Abstract
Topologically Associating Domains (TADs) are conserved during evolution and play roles in guiding and constraining long-range regulation of gene expression. Disruption of TAD boundaries results in aberrant gene expression by exposing genes to inappropriate regulatory elements. Recent studies have shown that TAD disruption is often found in cancer cells and contributes to oncogenesis through two mechanisms. One mechanism locally disrupts domains by deleting or mutating a TAD boundary leading to fusion of the two adjacent TADs. The other mechanism involves genomic rearrangements that break up TADs and creates new ones without directly affecting TAD boundaries. Understanding the mechanisms by which TADs form and control long-range chromatin interactions will therefore not only provide insights into the mechanism of gene regulation in general, but will also reveal how genomic rearrangements and mutations in cancer genomes can lead to misregulation of oncogenes and tumor suppressors.
Long-range gene regulation occurs within Topologically Associating Domains (TADs)
Each cell expresses a specific subset of all genes. Accurate regulation of gene expression is crucial for cell function and identity and is in large part determined by cell type specific gene regulatory elements such as enhancers and insulators. Dysregulation, e.g. as the result of inappropriate promoter-enhancer communication and genomic alterations, leads to altered gene expression patterns. Indeed, overexpression of oncogenes and downregulation of tumor suppressor genes can lead to cancer.
Promoters are regulated by enhancers that can sometimes be located hundreds of Kb from the start site of the gene. Enhancers act by first associating with a set of specific transcription factors that then engage through long-range interactions with target promoters. Why a given enhancer regulates one distal gene but not another has been puzzling for many years. Recent insights into the 3D folding of chromosomes are now providing new insights into the mechanisms that determine promoter-enhancer communication and how alterations in chromosome structure, as recently detected in cancer cells, can lead to inappropriate gene expression including aberrant activation of oncogenes.
Early studies using 3C, 4C, 5C and ChIA-PET showed that enhancers can physically associate with their target genes, through a process that either involves 3D looping, or some sort of tracking to link the enhancer to the promoter [1–4]. Although these studies showed that physical contacts between regulatory elements that can be located hundreds of Kb apart play key roles in gene regulation, they did not reveal what guides or determines these associations, and whether there is any specificity to these looping contacts. Subsequent more comprehensive 5C and genome-wide Hi-C studies showed that mammalian chromosomes are composed of a series of self-associating domains, often referred to as Topologically Associating Domains (TADs) [5,6]. Recently very high resolution Hi-C maps have shown that TADs are, at least in part, looped domains with associations between pairs of convergently positioned CTCF sites located at their boundaries [7–12]. Further, TADs can be composites of multiple nested CTCF-bounded self-interacting domains. Finally, although on average the CTCF sites flanking such domains interact relatively frequently with each other, it has been proposed this is a side effect of dynamic and randomly positioned loop extrusions that occur within TADs, perhaps by cohesin complexes, and that are blocked in a directional manner by CTCF-bound sites [13–15].
Although the concept of self-interacting domains is now well established [16], and in some cases a process of loop extrusion that drives their formation is proposed, it is not clear yet whether all TADs are equal and thus it remains speculative to molecularly define a TAD. In this review, we will call a TAD every domain that has preferential self-interactions, e.g. as determined by Hi-C, and that is clearly insulated from adjacent domains by boundaries. We will call a TAD boundary the genomic locus that is in between two adjacent TADs. This boundary could be formed by two mechanisms. First, a boundary could be formed by the binding of specific factors, e.g. CTCF. This appears to be the case for many boundaries. Second, we anticipate that some boundaries may also be created indirectly, e.g. by the aggregation of loci located within a TAD through other mechanisms, without explicit imposition of a boundary by localized binding a protein complex.
Interestingly, there are now multiple lines of evidence that indicate that TADs represent functional domains. Genes located in the same TAD display correlated expression across differentiation [6]. In a completely independent approach, Symmons et al. used a regulatory enhancer sensor composed of a weak promoter and a reporter gene [17]. This regulatory sensor was randomly inserted along mouse chromosomes. Regulatory sensors inserted within the same TAD displayed the same pattern of gene expression in mouse embryos. However, when regulatory sensors were inserted in different TADs or even in two neighboring TADs the pattern of gene expression was very different [17]. This striking result shows that enhancer capacities are insulated by TAD boundaries. Further evidence for this proposal comes from detailed studies of a set of TADs around the CFTR locus which showed that the CFTR gene touches different sets of enhancers in different cell types, but all these enhancers are located inside the same tissue invariant TAD [18,19]. Possibly loop extrusions occurring within TADs transiently juxtapose each promoter to each enhancer, and because CTCF sites at TAD boundaries block loop extrusion, promoters cannot become similarly juxtaposed to regulatory elements located outside the TAD [15]. Finally, the fact that TADs tend to be conserved between different species and throughout differentiation suggests a role for TADs in gene expression and not a role for gene expression in TAD formation [5]. Combined together these studies strongly suggest that TADs are chromosomal structural units that constrain, guide and facilitate enhancer-promoter interactions.
TAD disruption and altered gene expression
Given the important role of TADs in determining gene regulatory interactions, studying how TADs are formed and how their boundaries constrain looping interactions will give important insights into how genes are regulated in general, and how genes may become misregulated in disease. Several laboratories have deleted or relocated TAD boundaries and elements at or near TAD boundaries to assess the effect of these genetic manipulations on chromosome folding and gene expression. A first study in mouse embryos, has shown that deletion of a 35 Kb region in the hoxD locus increases chromatin interactions within a ‘compartment’ and increases the transcription of the HoxD11 gene whereas a smaller deletion (25 Kb) in the same locus does not change the interaction profile or activate gene expression [20]. This study highlights the fact that specific genetic determinants can create structural domains that might be important for gene expression. In a subsequent study from the same laboratory, these authors showed that an inversion of a 2 Mb region in the HoxD locus in mouse embryos changes the pattern of interactions within the locus and that precisely positioned insulators within the inverted 2 Mb DNA region determine the position of chromatin domains along the chromosome [21]. In another study from the Spitz laboratory, it was shown that the chromosomal locus containing the Tfap2c and Bmp7 genes is split in two structural and functional domains, with each gene located inside a separate TAD containing its dedicated set of enhancers. Inversions around the TAD boundary that relocate a heart enhancer for the Bmp7 gene into the TAD containing Tfap2c activated the latter in heart while Bmp7 was no longer expressed [22]. Similarly, it was shown that deletion of a TAD boundary can lead to overexpression of Lamin B1, probably by enhancers normally located outside the Lamin B1 containing TAD, which causes a rare neurological disorder with progressive central nervous system demyelination [23]. Finally, in an elegant series of experiments, it was shown that inversion, duplication or deletion of TAD boundaries in between the EPHA4 and PAX3 genes and near the WNT6 and IHH genes leads to inappropriate enhancer – promoter interactions that are linked to human limb malformation [24,25]. Together, these important studies show that TAD boundaries can restrict and guide enhancer activity towards genes located within the same TAD (Table 1).
Table 1.
TAD boundary disruptions and their effects.
| Study | Size of the deletion/inversion | Mentioned CTCF sites? | Effect | Reference |
|---|---|---|---|---|
| Mouse embryos, Hox locus 4C |
25 Kb and 35 Kb deletions | no | Increase interactions within a TAD Increase transcription of HoxD |
[20] |
| Mouse embryos Hox locus 4C |
2 Mb inversion | no | TAD boundary is moved. | [21] |
| Mouse embryos Tfap2c and Bmp7 locus 4C |
0.3 Mb inversions | no | TAD boundary is moved Tfap2c is activated. Bmp7 is no longer expressed. |
[22] |
| Mouse ESC 5C |
58 Kb deletion | no | Initial boundary is disrupted New boundary formed |
[6] |
| Human cells, Lamin B1 FISH, 4C |
660 Kb deletion | no | Neurological disorder | [23] |
| Human and mouse cells Wnt6, Ihh, Epha4, Pax3 4C |
Several deletion and inversions | no | Limb development malformation | [24] |
| Mouse β-globin locus 3C |
Mutation in the CTCF binding site | yes | Disruption of the chromatin loop of the β-globin gene | [26] |
| Human ESC ChIA-PET |
Deletion of CTCF site | yes | Dysregulation of genes | [32] |
| Human ESC and motor neurons 4C |
Deletion of CTCF site | yes | TAD boundary is moved | [33] |
| Human cells 4C |
Inversion of CTCF site | yes | Interactions are changed | [10] |
| Human cells 4C |
Deletion of CTCF site | yes | Interactions are changed | [8] |
| Human cells Capture-C |
Deletion of CTCF sites | yes | Chromatin loops are disrupted | [14] |
| Human cells PDGFRA 3C |
Deletion of CTCF site | yes | Oncogene expression, disruption of a loop | [36] |
| Human cells Tal1 and Lmo2 5C |
Tal1 locus 400 bp Lmo2 locus 27 Kb |
yes | Oncogene expression New enhancer-promoter interactions |
[39] |
The role of CTCF
Detailed Hi-C analyses and genetic perturbation studies have revealed that CTCF plays a critical role at TAD boundaries. First, a high resolution Hi-C study showed that TAD boundaries contain CTCF binding sites in convergent orientation and that these sites frequently interact to form self-interacting domains [7]. Earlier locus-specific 3C, 4C and 5C studies, e.g. at the beta-globin locus had also shown that CTCF sites often interact with each other [1,26,27]. The fact that CTCF interactions are mostly limited between pairs that are in a convergent orientation [7–12] has been one of the observations that led to the dynamic loop extrusion model for TADs [13–15].
CTCF is essential during development and it had long been suspected to play an important role in genome structure. Moreover, CTCF knock-down experiments showed that CTCF plays a role in genome organization [28–31]. A first pioneer study showed that the disruption of a CTCF binding site by changing four conserved nucleotides in the core CTCF binding site of the mouse β-globin locus prevents the binding of CTCF in vivo and destabilizes the large chromatin loop containing the globin genes [26]. Following this study, many other laboratories have disrupted CTCF binding sites [8,10,32,33]. Deletions of CTCF sites at TAD boundaries in embryonic stem cells affected the expression of neighboring genes and changed the interaction frequency between loci located in adjacent TADs [32]. Several other studies have shown that deletion or changing the orientation of CTCF binding sites at boundaries affect the frequency and direction of interaction with distal sites [8,10]. Together these studies show that CTCF binding, and site orientation plays a critical role in TAD boundary activity.
TAD disruption and cancer
Inappropriate activation of oncogenes, or repression of tumor suppressors can lead to cell proliferation, cellular transformation and cancer. Although oncogene activation can be caused by mutation or epigenetic modification that affect the gene itself or its regulatory elements [34,35], it is also possible that epigenetic or genetic alterations lead to inappropriate communication between otherwise unaltered genes and distal regulatory elements. For instance, as outlined in the examples above, deletion or inactivation of a TAD boundary can lead to inappropriate oncogene expression.
There are now several important studies that show that oncogene activation can be the result of TAD disruption. There are at least two mechanisms of TAD disruption. First, individual TAD boundaries could be mutated or epigenetically inactivated, affecting gene regulation in the two flanking TADs. Second, genomic rearrangements, with breakpoints within TADs, can lead to breakage and fusion of TADs without affecting the boundaries themselves, creating new regulatory domains where oncogenes can be activated by new sets of regulatory elements.
Local TAD boundary disruption
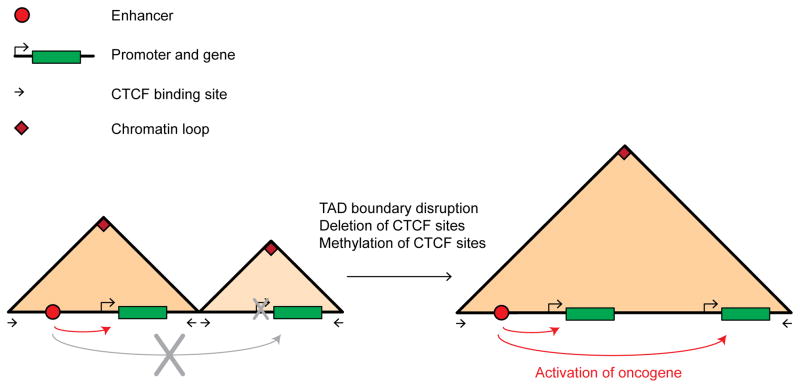
Gliomas can be caused by gain of function mutations in the IDH gene. Mutant IDH cells accumulate 2-hydroxyglutarate, which in turn represses TET proteins. As a result, these cells display elevated CpG methylation. How this causes cancer has remained unclear. A recent study found that the increased methylation of CTCF sites in mutant IDH cells leads to partial inactivation of TAD boundaries, and the concomitant activation of key cancer drivers such as PDGFRA by enhancers located outside the normal PDGFRA TAD. This study also showed that the inactivation of the TAD boundary is indeed DNA methylation dependent and leads to long-range interactions between the PDGFRA gene and a constitutive enhancer outside the TAD [36] (Figure 1).
Figure 1. Oncogene activation through local TAD boundary disruption.
In 5C/Hi-C interaction matrices TADs stand out as triangles along the diagonal of the interaction map. Here we show schematic TADs as triangles along the horizontal axis that represents the genome. Left: Before TAD boundary disruption, one TAD expresses a gene and the other TAD does not express the oncogene. Right: After TAD boundary disruption, a new fused TAD is formed that allows the activation of the oncogene by the enhancer that is now located in the same functional TAD. Enhancer is represented by a red dot. Genes are represented by green rectangles. Boundaries with CTCF sites in opposite directions are represented by black arrows and the chromatin loop formed by the CTCF sites is represented by a red rectangle at the corner of the TAD.
In a separate study Ji et al. found that many diseases the associated single nucleotide polymorphisms are often at or near gene regulatory elements, but rarely at TAD boundaries. In contrast, in cancer cells, TAD boundaries and especially CTCF DNA binding motifs at these boundaries are among the most altered factor binding sequences [37]. Further, they found that CTCF sites mutated in cancers are often adjacent to oncogenes and other genes known to be altered in expression in cancer. For example, in ovarian cancer, mutations in the CTCF motif at the boundary of the TAD that contains the NOTCH1 gene, lead to NOTCH1 misregulation, most likely through inappropriate enhancer action that is caused by TAD disruption [37].
A third example is provided by a very comprehensive mutation analysis in colorectal cancers [38]. These authors found that CTCF binding sites, especially those that also bind cohesin, are the most frequently mutated factor binding sites in these tumors. Although not shown directly in this study, it seems likely that many of these mutations lead to TAD disruption and associated miscommunication between genes and distal regulatory elements.
Finally, deletions found near the TAL1 and LMO2 oncogenes in T-cell acute lymphoblastic leukemia (T-ALL) eliminate TAD boundaries. The boundary deletions result in the activation of the oncogenes by creating new interactions between distal enhancers and oncogenes [39].
Global rearrangements of TADs
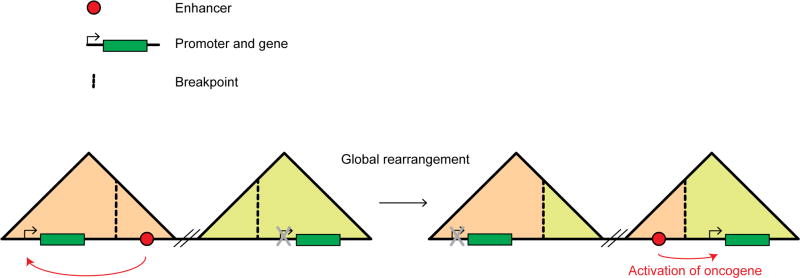
TAD organization is remarkably conserved between species [5,11]. This is probably at least in part related to the observation that CTCF binding sites, and their orientation, are highly conserved across syntenic regions. Interesting, when blocks do change genomic position across species, TADs are often maintained, indicating that large-scale domains are kept intact during genome evolution. This indicates that there is evolutionary pressure to conserve TAD related domains, as one would expect when these domains represent domains of gene regulation. Importantly, there is increasing evidence that in cancer genomes, that often are characterized by major intra- and inter-chromosomal rearrangements, TAD organization is often disrupted. In leukemia, genomic breakpoints tend to be in introns and in genomic regions containing open chromatin [40]. Another study showed that open chromatin associated histone marks can facilitate DNA breakage and thus translocations [41]. These studies highlight the fact that rearrangements happen at nonrandom positions and that TAD rearrangements in cancer genome might occur at these specific locations. In one study, Groschel and co-workers found that an inversion on chromosome 3 that is a cause of AML disrupts two TADs at the two inversion breakpoints [42]. One of these TADs contains the GATA2 gene, and the other contains the EVI oncogene. As a result of the inversion, an enhancer that normally is located in the GATA2 TAD and activates that gene, is now located in the EVI TAD and inappropriately activates this oncogene (Figure 2).
Figure 2. Oncogene activation through global rearrangements of TADs.
Two TADs that are located several megabases away are represented as triangles (as in Figure 1). The first TAD possesses an enhancer and an expressed gene. The second TAD does not express the proto-oncogene. Global rearrangements that occur at breakpoints (dashed black line) fuse the two far away TADs by inverting the sequence in between the two TADs and create two chimeric TADs. The gene in the first TAD is no longer expressed and the oncogene in the second TAD is now expressed by activation of the relocated enhancer. Enhancer is represented by a red dot and genes by green rectangles.
Similar TAD rearrangements may explain how genomic rearrangements that involve the MYC gene and the IGH locus can bring together this oncogene with strong immunoglobulin enhancers within the same TAD [43].
GFI1B has been identified as a medulloblastoma oncogene. Genomic rearrangements that juxtapose DNA elements that are normally located at 400 Kb from the GFI1B gene near its location might be responsible for its activation. At the rearrangement breakpoint near the GFI1B oncogene, increased enhancer signal has been detected and these elements show enhancer activity in luciferase reporter assays. These experiments suggest that through rearrangements, putative enhancers are placed near GFI1B and these can be responsible for its activation [44]. Another study in Chronic Lymphocytic Leukaemia (CLL) showed that an increased interaction frequency between the PAX locus and a potential enhancer located in the telomere region 330 Kb away is responsible for the overexpression of PAX5. Mutations in the enhancer region decrease the PAX5 expression restoring the wild-type phenotype [45]. However, these two last studies do not rule out that the identified enhancers and the activated oncogenes are already in the same TAD in normal cells.
Finally, another technique that simultaneously maps enhancer activity and proximal rearrangements (PEAR-ChIP) showed that in lymphoma cell lines, rearrangements drive activation of oncogenes by bringing together far away enhancers and oncogenes. The potential interactions were confirmed with 3C [46], suggesting that rearrangements have created new TADs where these enhancers activate the oncogenes.
Future perspective
TADs are emerging as a major feature of chromosome organization and gene regulation. Now that we are starting to understand how these domains are formed and how they regulate enhancer-promoter communication, we are starting to appreciate how 3D folding defects can lead to altered gene expression in disease. Given the increased genome instability of cancer genomes, we can expect TAD disruption to occur frequently in cancer, leading to major changes in gene expression that possibly even drive tumorigenesis. Further evidence that TAD formation and regulation is important in cancer is the fact that mutations in cohesin subunits, critical for TAD formation [28,31], are often associated with various cancers [47,48]. Molecular insights in the unique patterns of TADs in cancer cells may therefore contribute to a better understanding of the genetic basis of cancer cell gene expression, and possibly provide specific targets for treatment. We can imagine that in the future, genome editing with CRISPR-Cas9 in patient tumor cells could be employed to insert TAD boundaries in between the enhancer and the oncogene that drives the cancer. This insertion would result in the inactivation of the oncogene. The inserted TAD boundaries could be several CTCF binding sites in opposite directions. Obviously, there many hurdles, both experimental and ethical that need to be addressed before any such TAD engineering could be considered, but insights into the mechanism of TAD formation and disruption no doubt will lead to new ideas to treat disease.
Acknowledgments
Work in our lab is supported by the National Human Genome Research Institute (R01 HG003143, U54 HG007010, U01 HG007910), the National Cancer Institute (U54 CA193419), the NIH Common Fund (U54 DK107980, U01 DA 040588), the National Institute of General Medical Sciences (R01 GM 112720), and the National Institute of Allergy and Infectious Diseases (U01 R01 AI 117839). J.D. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 2.Gheldof N, Smith EM, Tabuchi TM, Koch CM, Dunham I, Stamatoyannopoulos JA, Dekker J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38:4325–4336. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Wit E, Vos ESM, Holwerda SJB, Valdes-Quezada C, Verstegen MJAM, Teunissen H, Splinter E, Wijchers PJ, Krijger PHL, de Laat W. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 9.Gómez-Marín C, Tena JJ, Acemel RD, López-Mayorga M, Naranjo S, de la Calle-Mustienes E, Maeso I, Beccari L, Aneas I, Vielmas E, et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci U S A. 2015;112:7542–7547. doi: 10.1073/pnas.1505463112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **13.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. 2015 doi: 10.1101/024620. bioRxiv. In this work the authors present loop extrusion, blocked by directional CTCF sites as a mechanism of TAD formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **14.Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–6465. doi: 10.1073/pnas.1518552112. In this work the authors show loop extrusion, blocked by directional CTCF sites can produce TAD structures as detected by Hi-C. Further, the authors show that this model explain effects of CTCF binding site mutation on TAD formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dekker J, Heard E. Structural and functional diversity of Topologically Associating Domains. FEBS Lett. 2015;589:2877–2884. doi: 10.1016/j.febslet.2015.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **17.Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. These authors use a functional reporter gene approach to show that TADs correspond to functional domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang R, Kerschner JL, Gosalia N, Neems D, Gorsic LK, Safi A, Crawford GE, Kosak ST, Leir S-H, Harris A. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith EM, Lajoie BR, Jain G, Dekker J. Invariant TAD Boundaries Constrain Cell-Type-Specific Looping Interactions between Promoters and Distal Elements around the CFTR Locus. Am J Hum Genet. 2016;98:185–201. doi: 10.1016/j.ajhg.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 21.Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- 22.Tsujimura T, Klein FA, Langenfeld K, Glaser J, Huber W, Spitz F. A discrete transition zone organizes the topological and regulatory autonomy of the adjacent tfap2c and bmp7 genes. PLoS Genet. 2015;11:e1004897. doi: 10.1371/journal.pgen.1004897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giorgio E, Robyr D, Spielmann M, Ferrero E, Di Gregorio E, Imperiale D, Vaula G, Stamoulis G, Santoni F, Atzori C, et al. A large genomic deletion leads to enhancer adoption by the lamin B1 gene: a second path to autosomal dominant adult-onset demyelinating leukodystrophy (ADLD) Hum Mol Genet. 2015;24:3143–3154. doi: 10.1093/hmg/ddv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **24.Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. This paper shows that TAD disruption leads to predictable novel enhancer-promoter interactions and alterations in gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupiáñez DG, Spielmann M, Mundlos S. Breaking TADs: How Alterations of Chromatin Domains Result in Disease. Trends Genet TIG. 2016 doi: 10.1016/j.tig.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Splinter E, Heath H, Kooren J, Palstra R-J, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sofueva S, Yaffe E, Chan W-C, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong C-T, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuin J, Dixon JR, van der Reijden MIJA, Ye Z, Kolovos P, Brouwer RWW, van de Corput MPC, van de Werken HJG, Knoch TA, van IJcken WFJ, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *33.Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. Transcription. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. This paper describes precise deletions of CTCF sites in the HoxA cluster and how leads to inappropriate gene expression and enlargement of TADs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heyn H, Vidal E, Ferreira HJ, Vizoso M, Sayols S, Gomez A, Moran S, Boque-Sastre R, Guil S, Martinez-Cardus A, et al. Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol. 2016;17:11. doi: 10.1186/s13059-016-0879-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C, McDaniel LD, Diamond M, Hart LS, Zhu S, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528:418–421. doi: 10.1038/nature15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2015 doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *37.Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, Shachar S, Weintraub AS, Hnisz D, Pegoraro G, Lee TI, et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell. 2015 doi: 10.1016/j.stem.2015.11.007. These authors show that CTCF bound sites demarcate domains of gene regulation, and that deletion of individual CTCF sites leads to novel long-range enhancer-promoter interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katainen R, Dave K, Pitkänen E, Palin K, Kivioja T, Välimäki N, Gylfe AE, Ristolainen H, Hänninen UA, Cajuso T, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–821. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- 39.Hnisz D, Weintraub AS, Day DS, Valton A-L, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016 doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair. 2006;5:1282–1297. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 41.Burman B, Zhang ZZ, Pegoraro G, Lieb JD, Misteli T. Histone modifications predispose genome regions to breakage and translocation. Genes Dev. 2015;29:1393–1402. doi: 10.1101/gad.262170.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *42.Gröschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BAM, Erpelinck C, van der Velden VHJ, Havermans M, Avellino R, van Lom K, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. This work shows that genomic rearrangement leads to novel chromatin domains and inappropriate enhancer - promoter pairing in AML. [DOI] [PubMed] [Google Scholar]
- 43.Walker BA, Wardell CP, Brioli A, Boyle E, Kaiser MF, Begum DB, Dahir NB, Johnson DC, Ross FM, Davies FE, et al. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J. 2014;4:e191. doi: 10.1038/bcj.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Northcott PA, Lee C, Zichner T, Stütz AM, Erkek S, Kawauchi D, Shih DJH, Hovestadt V, Zapatka M, Sturm D, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puente XS, Beà S, Valdés-Mas R, Villamor N, Gutiérrez-Abril J, Martín-Subero JI, Munar M, Rubio-Pérez C, Jares P, Aymerich M, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 46.Ryan RJH, Drier Y, Whitton H, Cotton MJ, Kaur J, Issner R, Gillespie S, Epstein CB, Nardi V, Sohani AR, et al. Detection of Enhancer-Associated Rearrangements Reveals Mechanisms of Oncogene Dysregulation in B-cell Lymphoma. Cancer Discov. 2015;5:1058–1071. doi: 10.1158/2159-8290.CD-15-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kon A, Shih L-Y, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 48.Leiserson MDM, Vandin F, Wu H-T, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2015;47:106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]