rs2278426, rs2116876, rs7248924, rs322132, and rs737337, which is concordant with rs4804576 in the CEU population) were analyzed in more than 2400 members of the SAFS using an additive measured genotype approach. As shown in Table 4, several markers showed association with TC concentration with rs2278426 yielding the strongest evidence (2.0% decrease per copy of the T allele; P=0.005842). In an analysis of the combined data from Pimas and SAMAFS participants, we continued to observe statistically significant evidence for association with several DOCK6 and ANGPTL8 variants (Table 5). Among these, rs2278426 showed the strongest estimated effect size of −2.7% per copy of the T allele (2.563 x 10−7).
Table 4.
Evidence for association between DOCK6/ANGPTL8 SNPs and TC concentration in SAMAFS
| SNP | 1 | 2 | Freq | 11 | 12 | 22 | TC11 | TC12 | TC22 | Effecta | P-valueb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs6511727 | C | A | 0.64 | 980 | 1133 | 300 | 194.3 | 195.7 | 197.2 | −0.8% | 0.255555 |
| rs2278426 | T | C | 0.25 | 155 | 916 | 1342 | 189.4 | 193.4 | 197.3 | −2.0% | 0.005842 |
| rs2116876 | A | G | 0.36 | 311 | 1170 | 932 | 191.1 | 194.4 | 197.7 | −1.7% | 0.010212 |
| rs7248924 | G | A | 0.33 | 250 | 1143 | 1020 | 190.5 | 194.1 | 197.7 | −1.8% | 0.006122 |
| rs322132 | A | G | 0.33 | 260 | 1143 | 1010 | 190.7 | 194.2 | 197.7 | −1.8% | 0.008235 |
| rs737337c | C | T | 0.28 | 185 | 989 | 1239 | 190.5 | 193.9 | 197.2 | −1.7% | 0.016029 |
Effect expressed as a multiplier (in %) per copy of the “1” allele;
adjusted for covariate effects described in the text;
in 100% LD with rs4804576 in CEU population
Table 5.
Meta-analysis of American Indian and SAMAFS data
| Marker | American Indians | SAMAFS | Combined | ||||
|---|---|---|---|---|---|---|---|
| Alleles (1/2) | Effecta | P-value | Effect | P-value | Effect | P-value | |
| TC | |||||||
| rs6511727 | (C/A) | −2.4% | 0.006193 | −0.8% | 0.255555 | −1.4% | 0.0120 |
| rs4804576b | (A/C) | −4.0% | 1.660 x 10−7 | −1.7% | 0.016029 | −2.7% | 1.242 x 10−7 |
| rs2278426 | (T/C) | −3.5% | 5.045 x 10−6 | −2.0% | 0.005842 | −2.7% | 2.563 x 10−7 |
| rs2116876 | (A/G) | −3.8% | 1.114 x 10−6 | −1.7% | 0.010212 | −2.5% | 3.684 x 10−7 |
| rs7248924 | (G/A) | −2.6% | 5.884 x 10−4 | −1.8% | 0.006122 | −2.2% | 1.514 x 10−5 |
| rs322122 | (A/G) | −2.5% | 0.000924 | −1.8% | 0.008235 | −2.1% | 3.049 x 10−5 |
| HDL-C | |||||||
| rs2278426 | (T/C) | −3.2% | 0.000741 | −4.5% | 0.00002 | −3.8% | 8.526 x 10−8 |
Effect is expressed as a multiplier (in %) per copy of the “1” allele;
Results for SAMAFS are given for rs737337, which is 100% concordant with rs4804576
Conditional analyses of rs2278426 and DOCK6/ANGPTL8 markers in American Indians
The rs2278426 marker, which showed the strongest association in the combined analysis, was in moderate linkage disequilibrium with other variants in the region in Pimas (Supplemental Figure 2). To evaluate potential independent effects we analyzed the association of the other DOCK6/ANGPTL8 markers conditional on the effect of rs2278426. The marker rs2278426 remained significantly associated with TC conditional on the effects of most other the markers, except for rs4804576 and rs2116876, with which it was highly concordant (r2=0.89 with each), and most other markers (except for rs4804576) were no longer significant conditional on the effect of rs2278426 (Supplemental Table 1).
Association of rs2278426 with HDL-C in Pima Indians and Mexican Americans
Previous studies (34, 47) reported association between the variant T allele of rs2278426 and reduced plasma levels of HDL-C. When we investigated this relationship in the two present populations, we found the variant T allele was associated with lower HDL-C levels in both Pima Indians (3.2% decrease per copy; P=0.000741) and Mexican Americans (4.5% decrease per copy; P=0.00002) as shown in Table 5. Meta-analysis of the combined Pima Indian and SAMAFS data found a 3.8% decrease in HDL-C levels per copy of the T allele (P=8.526 x 10−8). Because of these findings of association, combined with previous studies supporting a strong role for ANGPTL8 in lipid regulation, and the relative lack of evidence for DOCK6 in lipid-related biological pathways, we focused the remainder of our efforts on the rs2278426 locus.
ANGPTL8 transcript and protein levels are regulated by insulin and glucose
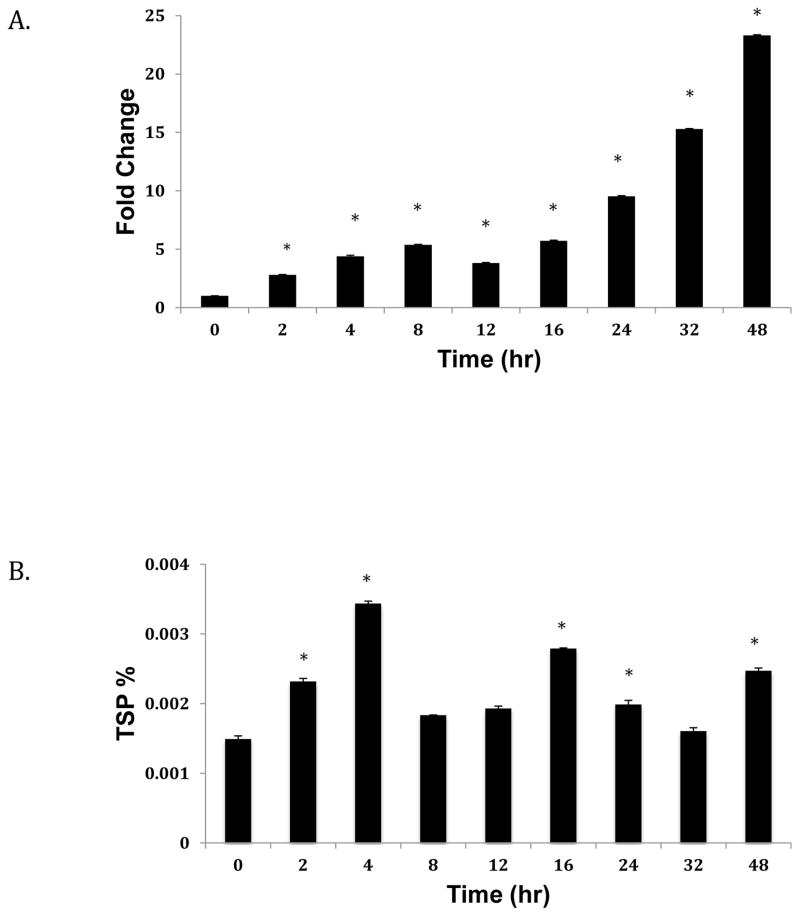
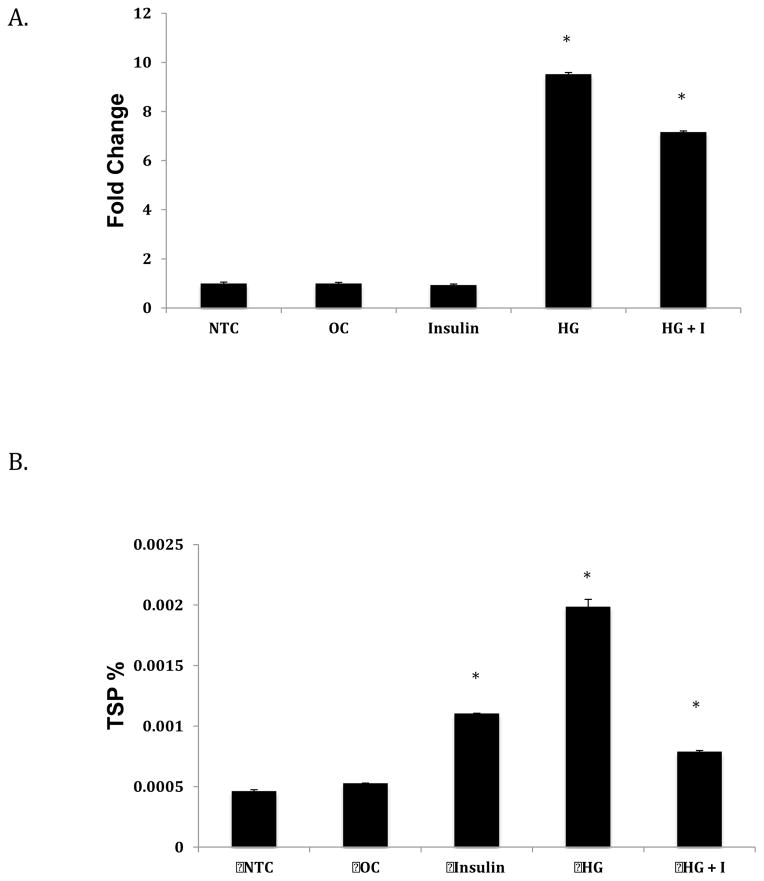
ANGPTL8 is upregulated by refeeding, and has also been identified as an insulin target gene in adipocytes (35). In 3T3-L1 cells, ANGPTL8 mRNA expression increases in response to glucose and insulin (35). To determine whether ANGPTL8 expression is regulated by these factors in liver, we assessed transcript and protein expression in response to glucose, insulin, and a combination of the two in Hep G2 cells. As shown in Figure 2A, we observed a significantly linear increase in ANGPTL8 transcript expression in response to 30 mM glucose. The highest increase corresponding to nearly 25-fold was at 48 hours. ANGPTL8 protein expression also increased in response to high glucose treatment, but in a non-linear manner. The highest increases in protein expression were observed at 4, 16, and 48 hours (P<0.05).
Figure 2. Effect of high glucose treatment on ANGPTL8 transcript (A) and protein (B) expression.
Hep G2 cells were grown to 80% confluence and starved overnight with serum-free media before treatment with conditioned media over 48 hrs. Total RNA and protein were extracted and expression levels assessed as described in the Methods section. mRNA levels were normalized against ACTB. Protein concentrations are shown as percentage of total soluble protein (TSP). Results represent averages from three independent experiments. Data are shown as means ± standard deviation; a two-tailed t-test was used to determine p-values; *p<0.05.
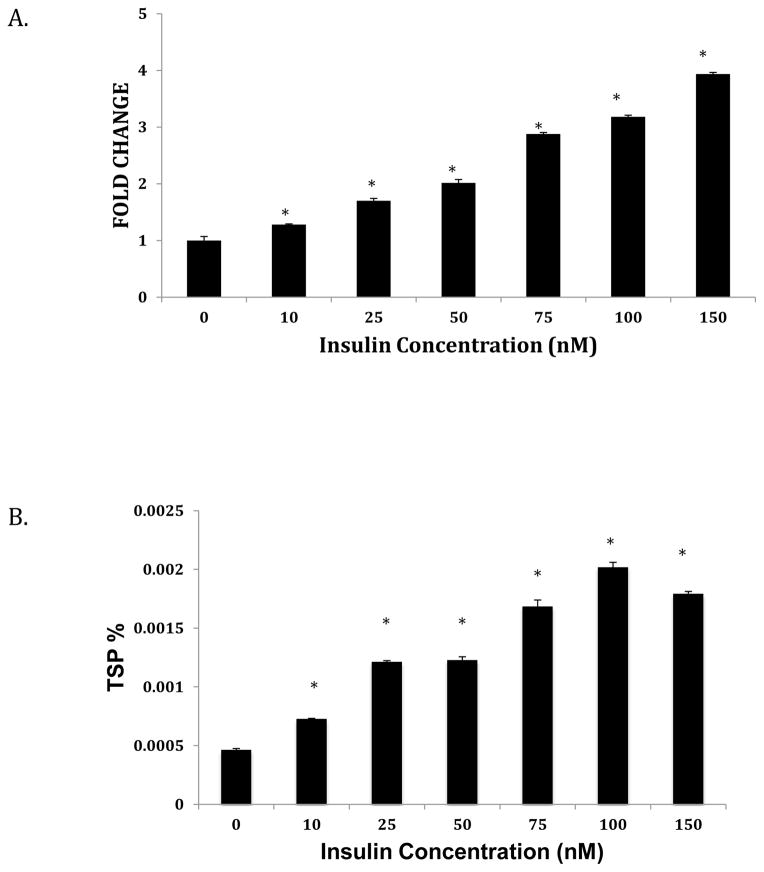
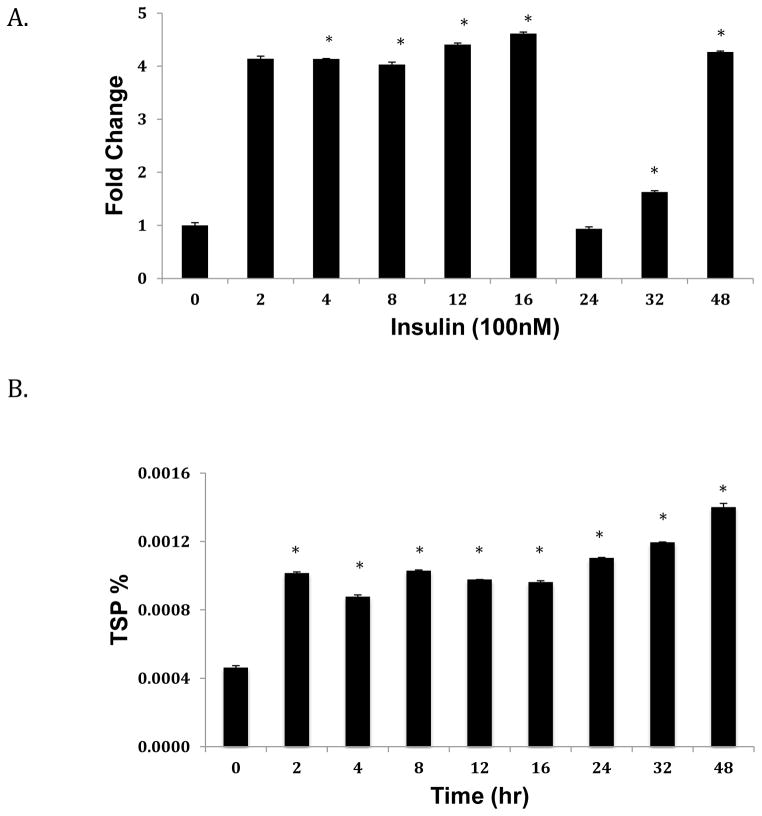
We next assessed the effect of increasing concentrations of insulin on ANGPTL8 transcript and protein expression. In response to insulin treatment, ANGPTL8 levels were upregulated compared to untreated controls (Fig. 3A and 3B). We also found that ANGPTL8 transcript levels significantly increased from 2–16 hours of treatment with 100 nM insulin, and then decreased sharply at 24 hours (Fig. 4A). In contrast, ANGPTL8 protein levels increased at 2 hours, and remained relatively stable for all time points (Fig. 4B). In response to treatment with 30 mM glucose and 100 nM insulin for 24 hours, ANGPTL8 transcript expression levels were intermediate between the two individual treatments (Fig. 5A). In contrast, levels of ANGPTL8 protein were reduced under combined insulin/glucose treatment relative to treatment with each component individually (Fig. 5B).
Figure 3. Effect of insulin concentration on ANGPTL8 transcript (A) and protein (B) expression.
Hep G2 Cells were grown to 80% confluence and starved overnight with serum-free media before treatment with varying concentrations of insulin over 12 hours. Total RNA and protein were extracted and expression levels assessed as described in the Methods section. mRNA levels were normalized against ACTB. Protein concentrations are shown as percentage of total soluble protein (TSP). Results represent averages from three independent experiments. Data are shown as means ± standard deviation; a two-tailed t-test was used to determine p-values; *p<0.05.
Figure 4. Effect of insulin over time on ANGPTL8 transcript (A) and protein (B) expression.
Hep G2 cells were grown to 80% confluence and starved overnight with serum-free media before treatment with 100 mM insulin over 48 hours. mRNA levels were normalized against ACTB. Protein concentrations are shown as percentage of total soluble protein (TSP). Results represent averages from three independent experiments. Data are shown as means ± standard deviation; the two-tailed t-test was used to determine p-values; *p<0.05.
Figure 5. Combined effect of insulin and glucose on ANGPTL8 transcript (A) and protein (B) expression.
Hep G2 cells were grown to 80% confluence and starved overnight with serum-free media before treatment with 30 mM glucose, 100 nM insulin, or both over 24 hrs. Total RNA and protein were extracted and expression levels assessed as described in the Methods section. mRNA levels were normalized against ACTB. Protein concentrations are shown as percentage of total soluble protein (TSP). Results represent averages from three independent experiments. Data are shown as means ± standard deviation; two-tailed t-test was used to determine p-values; *p<0.05.
Effect of rs2278426 on ANGPTL8:ANGPTL3 interaction
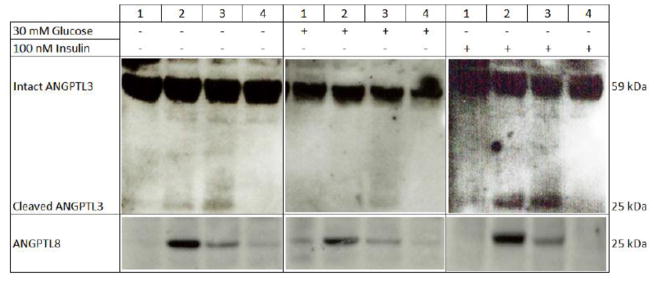
Recently, ANGPTL8 has emerged as a novel regulator of lipoprotein metabolism (33–37) and marker rs2278426, located within the coding sequence of ANGPTL8, has been associated with HDL-C and LDL-C levels in four independent populations (34, 47). ANGPTL8 activates ANGPTL3 by binding to its N-terminal domain and promoting cleavage of the full-length protein (34). To evaluate the potential functional significance of rs2278426, which causes an arginine to tryptophan substitution at position 59 (R59W), we investigated the allele-specific effects on ANGPTL3 cleavage by co-expressing ANGPTL3 with expression vectors containing either the wild type (C allele) or variant (T allele) of rs2278426 in HEK293 cells. As shown in Figure 6, we observed increased levels of the cleaved ANGPTL3 terminus in the presence of the ANGPTL8 variant allele compared to the wild type control. To determine whether levels of the ANGPTL3 cleaved product in the presence of ANGPTL8 were modulated by metabolic factors, we treated cells with 30 mM glucose or 100 nM insulin. In the presence of 30 mM glucose, levels of cleaved ANGPTL3 product were reduced relative to normal glucose conditions, but the extent of reduction did not vary by allele. Under conditions of insulin treatment, levels of the ANGPTL3 cleaved product were higher in the presence of the variant allele compared to the wild-type allele. Across each set of conditions, levels of the variant form of ANGPTL8 in cell lysates were lower compared to the wild-type protein.
Figure 6. Differential expression of ANGPTL3 and ANGPTL8 in conditioned media and cell lysates.
HEK293 cells were grown in 60 mm dishes to 90% confluence and then serum-starved for 24 hours prior to transfection with the appropriate vector for 6 hours with Lipofectamine 2000. Cells were incubated with either glucose (30 mM) or insulin (100 nM) for an additional 8 hours. Lane 1: ANGPTL3 only; Lane 2: ANGPTL3 plus the C allele (wild-type) of rs2278426; Lane 3: ANGPTL3 plus the T allele (variant) of rs2278426; and Lane 4: empty vector. The top panels for each set of conditions represent ANGPTL3 levels in conditioned media, while the bottom panels represent ANGPTL8 levels in cell lysates.
Discussion
Our findings demonstrate that marker rs2278426 contributes to variability in TC and HDL-C concentrations in Pima Indians and Mexican Americans. Our results provide evidence that the variant T allele, which encodes a tryptophan residue at position 59 of the ANGPTL8 protein, is associated with decreased levels of TC and HDL-C in both populations. We also show that the variant T allele corresponds with an increase in levels of the cleaved form of ANGPTL3, and that this interaction is modulated by insulin.
Marker rs2278426 has been previously associated with altered concentrations of blood lipids. In Hispanic and African American members of the Dallas Heart Study, the variant T allele was associated with lower plasma levels of LDL-C and HDL-C (34). Findings of association were replicated in African American participants of the Atherosclerosis Risk in Communities Study (ARIC) and the Dallas Biobank (34), and a recent GWAS for lipid traits in Mexicans also observed association between rs2278426 and HDL-C (47). In individuals of European ancestry, however, association findings reported to date have been less consistent. For example, in European American participants of ARIC, the variant allele was associated with significantly lower levels of HDL-C, but not LDL-C (34), while in an analysis using imputed genotypes from a genome-wide association study (48), the T allele was associated with levels of both HDL-C and LDL-C, although for the latter only at the nominal significance threshold (34). In the Dallas Heart Study, rs2278426 was not associated with either HDL-C or LDL-C in European American participants (34). The frequency of the variant T allele in Caucasians is approximately 5%, which is far less common than it is in the Mexican American (25%) and American Indian (50%) populations studied here. Significant differences in allele frequency may reflect historical differences in selective pressures among various ethnic groups, which could account for the discrepancies in findings of association among populations with European ancestry.
In addition to rs2278426, another ANGPTL8 variant, rs145464906, was recently identified in a study of blood lipid levels in ~14,000 and over 42,000 individuals of African and European ancestry, respectively; Caucasian, but not African American, carriers of the variant allele had higher levels of HDL-C and lower levels of TGs, and a trend toward lower LDL-C was observed, but at levels that did not reach statistical significance (49). The variant allele predicts a premature stop codon, and is relatively rare (MAF 0.01–0.1%). The correlation between rs145464906 and rs2278426 in these individuals was low and conditional analyses determined that the two variants are independent association signals (49). Based on whole genome sequencing conducted in 335 Pima Indians, the variant allele at rs145464906 was not observed (unpublished data).
ANGPTL8 belongs to a family of secreted proteins, including two closely related members, ANGPTL3 and ANGPTL4, located on chromosomes 1 and 19, respectively, which also play key roles in lipid trafficking and metabolism (50–57). Like ANGPTL8, variants in ANGPTL3 and ANGPTL4 have been associated with lipid traits in genome-wide association analyses (48), and low frequency, loss-of-function mutations in both genes are linked with changes in lipid profiles. For example, nonsense mutations in ANGPTL3 correspond with very low plasma levels of HDL-C, LDL-C, and TG in families with combined hypolipidemia (58), while the E40K variant in ANGPTL4 is associated with lower plasma TG and HDL-C concentrations (59, 60). Similarly, Romeo et al (61) found rare loss-of-function mutations in ANGPTL3, ANGPTL4, and ANGPTL5 in 1% of the Dallas Heart Study population and 4% of these individuals had fasting plasma TG levels in the lowest quartile. Together, these findings implicate a role for genetic variation in ANGPTL family members, including ANGPTL8, in the regulation of trafficking and metabolism of lipids.
In this study, we report evidence supporting a potential allele-specific effect of rs2278426 on ANGPTL3 cleavage. Several studies have shown that ANGPTL3 inhibits lipoprotein lipase (LPL) activity in vitro (53, 57, 61, 62), and ANGPTL3−/− mice have higher LPL activity (52, 57, 63). Additional studies demonstrated that ANGPTL3 also modulates the activity of endothelial lipase (56, 64). In ANGPTL3-deficient mice, plasma HDL-C levels were markedly decreased, and in humans, ANGPTL3 levels were significantly correlated with plasma HDL-C (56). Cleavage of ANGPTL3, which releases the N-terminal domain, is necessary for ANGPTL3-mediated lipase inhibition (65). Previous studies in cultured hepatocytes demonstrated that ANGPTL8 expression corresponded with increased levels of the ANGPTL3 N-terminal domain in the medium, suggesting that ANGPTL8 may activate ANGPTL3 (34). Mice overexpressing both ANGPTL3 and ANGPTL8 in the liver developed hypertriglyceridemia, although circulating levels of ANGPTL3 were reduced. In these animals, ANGPTL8 coimmunoprecipitated with the N-terminal domain of ANGPTL3, demonstrating an interaction between the two proteins in vivo.
In the results reported here, we found that the variant ANGPTL8 protein was associated with an increase in levels of cleaved ANGPTL3 protein, an interaction that was enhanced in the presence of insulin, but not high glucose. We do not currently know why ANGPTL8 levels are decreased with the variant allele of rs2278426, although we speculate that the protein may be consumed more quickly through increased cleavage of ANGPTL3. Despite the finding that ANGPTL8-deficient mice have lower plasma TG levels and higher post-heparin LPL-activity (66), we did not observe an association between rs2278426 and TG levels in Pima Indians or Mexican Americans. These results are in accord with other genetic studies in which no evidence for association between rs2278426 and TG levels was reported (34, 47, 48). Shimamura et al (56) reported that ANGPTL3-deficient mice showed low plasma HDL-C levels and that plasma ANGPTL3 levels in humans correlated with HDL-C levels, thus establishing a potential link between ANGPTL8 and HDL-C. It is possible that in humans, in contrast to mice (66), ANGPTL8 is linked with a reduced endothelial lipase function. Additional studies will be necessary to clarify this relationship.
Unlike ANGPTL3, ANGPTL8 is strongly regulated by food uptake (34–36, 66, 67). ANGPTL8 has also been identified as an insulin target gene in human and mice adipocytes (34, 35, 68). Our findings here now show that ANGPTL8 is also regulated by insulin in liver cells. Here, we found a significant increase of ANGPTL8 transcript levels over eight hours, which is in agreement with results obtained in 3T3-L1 cells (35). However, after eight hours, we observed stable transcript levels over 48 hours, in contrast to the linear increase reported in 3T3-L1 cells. We did not observe an upregulation of ANGPTL8 expression at lower levels of insulin treatment (i.e., 10–150 nM) as seen in 3T3-L1 cells (35), nor did we find an increase in ANGPTL8 protein in response to glucose treatment as also seen in 3T3-L1 cells (33), suggesting differential regulation of the gene by insulin in different cell types.
In addition to its role in lipoprotein metabolism, ANGPTL8 has been shown to promote pancreatic β-cell proliferation, expand β-cell mass, and improve glucose tolerance (38), suggesting that decreases in ANGPTL8 levels or function may worsen glucose tolerance. In genetic studies, however, the variant alleles at rs2278426 and rs146464906, both of which predict loss of function consequences, were not associated with either fasting plasma glucose levels (34, 49) or homeostatic model assessment insulin resistance (34) in any ethnic group. Similarly, betatrophin concentrations did not differ between healthy individuals and those with T2D and were not associated with variables of β-cell function and glucose homeostasis (69). However, betatrophin concentration was correlated significantly with levels of total cholesterol, LDL-C, and apolipoprotein B in both morbidly obese individuals and patients with T2D, but not in healthy, unaffected controls (69). Conversely, individuals with both insulin resistance and hypercholesterolemia had significantly higher betatrophin concentrations compared to those with normal cholesterol levels (69). These results suggest that ANGPTL8/betatrophin may preferentially impact lipid metabolism in metabolically compromised patients, including those with insulin resistance and T2D.
Despite the importance of these findings to an enhanced understanding of ANGPTL8, we acknowledge some limitations of this study. Although the association of betatrophin levels with the rs2278426 variant is of interest, measures of betatrophin are not currently available in the populations comprising this study. In addition, while our association results implicate variants in the genes encoding both DOCK6 and ANGPTL8, our findings only provide functional evidence supporting a role for ANGPTL8 in lipoprotein metabolism. We based our decision to focus on the rs2278426 locus on the combined findings of association in Pima Indians and members of the SAMAFS cohorts, prior evidence supporting a role for ANGPTL8 in lipid regulation, and the relative lack of evidence for DOCK6 in lipid-related biological pathways. However, we acknowledge that DOCK6 may contribute to variability in lipid traits and additional functional studies will be necessary to address this possibility.
Conclusions
In summary, we provide evidence confirming genetic association between rs2278426 and lipid traits in two ethnic minority populations; these findings may reflect historical differences in selective pressures among various ethnic groups and suggest that the variant allele may have a greater effect in populations other than those of European Caucasian ancestry. We demonstrate an allele-specific effect of rs2278426 on levels of the cleaved form of ANGPTL3, thus providing support for functional consequences of the rs2278426 variant. However, given that the T allele of rs2278426 is associated in a fashion that might be expected to produce opposite effects on cardiovascular risk (low TC, and low HDL), the therapeutic implications of the finding are unclear. These results add to the growing literature supporting ANGPTL8 as an important determinant of lipid metabolism.
Supplementary Material
To narrow the region of linkage, we selected a panel of 346 SNPs from the HapMap database, and genotyped them in the Pima Indian families who participated in the original genome scan. Markers common to all four populations available in the HapMap database (i.e., Caucasian, African, Chinese, and Japanese) were selected based upon physical position using a minor allele frequency ≥0.10 and r2=0.80. The solid line depicts the original region of linkage and the dotted line reflects the reduced linkage interval following additional SNP genotyping.
Supplemental Figure 2. Linkage disequilibrium plot of region surrounding rs2278426 in Pima Indians. 49 SNPs from a 200 kb region surrounding rs2278426 (denoted with *) are shown. The numbers are r2 values and shading represents the magnitude of r2.
Highlights.
Marker rs2278426 contributes to variability in total and high density lipoprotein (HDL) cholesterol in American Indians and Mexican Americans
The variant allele of rs2278426 corresponds with an increased in levels of cleaved ANGPTL3
The relationship between the variant rs2278426 allele and cleaved ANGPTL3 is modulated by insulin
Acknowledgments
This work was supported by the National Institutes of Health HL093042 (JKD) and the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (RLH, SK). The San Antonio Family Heart Study (SAFHS) and San Antonio Family Diabetes/Gallbladder Study (SAFDGS) were supported by NIH grants P01 HL045222, R01 DK047482, and R01 DK053889. We appreciate the technical assistance of Mark Zhang, Mahdieh Khosroheidari, and Matthew Taila at TGen. We thank Dr. William C. Knowler for his support and advice, the staff of the Phoenix Epidemiology and Clinical Research Branch for their significant contribution to this study, and all Pima study volunteers and participants of the SAFHS and SAFDGS for their cooperation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thom T, Haase N, Rosamond W, Howard VJ, Rumsfeld J, Manolio T, Zheng ZJ, Flegal K, O’Donnell C, Kittner S, Lloyd-Jones D, Goff DC, Jr, Hong Y, Adams R, Friday G, Furie K, Gorelick P, Kissela B, Marler J, Meigs J, Roger V, Sidney S, Sorlie P, Steinberger J, Wasserthiel-Smoller S, Wilson M, Wolf P. Heart disease and stroke statistics--2006 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2006;113:e85–151. doi: 10.1161/CIRCULATIONAHA.105.171600. [DOI] [PubMed] [Google Scholar]
- 2.Nilsson PM, Cederholm J, Gudbjornsdottir S, Eliasson B. Predictors of successful long-term blood pressure control in type 2 diabetic patients: data from the Swedish National Diabetes Register (NDR) J Hypertens. 2005;23:2305–2311. doi: 10.1097/01.hjh.0000188733.60345.78. [DOI] [PubMed] [Google Scholar]
- 3.Cooper R. The role of genetic and environmental factors in cardiovascular disease in African Americans. Am J Med Sci. 1999;317:208–213. doi: 10.1097/00000441-199903000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Potts JL, Thomas J. Traditional coronary risk factors in African Americans. Am J Med Sci. 1999;317:189–192. doi: 10.1097/00000441-199903000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Kannel WB, Castelli WP, Gordon T, McNamara PM. Serum cholesterol, lipoproteins, and the risk of coronary heart disease. The Framingham study. Ann Intern Med. 1971;74:1–12. doi: 10.7326/0003-4819-74-1-1. [DOI] [PubMed] [Google Scholar]
- 6.Bigazzi F, Pino BD, Forastiere F, Pistelli R, Rossi G, Simoni M, Baldacci S, Viegi G, Bionda A, Sampietro T. HDL and clinical and biochemical correlates in Italian non-smoker women. Clin Chem Lab Med. 2004;42:1408–1416. doi: 10.1515/CCLM.2004.262. [DOI] [PubMed] [Google Scholar]
- 7.Freiberg MS, Cabral HJ, Heeren TC, Vasan RS, Curtis Ellison R. Alcohol consumption and the prevalence of the Metabolic Syndrome in the US.: a cross-sectional analysis of data from the Third National Health and Nutrition Examination Survey. Diabetes Care. 2004;27:2954–2959. doi: 10.2337/diacare.27.12.2954. [DOI] [PubMed] [Google Scholar]
- 8.Lewis V, Hoeger K. Prevention of coronary heart disease: a nonhormonal approach. Semin Reprod Med. 2005;23:157–166. doi: 10.1055/s-2005-869483. [DOI] [PubMed] [Google Scholar]
- 9.Davignon J, Genest J., Jr Genetics of lipoprotein disorders. Endocrinol Metab Clin North Am. 1998;27:521–550. doi: 10.1016/s0889-8529(05)70024-4. [DOI] [PubMed] [Google Scholar]
- 10.Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, Loubser O, Ouelette BF, Fichter K, Ashbourne-Excoffon KJ, Sensen CW, Scherer S, Mott S, Denis M, Martindale D, Frohlich J, Morgan K, Koop B, Pimstone S, Kastelein JJ, Genest J, Jr, Hayden MR. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 11.Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, Drobnik W, Barlage S, Buchler C, Porsch-Ozcurumez M, Kaminski WE, Hahmann HW, Oette K, Rothe G, Aslanidis C, Lackner KJ, Schmitz G. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 12.Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette JC, Deleuze JF, Brewer HB, Duverger N, Denefle P, Assmann G. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 13.Bosse Y, Chagnon YC, Despres JP, Rice T, Rao DC, Bouchard C, Perusse L, Vohl MC. Compendium of genome-wide scans of lipid-related phenotypes: adding a new genome-wide search of apolipoprotein levels. J Lipid Res. 2004;45:2174–2184. doi: 10.1194/jlr.R400008-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Adeyemo AA, Johnson T, Acheampong J, Oli J, Okafor G, Amoah A, Owusu S, Agyenim-Boateng K, Eghan BA, Jr, Abbiyesuku F, Fasanmade O, Rufus T, Doumatey A, Chen G, Zhou J, Chen Y, Furbert-Harris P, Dunston G, Collins F, Rotimi C. A genome wide quantitative trait linkage analysis for serum lipids in type 2 diabetes in an African population. Atherosclerosis. 2005;181:389–397. doi: 10.1016/j.atherosclerosis.2004.12.049. [DOI] [PubMed] [Google Scholar]
- 15.Beekman M, Heijmans BT, Martin NG, Whitfield JB, Pedersen NL, DeFaire U, Snieder H, Lakenberg N, Suchiman HE, de Knijff P, Frants RR, van Ommen GJ, Kluft C, Vogler GP, Boomsma DI, Slagboom PE. Evidence for a QTL on chromosome 19 influencing LDL cholesterol levels in the general population. Eur J Hum Genet. 2003;11:845–850. doi: 10.1038/sj.ejhg.5201053. [DOI] [PubMed] [Google Scholar]
- 16.Bielinski SJ, Tang W, Pankow JS, Miller MB, Mosley TH, Boerwinkle E, Olshen RA, Curb JD, Jaquish CE, Rao DC, Weder A, Arnett DK. Genome-wide linkage scans for loci affecting total cholesterol, HDL-C, and triglycerides: the Family Blood Pressure Program. Hum Genet. 2006;120:371–380. doi: 10.1007/s00439-006-0223-0. [DOI] [PubMed] [Google Scholar]
- 17.Bosse Y, Chagnon YC, Despres JP, Rice T, Rao DC, Bouchard C, Perusse L, Vohl MC. Genome-wide linkage scan reveals multiple susceptibility loci influencing lipid and lipoprotein levels in the Quebec Family Study. J Lipid Res. 2004;45:419–426. doi: 10.1194/jlr.M300401-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Li S, Srinivasan SR, Boerwinkle E, Berenson GS. A genome scan for loci influencing levels and trends of lipoprotein lipid-related traits since childhood: The Bogalusa Heart Study. Atherosclerosis. 2006 doi: 10.1016/j.atherosclerosis.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Elbein SC, Hasstedt SJ. Quantitative trait linkage analysis of lipid-related traits in familial type 2 diabetes: evidence for linkage of triglyceride levels to chromosome 19q. Diabetes. 2002;51:528–535. doi: 10.2337/diabetes.51.2.528. [DOI] [PubMed] [Google Scholar]
- 20.Feitosa MF, Rice T, North KE, Kraja A, Rankinen T, Leon AS, Skinner JS, Blangero J, Bouchard C, Rao DC. Pleiotropic QTL on chromosome 19q13 for triglycerides and adiposity: the HERITAGE Family Study. Atherosclerosis. 2006;185:426–432. doi: 10.1016/j.atherosclerosis.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 21.Imperatore G, Knowler WC, Pettitt DJ, Kobes S, Fuller JH, Bennett PH, Hanson RL. A locus influencing total serum cholesterol on chromosome 19p: results from an autosomal genomic scan of serum lipid concentrations in Pima Indians. Arterioscler Thromb Vasc Biol. 2000;20:2651–2656. doi: 10.1161/01.atv.20.12.2651. [DOI] [PubMed] [Google Scholar]
- 22.Kullo IJ, Ding K, Boerwinkle E, Turner ST, de Andrade M. Quantitative trait loci influencing low density lipoprotein particle size in African Americans. J Lipid Res. 2006;47:1457–1462. doi: 10.1194/jlr.M600078-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Malhotra A, Wolford JK. Analysis of quantitative lipid traits in the genetics of NIDDM (GENNID) study. Diabetes. 2005;54:3007–3014. doi: 10.2337/diabetes.54.10.3007. [DOI] [PubMed] [Google Scholar]
- 24.Pollin TI, Hsueh WC, Steinle NI, Snitker S, Shuldiner AR, Mitchell BD. A genome-wide scan of serum lipid levels in the Old Order Amish. Atherosclerosis. 2004;173:89–96. doi: 10.1016/j.atherosclerosis.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Rainwater DL, Almasy L, Blangero J, Cole SA, VandeBerg JL, MacCluer JW, Hixson JE. A genome search identifies major quantitative trait loci on human chromosomes 3 and 4 that influence cholesterol concentrations in small LDL particles. Arterioscler Thromb Vasc Biol. 1999;19:777–783. doi: 10.1161/01.atv.19.3.777. [DOI] [PubMed] [Google Scholar]
- 26.Malhotra A, Elbein SC, Ng MC, Duggirala R, Arya R, Imperatore G, Adeyemo A, Pollin TI, Hsueh WC, Chan JC, Rotimi C, Hanson RL, Hasstedt SJ, Wolford JK. Meta-analysis of genome-wide linkage studies of quantitative lipid traits in families ascertained for type 2 diabetes. Diabetes. 2007;56:890–896. doi: 10.2337/db06-1057. [DOI] [PubMed] [Google Scholar]
- 27.Kathiresan S, Manning AK, Demissie S, D’Agostino RB, Surti A, Guiducci C, Gianniny L, Burtt NP, Melander O, Orho-Melander M, Arnett DK, Peloso GM, Ordovas JM, Cupples LA. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC medical genetics. 2007;8(Suppl 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O’Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, Song K, Yuan X, Johnson T, Ashford S, Inouye M, Luben R, Sims M, Hadley D, McArdle W, Barter P, Kesaniemi YA, Mahley RW, McPherson R, Grundy SM, Bingham SA, Khaw KT, Loos RJ, Waeber G, Barroso I, Strachan DP, Deloukas P, Vollenweider P, Wareham NJ, Mooser V. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marcano AC, Hajat C, Burton P, Deloukas P, Brown M, Connell JM, Dominiczak A, Lathrop GM, Webster J, Farrall M, Spector T, Samani NJ, Caulfield MJ, Munroe PB. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu Z, Yao F, Abou-Samra AB, Zhang R. Lipasin, thermoregulated in brown fat, is a novel but atypical member of the angiopoietin-like protein family. Biochem Biophys Res Commun. 2013;430:1126–1131. doi: 10.1016/j.bbrc.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 34.Quagliarini F, Wang Y, Kozlitina J, Grishin NV, Hyde R, Boerwinkle E, Valenzuela DM, Murphy AJ, Cohen JC, Hobbs HH. Atypical angiopoietin-like protein that regulates ANGPTL3. Proc Natl Acad Sci U S A. 2012;109:19751–19756. doi: 10.1073/pnas.1217552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren G, Kim JY, Smas CM. Identification of RIFL, a novel adipocyte-enriched insulin target gene with a role in lipid metabolism. American journal of physiology. 2012;303:E334–351. doi: 10.1152/ajpendo.00084.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang R. Lipasin, a novel nutritionally-regulated liver-enriched factor that regulates serum triglyceride levels. Biochem Biophys Res Commun. 2012;424:786–792. doi: 10.1016/j.bbrc.2012.07.038. [DOI] [PubMed] [Google Scholar]
- 37.Zhang R, Abou-Samra AB. Emerging roles of Lipasin as a critical lipid regulator. Biochem Biophys Res Commun. 2013;432:401–405. doi: 10.1016/j.bbrc.2013.01.129. [DOI] [PubMed] [Google Scholar]
- 38.Yi P, Park JS, Melton DA. Betatrophin: a hormone that controls pancreatic beta cell proliferation. Cell. 2013;153:747–758. doi: 10.1016/j.cell.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Knowler WC, Bennett PH, Hamman RF, Miller M. Diabetes incidence and prevalence in Pima Indians: a 19-fold greater incidence than in Rochester, Minnesota. Am J Epidemiol. 1978;108:497–505. doi: 10.1093/oxfordjournals.aje.a112648. [DOI] [PubMed] [Google Scholar]
- 40.Allain CC, Poon LS, Chan CS, Richmond W, Fu PC. Enzymatic determination of total serum cholesterol. Clin Chem. 1974;20:470–475. [PubMed] [Google Scholar]
- 41.MacCluer JW, Stern MP, Almasy L, Atwood LA, Blangero J, Comuzzie AG, Dyke B, Haffner SM, Henkel RD, Hixson JE, Kammerer CM, Mahaney MC, Mitchell BD, Rainwater DL, Samollow PB, Sharp RM, VandeBerg JL, Williams JT. Genetics of atherosclerosis risk factors in Mexican Americans. Nutrition reviews. 1999;57:S59–65. doi: 10.1111/j.1753-4887.1999.tb01790.x. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, VandeBerg JL, Stern MP, MacCluer JW. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- 43.Hunt KJ, Lehman DM, Arya R, Fowler S, Leach RJ, Goring HH, Almasy L, Blangero J, Dyer TD, Duggirala R, Stern MP. Genome-wide linkage analyses of type 2 diabetes in Mexican Americans: the San Antonio Family Diabetes/Gallbladder Study. Diabetes. 2005;54:2655–2662. doi: 10.2337/diabetes.54.9.2655. [DOI] [PubMed] [Google Scholar]
- 44.Blackburn A, Goring HH, Dean A, Carless MA, Dyer T, Kumar S, Fowler S, Curran JE, Almasy L, Mahaney M, Comuzzie A, Duggirala R, Blangero J, Lehman DM. Utilizing extended pedigree information for discovery and confirmation of copy number variable regions among Mexican Americans. Eur J Hum Genet. 2013;21:404–409. doi: 10.1038/ejhg.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanson RL, Muller YL, Kobes S, Guo T, Bian L, Ossowski V, Wiedrich K, Sutherland J, Wiedrich C, Mahkee D, Huang K, Abdussamad M, Traurig M, Weil EJ, Nelson RG, Bennett PH, Knowler WC, Bogardus C, Baier LJ. A genome-wide association study in American Indians implicates DNER as a susceptibility locus for type 2 diabetes. Diabetes. 2014;63:369–376. doi: 10.2337/db13-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 47.Weissglas-Volkov D, Aguilar-Salinas CA, Nikkola E, Deere KA, Cruz-Bautista I, Arellano-Campos O, Munoz-Hernandez LL, Gomez-Munguia L, Ordonez-Sanchez ML, Reddy PM, Lusis AJ, Matikainen N, Taskinen MR, Riba L, Cantor RM, Sinsheimer JS, Tusie-Luna T, Pajukanta P. Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J Med Genet. 2013;50:298–308. doi: 10.1136/jmedgenet-2012-101461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, Stitziel NO, Brody JA, Khetarpal SA, Crosby JR, Fornage M, Isaacs A, Jakobsdottir J, Feitosa MF, Davies G, Huffman JE, Manichaikul A, Davis B, Lohman K, Joon AY, Smith AV, Grove ML, Zanoni P, Redon V, Demissie S, Lawson K, Peters U, Carlson C, Jackson RD, Ryckman KK, Mackey RH, Robinson JG, Siscovick DS, Schreiner PJ, Mychaleckyj JC, Pankow JS, Hofman A, Uitterlinden AG, Harris TB, Taylor KD, Stafford JM, Reynolds LM, Marioni RE, Dehghan A, Franco OH, Patel AP, Lu Y, Hindy G, Gottesman O, Bottinger EP, Melander O, Orho-Melander M, Loos RJ, Duga S, Merlini PA, Farrall M, Goel A, Asselta R, Girelli D, Martinelli N, Shah SH, Kraus WE, Li M, Rader DJ, Reilly MP, McPherson R, Watkins H, Ardissino D, Zhang Q, Wang J, Tsai MY, Taylor HA, Correa A, Griswold ME, Lange LA, Starr JM, Rudan I, Eiriksdottir G, Launer LJ, Ordovas JM, Levy D, Chen YD, Reiner AP, Hayward C, Polasek O, Deary IJ, Borecki IB, Liu Y, Gudnason V, Wilson JG, van Duijn CM, Kooperberg C, Rich SS, Psaty BM, Rotter JI, O’Donnell CJ, Rice K, Boerwinkle E, Kathiresan S, Cupples LA. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94:223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desai U, Lee EC, Chung K, Gao C, Gay J, Key B, Hansen G, Machajewski D, Platt KA, Sands AT, Schneider M, Van Sligtenhorst I, Suwanichkul A, Vogel P, Wilganowski N, Wingert J, Zambrowicz BP, Landes G, Powell DR. Lipid-lowering effects of anti-angiopoietin-like 4 antibody recapitulate the lipid phenotype found in angiopoietin-like 4 knockout mice. Proc Natl Acad Sci U S A. 2007;104:11766–11771. doi: 10.1073/pnas.0705041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koishi R, Ando Y, Ono M, Shimamura M, Yasumo H, Fujiwara T, Horikoshi H, Furukawa H. Angptl3 regulates lipid metabolism in mice. Nat Genet. 2002;30:151–157. doi: 10.1038/ng814. [DOI] [PubMed] [Google Scholar]
- 52.Koster A, Chao YB, Mosior M, Ford A, Gonzalez-DeWhitt PA, Hale JE, Li D, Qiu Y, Fraser CC, Yang DD, Heuer JG, Jaskunas SR, Eacho P. Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: regulation of triglyceride metabolism. Endocrinology. 2005;146:4943–4950. doi: 10.1210/en.2005-0476. [DOI] [PubMed] [Google Scholar]
- 53.Lee EC, Desai U, Gololobov G, Hong S, Feng X, Yu XC, Gay J, Wilganowski N, Gao C, Du LL, Chen J, Hu Y, Zhao S, Kirkpatrick L, Schneider M, Zambrowicz BP, Landes G, Powell DR, Sonnenburg WK. Identification of a new functional domain in angiopoietin-like 3 (ANGPTL3) and angiopoietin-like 4 (ANGPTL4) involved in binding and inhibition of lipoprotein lipase (LPL) J Biol Chem. 2009;284:13735–13745. doi: 10.1074/jbc.M807899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lichtenstein L, Mattijssen F, de Wit NJ, Georgiadi A, Hooiveld GJ, van der Meer R, He Y, Qi L, Koster A, Tamsma JT, Tan NS, Muller M, Kersten S. Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell metabolism. 2010;12:580–592. doi: 10.1016/j.cmet.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimamura M, Matsuda M, Kobayashi S, Ando Y, Ono M, Koishi R, Furukawa H, Makishima M, Shimomura I. Angiopoietin-like protein 3, a hepatic secretory factor, activates lipolysis in adipocytes. Biochem Biophys Res Commun. 2003;301:604–609. doi: 10.1016/s0006-291x(02)03058-9. [DOI] [PubMed] [Google Scholar]
- 56.Shimamura M, Matsuda M, Yasumo H, Okazaki M, Fujimoto K, Kono K, Shimizugawa T, Ando Y, Koishi R, Kohama T, Sakai N, Kotani K, Komuro R, Ishida T, Hirata K, Yamashita S, Furukawa H, Shimomura I. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol. 2007;27:366–372. doi: 10.1161/01.ATV.0000252827.51626.89. [DOI] [PubMed] [Google Scholar]
- 57.Shimizugawa T, Ono M, Shimamura M, Yoshida K, Ando Y, Koishi R, Ueda K, Inaba T, Minekura H, Kohama T, Furukawa H. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. J Biol Chem. 2002;277:33742–33748. doi: 10.1074/jbc.M203215200. [DOI] [PubMed] [Google Scholar]
- 58.Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, Fennell T, Banks E, Ambrogio L, Cibulskis K, Kernytsky A, Gonzalez E, Rudzicz N, Engert JC, DePristo MA, Daly MJ, Cohen JC, Hobbs HH, Altshuler D, Schonfeld G, Gabriel SB, Yue P, Kathiresan S. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Talmud PJ, Smart M, Presswood E, Cooper JA, Nicaud V, Drenos F, Palmen J, Marmot MG, Boekholdt SM, Wareham NJ, Khaw KT, Kumari M, Humphries SE. ANGPTL4 E40K and T266M: effects on plasma triglyceride and HDL levels, postprandial responses, and CHD risk. Arterioscler Thromb Vasc Biol. 2008;28:2319–2325. doi: 10.1161/ATVBAHA.108.176917. [DOI] [PubMed] [Google Scholar]
- 61.Romeo S, Yin W, Kozlitina J, Pennacchio LA, Boerwinkle E, Hobbs HH, Cohen JC. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shan L, Yu XC, Liu Z, Hu Y, Sturgis LT, Miranda ML, Liu Q. The angiopoietin-like proteins ANGPTL3 and ANGPTL4 inhibit lipoprotein lipase activity through distinct mechanisms. The Journal of biological chemistry. 2009;284:1419–1424. doi: 10.1074/jbc.M808477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujimoto K, Koishi R, Shimizugawa T, Ando Y. Angptl3-null mice show low plasma lipid concentrations by enhanced lipoprotein lipase activity. Experimental animals/Japanese Association for Laboratory Animal Science. 2006;55:27–34. doi: 10.1538/expanim.55.27. [DOI] [PubMed] [Google Scholar]
- 64.Jin W, Wang X, Millar JS, Quertermous T, Rothblat GH, Glick JM, Rader DJ. Hepatic proprotein convertases modulate HDL metabolism. Cell metabolism. 2007;6:129–136. doi: 10.1016/j.cmet.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ono M, Shimizugawa T, Shimamura M, Yoshida K, Noji-Sakikawa C, Ando Y, Koishi R, Furukawa H. Protein region important for regulation of lipid metabolism in angiopoietin-like 3 (ANGPTL3): ANGPTL3 is cleaved and activated in vivo. The Journal of biological chemistry. 2003;278:41804–41809. doi: 10.1074/jbc.M302861200. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Quagliarini F, Gusarova V, Gromada J, Valenzuela DM, Cohen JC, Hobbs HH. Mice lacking ANGPTL8 (Betatrophin) manifest disrupted triglyceride metabolism without impaired glucose homeostasis. Proc Natl Acad Sci U S A. 2013;110:16109–16114. doi: 10.1073/pnas.1315292110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ge H, Cha JY, Gopal H, Harp C, Yu X, Repa JJ, Li C. Differential regulation and properties of angiopoietin-like proteins 3 and 4. Journal of lipid research. 2005;46:1484–1490. doi: 10.1194/jlr.M500005-JLR200. [DOI] [PubMed] [Google Scholar]
- 68.Ebert T, Kralisch S, Hoffmann A, Bachmann A, Lossner U, Kratzsch J, Bluher M, Stumvoll M, Tonjes A, Fasshauer M. Circulating angiopoietin-like protein 8 is independently associated with fasting plasma glucose and type 2 diabetes mellitus. The Journal of clinical endocrinology and metabolism. 2014;99:E2510–2517. doi: 10.1210/jc.2013-4349. [DOI] [PubMed] [Google Scholar]
- 69.Fenzl A, Itariu BK, Kosi L, Fritzer-Szekeres M, Kautzky-Willer A, Stulnig TM, Kiefer FW. Circulating betatrophin correlates with atherogenic lipid profiles but not with glucose and insulin levels in insulin-resistant individuals. Diabetologia. 2014 doi: 10.1007/s00125-014-3208-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
To narrow the region of linkage, we selected a panel of 346 SNPs from the HapMap database, and genotyped them in the Pima Indian families who participated in the original genome scan. Markers common to all four populations available in the HapMap database (i.e., Caucasian, African, Chinese, and Japanese) were selected based upon physical position using a minor allele frequency ≥0.10 and r2=0.80. The solid line depicts the original region of linkage and the dotted line reflects the reduced linkage interval following additional SNP genotyping.
Supplemental Figure 2. Linkage disequilibrium plot of region surrounding rs2278426 in Pima Indians. 49 SNPs from a 200 kb region surrounding rs2278426 (denoted with *) are shown. The numbers are r2 values and shading represents the magnitude of r2.