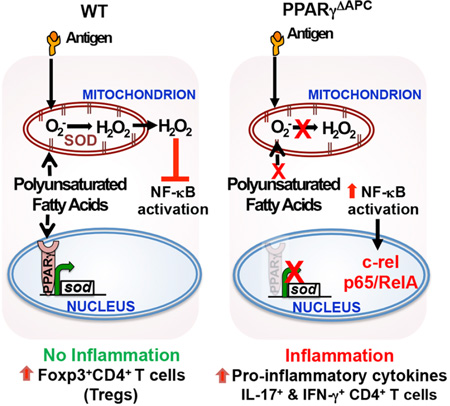
Summary
Inhalation of environmental antigens including allergens does not always induce inflammation in the respiratory tract. While antigen-presenting cells (APCs) including dendritic cells and macrophages take up inhaled antigens, the cell-intrinsic molecular mechanisms that prevent an inflammatory response during this process, such as activation of the transcription factor NF-κB, are not well understood. Here, we show that the nuclear receptor PPARγ plays a critical role in blocking NF-κB activation in response to inhaled antigens to preserve immune tolerance. Tolerance induction promoted mitochondrial respiration, generation of H2O2, and suppression of NF-κB activation in WT but not PPARγ-deficient APCs. Forced restoration of H2O2 in PPARγ-deficient cells suppressed IκBα degradation and NF-κB activation. Conversely, scavenging reactive oxygen species from mitochondria promoted IκBα degradation with loss of regulatory and promotion of inflammatory T cell responses in vivo. Thus, communication between PPARγ and the mitochondria maintains immune quiescence in the airways.
Graphical Abstract
Introduction
Inhalation of antigen/allergen is a natural and spontaneous process, which normally maintains immune tolerance in the airways (Curotto de Lafaille et al., 2008; Khare et al., 2015; Khare et al., 2013; McMenamin et al., 1994; Ostroukhova et al., 2004). This process of tolerance prevents inflammatory immune responses to inhaled antigens that in susceptible individuals can lead to allergic diseases such as asthma (Lambrecht and Hammad, 2012). Immune tolerance also prevents autoimmune diseases and transplant rejections. Antigen presenting cells (APCs) such as dendritic cells (DCs) play a central role in the decision-making process between immune activation and tolerance (Steinman, 2012). It is, therefore, important to understand the molecular mechanisms by which APCs mediate immune tolerance to be able to use their full potential for suppression of undesirable immune activation.
Recent literature highlights cross-talk between cellular metabolism and immune function (Odegaard et al., 2007; Tschopp, 2011). One example is metabolic syndrome, which is often associated with chronic unregulated inflammation in various organs (Odegaard et al., 2007; Tschopp, 2011). It is suggested that dysregulated production of reactive oxygen species (ROS) in mitochondria contributes to metabolic syndrome (James et al., 2012). More than 30 years ago, the ability of isolated mitochondria to produce the ROS, H2O2, was demonstrated (Chance et al., 1979). Subsequent studies showed that H2O2 is generated by dismutation of superoxide by the action of a superoxide dismutase (SOD) within mitochondria (Forman and Kennedy, 1974; Loschen et al., 1974). These discoveries collectively established mitochondria as an important source of cellular H2O2. Given that mitochondria have emerged as important regulators of multiple cellular functions (Galluzzi et al., 2012), it seems equally plausible that regulated mitochondrial ROS production contributes to immune homeostasis.
Peroxisome proliferator-activated receptor gamma (PPARγ), a member of the nuclear receptor superfamily, not only promotes adipocyte differentiation and glucose homeostasis, but it also exerts anti-inflammatory effects (Wahli and Michalik, 2012). PPARγ deletion in myeloid cells was shown to impair generation of alternatively activated macrophages and induce insulin resistance suggesting a beneficial role of PPARγ in controlling metabolic diseases such as type 2 diabetes (Odegaard et al., 2007; Tschopp, 2011). In the lung, PPARγ is expressed by multiple cell types including CD11c+ cells, which include the APCs DCs and macrophages (Belvisi et al., 2006). We recently reported that conditional deletion of PPARγ in the CD11c+ APCs in mice induces an inflammatory response in the airways of mice (Khare et al., 2015). However, the molecular mechanism by which PPARγ expression in CD11c+ cells efficiently suppresses airway inflammation despite constant provocation of the lungs by environmental antigens remains poorly understood. Here we show that in the absence of PPARγ, NF-κB is recruited to the promoters of the pro-inflammatory cytokine genes, IL-6 and the p19 subunit of IL-23 in lung APCs in keeping with increased production of these cytokines in these cells (Khare et al., 2015). Under tolerizing conditions, PPARγ-sufficient CD11c+ cells displayed higher oxygen consumption rate (OCR) than PPARγ-deficient CD11c+ cells, which was sensitive to Cpt1 blockade. Using two independent H2O2 detection methods, we identified H2O2 in WT but not PPARγ-deficient cells from tolerized mice, which involved mitochondrial Complex I but not Complex III activity. PPARγ was essential for increased SOD activity in the cells. Forced restoration of H2O2 in PPARγ-deficient cells suppressed IκBα degradation. Conversely use of a mitochondrially-targeted H2O2 scavenger, Mito-Tempo (Dikalova et al., 2010; Murphy, 1997), promoted IκBα degradation and airway tolerance was replaced by an inflammatory response, as observed in mice devoid of PPARγ in CD11c+ cells (Khare et al., 2015). Taken together, these findings establish a communication axis between the nucleus, mitochondria and the cell cytoplasm to prevent unnecessary immune activation in the airways under conditions of constant antigenic provocation.
Results
Increased recruitment of NF-κB to promoters of pro-inflammatory cytokine genes in PPARγ-deficient CD11c+ cells and PPARγ-dependent expression of genes involved in fatty acid metabolism
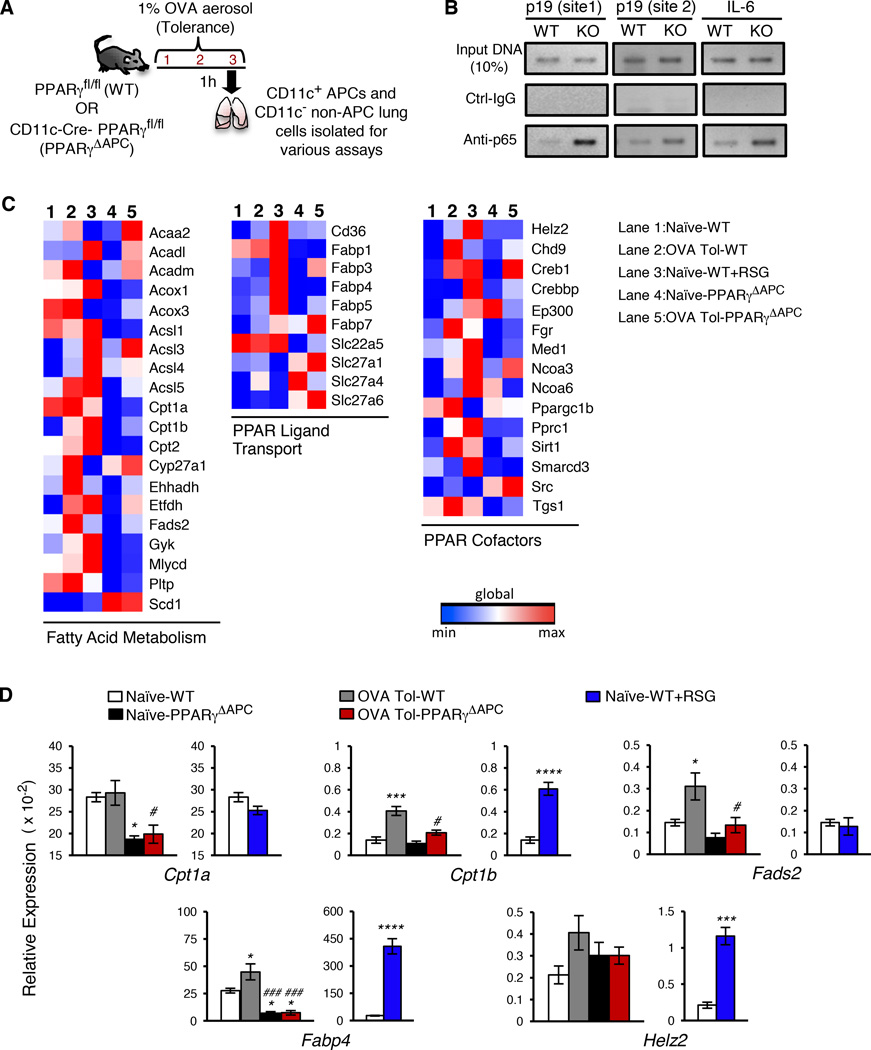
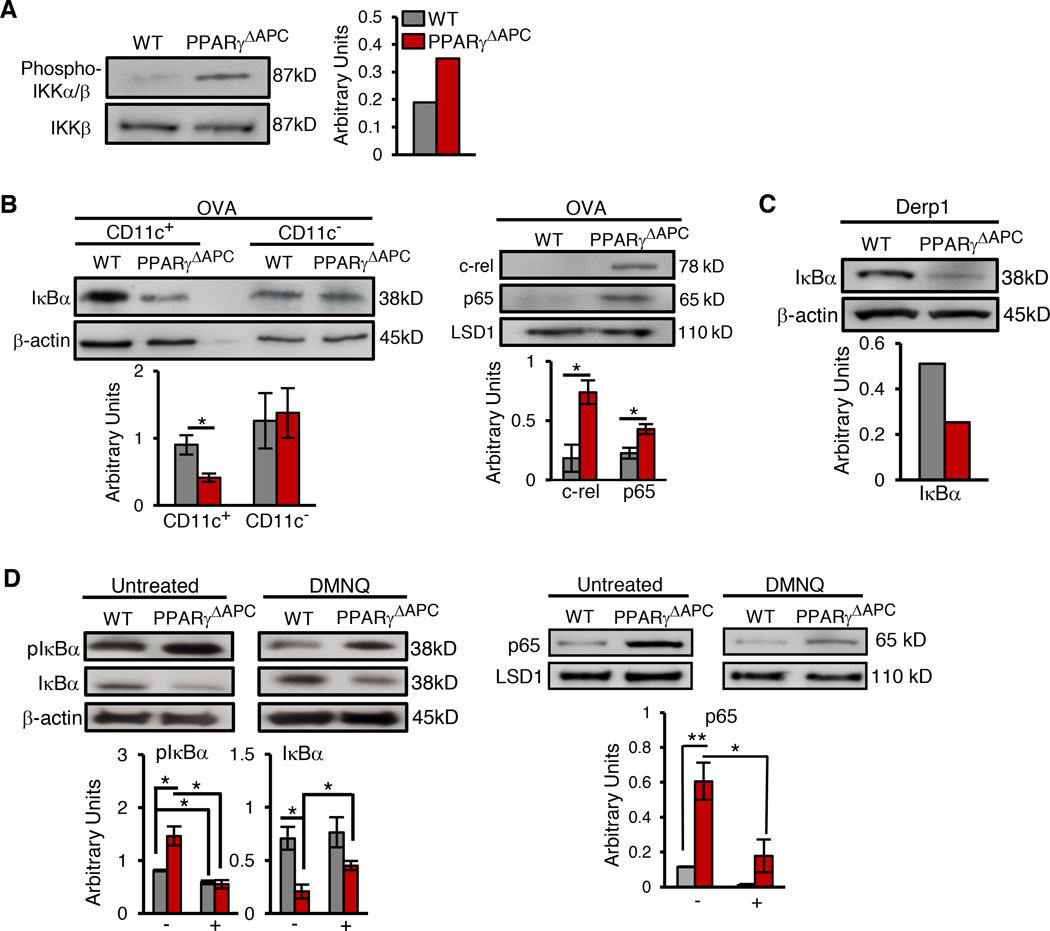
We recently reported that conditional deletion of PPARγ in CD11c+ cells, which include the APCs DCs and macrophages in the lungs, abolishes tolerance in the airways induced by low doses of inhaled antigen with induction of inflammation although the underlying molecular mechanism was not described (Khare et al., 2015). These CD11c-Cre-PPARγfl/fl mice (referred to hereafter as PPARγΔAPC mice) showed a minimal change in the numbers of lung DCs as compared to the control PPARγfl/fl mice (referred to hereafter as WT mice) but showed enhanced expression of pro-inflammatory cytokine genes under tolerizing conditions in all CD11c+ cells that included CD103+ DCs, CD103− DCs and macrophages (Khare et al., 2015). Since NF-κB is a central regulator of expression of pro-inflammatory cytokine genes (Hayden and Ghosh, 2008; Pasparakis, 2009), we investigated whether deletion of PPARγ in CD11c+ cells aberrantly triggers NF-κB activation in response to a low dose of inhaled antigen, which normally maintains airway tolerance (Khare et al., 2015; Khare et al., 2013; Ostroukhova et al., 2004). We used a shorter protocol for tolerance induction, which involved exposure of the mice to 3 consecutive days of aerosolized ovalbumin (OVA) (Figure 1A) instead of 10 days as previously described by us (Khare et al., 2015; Khare et al., 2013; Ostroukhova et al., 2004). Both protocols blunted induction of airway inflammation to the same degree when tested for tolerance induction by subsequently immunizing the mice with OVA plus adjuvant and then repeatedly challenging by aerosolized OVA (Figure S1A). In both protocols, we observed minimal recruitment of the p65 subunit (Rel A) of NF-κB to the different NF-κB sites in the p19 and IL-6 promoters in PPARγ-sufficient cells while p65 recruitment was readily detectable when cells were PPARγ-deficient (Figure 1B and S1B).
Figure 1. Inhaled antigen-activated PPARγ regulates genes involved in regulation of inflammation and fatty acid metabolism.
(A) Schematic describing the 3-day OVA-exposure protocol for tolerance. (B) ChIP assay of p19 and IL-6 promoters in WT CD11c+ cells sorted from indicated conditions using anti-p65 and control antibodies. (C) Heat map representation of PPAR target array experiments for the indicated genes clustered based on function. mRNA was isolated from CD11c+ cells from naïve (lane 1), tolerized (lane 2), and ex-vivo RSG-treated cells from WT mice (lane 3) and also from naïve (lane 4) and tolerized PPARγΔAPC (lane 5) mice. (D) Graphical representation of relative expression in CD11c+ cells of indicated genes in specific groups present in the PPAR target array. Data shown are mean ± SEM and are representative of two independent experiments (n=3–4 mice/group). One-way ANOVA with Tukey’s post hoc test was used for comparisons between multiple groups: *p< 0.05; ***p< 0.001, ****p< 0.0001 reflect comparisons to expression in naïve PPARγfl/fl and #p< 0.05; ###p< 0.001 correspond to comparisons with OVA tol PPARγfl/fl. See also Figures S1, S2 and S3.
Of note, in a study published recently in which PPARγ was similarly deleted in CD11c+ cells, a complete arrest in the development of alveolar macrophages (AMs) characterized as CD11c+CD11blo cells in bronchoalveolar lavage (BAL) and lung cells was reported (Schneider et al., 2014). Instead, a significant increase in CD11chi cells that were CD11bhi, autofluorescent and expressed F4/80 was noted; these cells were described as AM-like cells (Schneider et al., 2014). Using a similar AM characterization strategy, we found that while CD11chiCD11bhi AM-like cells are present in BAL and lung cells of PPARγΔAPC mice generated by us (Figure S2A), CD11c+CD11blo cells resembling AMs were only reduced by ∼50% in these mice (Figure S2A) unlike their complete absence in the PPARγ conditional knockouts described in the earlier study (Schneider et al., 2014). We then focused on Siglec-F expression on CD11c+F4/80+ cells, the co-expression of these molecules being restricted to only macrophages in the lungs. We observed a slight reduction in CD11c+F4/80+ cells in the lungs of PPARγΔAPC mice as compared to those in PPARγfl/fl mice (Figure S2B). When gated on these cells, 15% were CD11bhiSiglec-F+ cells in PPARγΔAPC mice as compared to 5% in WT mice with no downregulation of Siglec-F expression in any subset, unlike what was observed in the earlier study (Schneider et al., 2014). Thus, collectively, the mice used by us had no difference in number of CD11c+ autofluorescencelo DCs or CD11c+ autofluorescencehi macrophages (AM and AM-like cells together) (Khare et al., 2015). The partial reduction in AM numbers in the PPARγΔAPC mice (Khare et al., 2015) did not explain either the increase in expression of pro-inflammatory cytokine genes in the CD11c+ cells derived from these mice or the inability of these mice to support Treg development in response to tolerance-inducing conditions (Khare et al, 2015). Further, the increased presence of CD11bhi macrophages in the lungs of the PPARγ conditional knockout mice suggested a shift in the population towards an activated phenotype (Guth et al., 2009; Striz et al., 1993), as also suggested by increased production of cytokines such as IL-6 and the p19 subunit of IL-23 by these cells (Khare et al., 2015).
Our next task was to determine how presence of PPARγ blocks NF-κB recruitment to the promoters of cytokine genes in CD11c+ cells. PPARγ-mediated immune regulation requires activation by either synthetic agonists like thiazolidinediones (TZDs) or by endogenous ligands such as fatty acids. Our model did not employ any synthetic ligand and yet involvement of PPARγ in regulating the activation status of CD11c+ cells was evident. Using a focused gene array we compared the mRNA expression profile of PPAR-regulated genes in different sets of lung CD11c+ cells-those stimulated by the TZD Rosiglitazone (RSG) ex vivo and those isolated from naïve or antigen (ovalbumin)-tolerized (OVA-Tol) WT or PPARγΔAPC mice (Figure 1C, 1D and S3). The enzymes Cpt1a and Cpt1b are involved in fatty acid translocation across the mitochondrial membrane and expression of the former is typically higher in immune cells (Wakil and Abu-Elheiga, 2009). The expression of Cpt1a was significantly reduced in PPARγ-deficient cells as compared to that in WT cells (Figure 1C and 1D). Albeit expressed at a lower level, the relative increase in Cpt1b expression was also less in cells without PPARγ (Fig. 1C and 1D). Fatty acid desaturase-2 (Fads-2) is an important enzyme that catalyzes the generation of polyunsaturated fatty acids (PUFAs) (Guillou et al., 2010). In response to inhaled antigen, the increase in Fads-2 expression was higher in WT cells as compared to that in PPARγ-deficient cells. As expected from previous publications, mRNA level of fatty acid binding protein-4 (Fabp4) increased several fold in RSG-treated treated cells (Odegaard et al., 2007) and the expression of helicase with zinc finger 2 (helz2), which is a co-activator of PPARγ and promotes optimal expression of Fabp4 (Katano-Toki et al., 2013), was also increased (Figure 1C and 1D). The differential response of cells to inhaled antigen versus RSG, both mediated by PPARγ, suggested a distinct molecular mechanism of PPARγ action under conditions of tolerance. Since Fabp4 binds and sequesters FFAs in the cytoplasm of cells (Furuhashi and Hotamisligil, 2008), and its expression was not upregulated in the CD11c+ cells from tolerized mice to the extent it was upon RSG treatment, we next examined the status of FFAs in the CD11c+ cells.
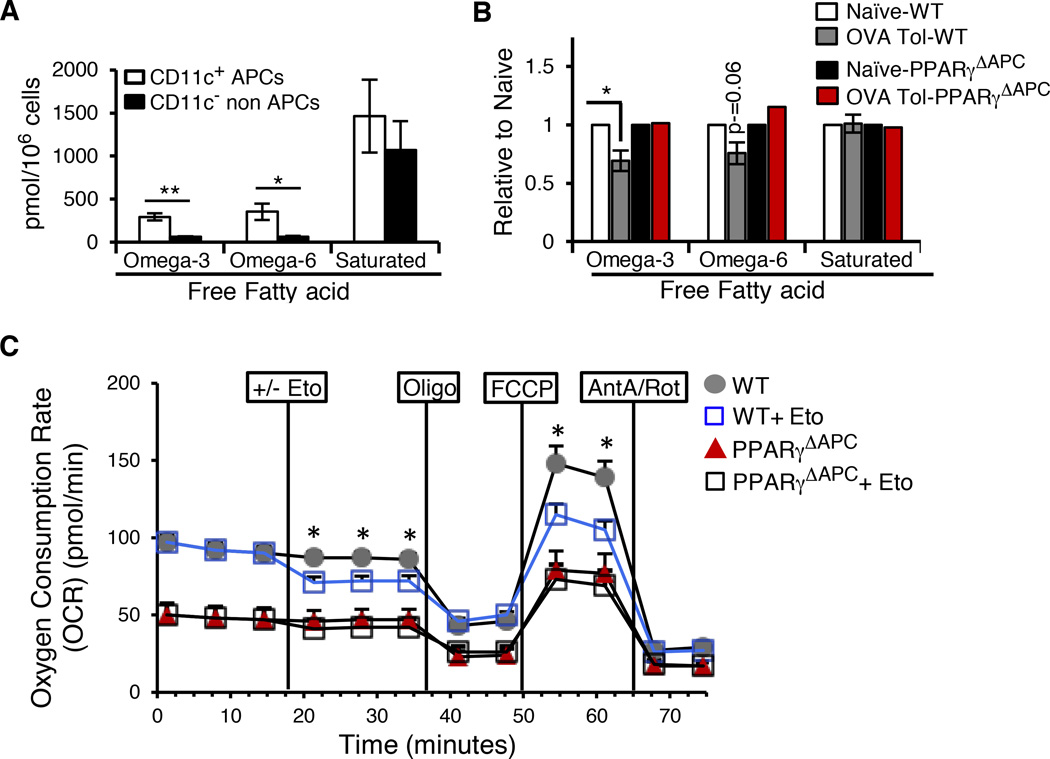
Lung CD11c+ cells as well as CD11c− cells (minus B cells) sorted from naïve and tolerized WT and PPARγΔAPC mice were subjected to FFA analysis employin gas chromatography coupled with mass spectrometry (GC/MS) (Figure 2A and 2B) (Quehenberger et al., 2011). CD11c+ APCs from naïve WT mice contained significantly higher levels of free ω-3 and ω-6 PUFAs compared to CD11c- non-APCs, in which these FFAs were barely detectable (Figure 2A). The levels of free saturated fatty acids were however comparable between CD11c+ APCs and CD11c- non-APCs (Figure 2A). Under PPARγ-sufficient conditions, the levels of both ω-3 and ω-6 PUFAs were lower in CD11c+ cells from tolerized mice as compared to cells from naïve mice with the decrease in ω-3 PUFAs reaching statistical significance. However, the levels of saturated fatty acids were similar in cells from naïve and tolerized WT mice (Figure 2B). In accordance with the gene expression data showing lower Cpt1 expression in PPARγ-deficient cells, there was no decrease in ω-3 and ω-6 PUFAs in these cells isolated from tolerized PPARγΔAPC mice (Figure 2B). We next assessed oxygen consumption rate (OCR) of CD11c+ cells isolated from tolerized WT and PPARγΔAPC mice. OCR was measured at the basal level and then one batch of cells in both groups was treated with Etomoxir (Eto) and then sequentially treated with oligomycin (Oligo), FCCP, and rotenone plus antimycin (Ant A+Rot). Basal OCR in WT cells was inhibited upon treatment with Eto, a specific inhibitor of Cpt1 (Wolf and Brenner, 1988). PPARγ-deficient CD11c+ cells showed lower OCR overall as compared to PPARγ-sufficient cells, which was unchanged upon Eto treatment (Figure 2C). The maximal OCR reached after FCCP injection of WT cells previously treated with Eto remained lower than those maintained without Eto suggesting fatty acid oxidation (FAO) to be an important component of OCR in tolerized WT cells. These results suggested that conditions of tolerance or even basal conditions that regulate immune homeostasis favor maintenance of a relatively high level of endogenous PPARγ ligands such as PUFAs in lung APCs but not in non-APCs. Also, the lower levels of free PUFAs in CD11c+ cells from tolerized WT mice combined with higher Fads2 and Cpt1 expression together with higher Eto-sensitive OCR in these cells suggested β-oxidation of fatty acids in mitochondria with increased mitochondrial ROS production in the cells.
Figure 2. Antigen-activated PPARγ regulates fatty acid metabolism.
(A) Total omega-3, omega-6 and saturated free fatty acid quantification (pmol/106 cells) as estimated by GC/MS analysis performed on CD11c+ APCs and CD11cB220- non-APCs sorted from the lungs of naïve WT mice. Data shown are composite of 3 independent experiments and represent mean values ± SEM. (B) Presence of omega-3, omega-6 and saturated free fatty acids in CD11c+ cells sorted from the lungs of naïve and tolerized WT and PPARγΔAPC mice as observed in 2 independent experiments, expressed as fold change (relative to naïve of the respective strain). Data shown are composite of 2 independent experiments and represent mean values ± SD. (C) OCR of CD11c+ cells isolated from tolerized WT and PPARγΔAPC mice was measured at the basal level and then after-sequential treatment with DMEM/200 µM Etomoxir (Eto) followed by 1 µM oligomycin (Oligo), 1.5 µM FCCP, and 0.1 µM rotenone plus 1 µM antimycin (Ant A+Rot) using the XF-96 Seahorse system. The assay is representative of 2 independent experiments, run in triplicates and shown as mean values ± SEM *p< 0.05, **p< 0.01.
Increased mitochondrial H2O2 production under conditions of tolerance
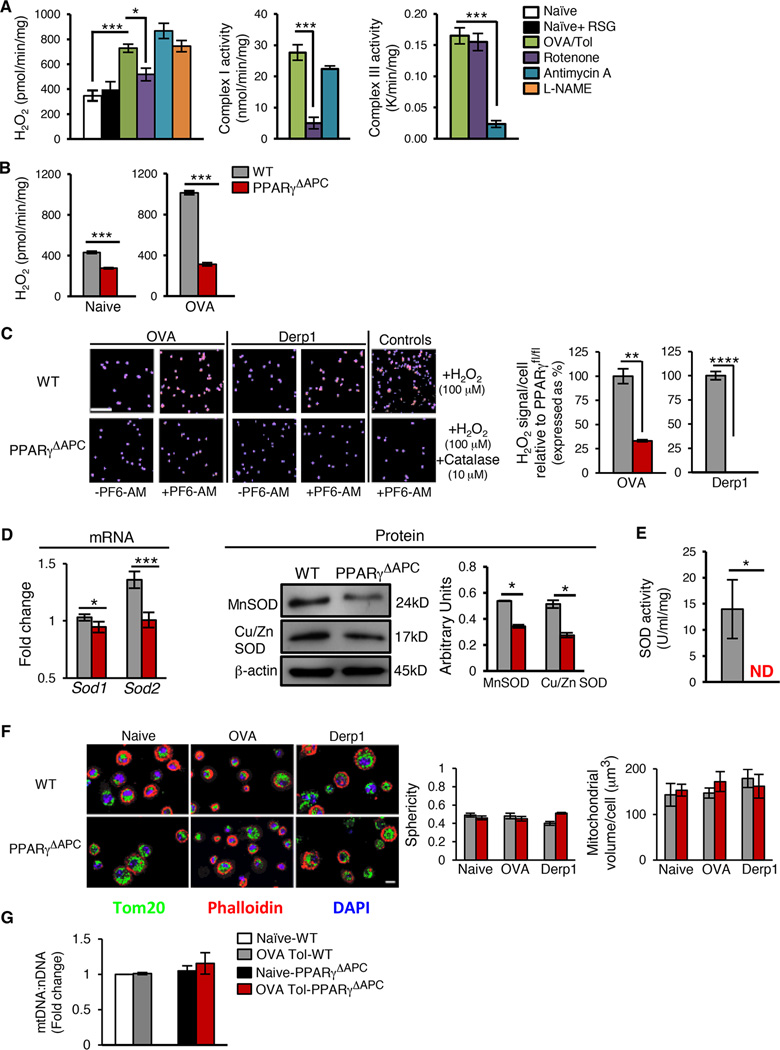
The fact that PUFAs are not only ligands of PPARγ but are also substrates for mitochondrial electron transport (Lombardi et al., 2008; Nethery et al., 2000), led us to examine the production of ROS in lung CD11c+ cells. When generated in small quantities via the mitochondrial electron transport chain (mETC) (Chance et al., 1979), H2O2 has been shown to be important for tissue homeostasis (Poole et al., 1997). Surprisingly, the role of ROS in regulation of immune homeostasis including activation of important transcription factors such as NF-κB under physiological conditions remains unclear. H2O2 is a diffusible molecule and can be assayed in the extracellular medium by the Amplex Red assay, a suitable method for quantitation of H2O2 production (Kalyanaraman et al., 2012). H2O2 generated in CD11c+ cells from tolerized PPARγfl/fl mice was greater when compared to cells from naïve mice (Figure 3A). Inhibition of H2O2 generation in PPARγ-sufficient CD11c+ cells from tolerized mice by rotenone, a specific inhibitor of mitochondrial complex I (Figure 3A) (Lombardi et al., 2008; Murphy, 2009), but not by antimycin A, a specific inhibitor of complex III (Figure 3A) or L-NAME (NOS2 inhibitor) (Xia et al., 1996), confirmed the involvement of the mETC in increased H2O2 levels in response to antigen (Figure 3A). Furthermore, inhibition by rotenone suggested the involvement of reverse electron transport (RET) in mitochondrial ROS generation (Murphy, 2009). In other studies in skeletal muscle cells, mitochondrial H2O2 generation induced by PUFA (arachidonic acid) was also inhibited by rotenone (Lombardi et al., 2008). We also observed that PPARγ-sufficient CD11c+ cells treated with RSG did not generate H2O2 (Figure 3A), which reaffirmed that RSG-mediated and low dose antigen-mediated PPARγ activation induce different responses in the CD11c+ cells. Figure 3A shows specificity of the mETC inhibitors with selective inhibition of complex I by rotenone and of complex III by antimycin A (Shiva et al., 2007).
Figure 3. PPARγ expression regulates mitochondrial ROS generation.
(A) Amplex Red Assay was used to quantitate H2O2 generation in sorted lung CD11c+ cells from naïve WT mice in the presence or absence of RSG (10 µM) and tolerized WT mice in the presence or absence of Rotenone (10 µM), Antimycin A (100 µM) or L-NAME (50 µM). Complex I and III activities were measured to assess specificity and dose efficiency of Rotenone and Antimycin A, respectively. (B) Quantification of H2O2 generation in sorted lung CD11c+ cells from naïve and tolerized WT and PPARγΔAPC mice. Data for panel A and B represents one of 3 independent experiments and are shown as mean ± SEM. (C) Intracellular H2O2 detection in sorted lung CD11c+ cells from tolerized WT and PPARγΔAPC mice using the H2O2-specific probe PF6-AM. During the incubation with PF6-AM, OVA (10 µg/ml) or Der p1 (0.5 µg/ml) was included. In the absence of antigen in vitro, a similar profile was obtained with lower signal intensity in the WT cells and none detected in the PPARγ-deficient cells (data not shown). As a positive control, exogenous H2O2 (100 µM) was added to the CD11c+ cells in vitro ± catalase (10 µM). Representative immunofluorescence images (left) and fluorescence quantification (right) represent cytosolic H2O2 signal per cell per field (5–6 fields chosen randomly using DAPI as a guide and then assessed for PF6-AM-specific signal). Scale bar-100 µm and magnification 20X. Bar graphs show data as mean values ± SEM. (D–E) Lung CD11c+ cell from tolerized WT and PPARγΔAPC mice were analyzed for (D) MnSOD and Cu/ZnSOD mRNA (left panel) and protein expression (right panel) and (E) SOD activity. Data are shown as mean values ± SD. (F) Representative immunofluorescence images (left) and sphericity and mitochondrial volume/cell estimation (right) of lung CD11c+ cells stained with Tom20 (green), DAPI (blue), and phalloidin (red). Scale bar- 10 µm; magnification 60X. (G) Quantification of mitochondrial DNA (mtDNA) and nuclear (nDNA) in purified lung CD11c+ cells from naïve and tolerized WT and PPARγΔAPC mice. Data in panels (F) and (G) represent one of two independent experiments and shown as mean values ± SD. ND: Not detected, NS: Not significant, *p< 0.05, ***p< 0.001, ****p<0.0001. See also Figure S4.
We next assessed whether the increase in H2O2 production from CD11c+ cells from tolerized WT mice depends on PPARγ, which was found to be the case (Figure 3B). Essentially, similar results were obtained by imaging when the boronate compound peroxyfluor-6 acetoxymethyl ester (PF6-AM), a selective fluorescent detector of H2O2 (Basu et al., 2014; Dickinson et al., 2011; Lin et al., 2013), was used with H2O2 readily detected in CD11c+ cells from WT but not PPARγ-deficient cells (Figure 3C). Similar results were obtained when the mice were exposed to a different allergen, Derp1, of house dust mite (Cates et al., 2004) (Figure 3C) demonstrating the generality of effect of PPARγ-deficiency in CD11c+ cells on H2O2 generation in the cells. The results described above raised the question of how PPARγ promotes H2O2 generation in the CD11c+ cells. Cellular H2O2 is generated following dismutation of superoxide radical generated during mitochondrial electron transport by SOD enzymes (Buettner, 2011). The expression of both MnSOD (SOD2), which is targeted to mitochondria, and of CuZnSOD (SOD1), which is cytosolic, at both mRNA and protein levels, was reduced in PPARγ-deficient cells as compared to that in WT cells (Figure 3D). PPARγ-sufficient but not PPARγ-deficient CD11c+ cells showed increased SOD activity (Figure 3E). PPARγ-induced SOD activity under tolerizing conditions in WT cells can therefore be expected to cause an increase in cytosolic H2O2 level due to diffusion of H2O2 across the mitochondrial membrane (Brand, 2010).
We next examined the morphology of mitochondria in CD11c+ cells derived from naïve and tolerized WT and PPARγΔAPC mice by staining the mitochondrial membrane with Tom20. Mitochondria are extremely dynamic organelles that fragment and fuse continuously; the morphology of mitochondria (fragmented versus extensive fused networks) strongly influences function (da Silva et al., 2014). Mitochondrial abundance was measured using confocal microscopy followed by 3-dimensional surface rendering and calculation of mitochondrial volume (abundance). Mitochondrial fragmentation was assessed using the sphericity parameter. The closer the sphericity value is to 1, the more spherical and fragmented the mitochondria. The average sphericity in our samples was low, ranging around 0.5, indicating that the mitochondria are highly networked. Both volume and sphericity calculations showed no difference whether cells were examined from WT or PPARγ-deficient cells (Figure 3F and Figure S4). Similar data were obtained by quantifying the mitochondrial DNA (mtDNA) copy number in WT and PPARγ-deficient cells (Figure 3G). These observations ruled out a difference in mitochondrial abundance or health between the WT and PPARγΔAPC mice as contributing to the increased inflammatory phenotype of the latter and instead focused our attention on regulation of gene expression by H2O2.
PPARγ prevents NF-κB activation in tolerized mice
The physiological relevance of effects of high concentrations of H2O2 on NF-κB activation in vitro is difficult to ascertain without in vivo supportive evidence (Oliveira-Marques et al., 2009). Unlike most studies that have shown mild NF-κB activation in response to large doses of H2O2 (Oliveira-Marques et al., 2009), one prior study using mouse alveolar epithelial cells showed that H2O2 can inhibit NF-κactivation by limiting IKK phosphorylation (Korn et al., 2001). Consistent with this finding, we observed greater IKK phosphorylation in tolerized PPARγ-deficient CD11c+ cells compared to PPARγ-sufficient cells (Figure 4A), the former having lower H2O2 levels (Figure 3B and 3C). IKK-mediates phosphorylation of specific Ser residues in IκBα leading to ubiquitination and proteasomal degradation (reviewed in (Karin and Ben-Neriah, 2000). In accordance with the IKK phosphorylation data, lower expression level of IκBα in the cytoplasmic extracts of PPARγ-deficient CD11c+ cells was found as compared to that in control CD11c+ cells. In lung CD11c− cells, in which PPARγ was not deleted, the level of IκBα was comparable between PPARγΔAPC and WT mice (Figure 4A). Conversely, a higher expression of c-rel and the p65 subunit of NF-κB (RelA) was observed in the nuclear extracts of PPARγ-deficient CD11c+ cells when compared to control extracts (Figure 4B). Similar observations were made when mice were exposed to Derp1 (Figure 4C) demonstrating the generality of effect of PPARγ-deficiency in CD11c+ cells on NF-κB activation. Both chymotrypsin-like and trypsin-like proteasomal activities were similar in CD11c+ cells from PPARγΔAPC and WT mice irrespective of whether cells were isolated from naïve or OVA-exposed mice (Figure S5A) ruling out a difference in proteasomal activity between PPARγ-sufficient and PPARγ-deficient cells as playing any role in differential NF-κB activation in these cells. If PPARγ-dependent H2O2 generation indeed inhibits phosphorylation of IκBα, we hypothesized that incubation of PPARγ-deficient CD11c+ cells with 2, 3-dimethoxy-1,4-naphthoquinone (DMNQ), a non-thiol-capturing and non-alkylating redox-cycling quinone that causes continuous intracellular generation of H2O2 (Parry et al., 2009), would reverse the effects on IκBα. Importantly, forced generation of H2O2 using DMNQ does not require SOD, which provided an opportunity to bypass PPARγ. Indeed, DMNQ treatment resulted in increased intracellular H2O2 levels (Figure S5B) reducing pIκBα and increasing IκBα levels as compared to that in untreated cells (Figure 4D). DMNQ caused a similar differential in pIκBα and IκBα, albeit lower in magnitude, in cells from tolerized WT mice that to begin with harbored a higher level of H2O2 (Figure 4D). Correspondingly, reduced nuclear translocation of p65 was observed in DMNQ-treated cells as compared to that in untreated cells (Figure 4D).
Figure 4. Absence of mitochondrial ROS promotes NF-κB activation.
Representative western blot and densitometry quantification to detect expression of (A) phospho IKKα/IKKβ levels in sorted CD11c+ cells, (B) IκBα in cytoplasmic extracts and c-rel, p65 subunit (RelA) of NF-κB in nuclear extracts prepared from lung CD11c+ and CD11c- cells from PPARγΔAPC and WT mice 1 hr after the last exposure to OVA, (C) IκBα in lung CD11c+ cells sorted from PPARγΔAPC and WT mice 1 hr after the last exposure to Derp1 and (D) phospho IκBα (pIκBa) and total IκBα levels in whole cell extract and p65 levels in nuclear extracts from lung CD11c+ cells (sorted from tolerized mice), cultured for 1 hr in the presence or absence of DMNQ (200 µM). For all western blot analyses, expression of β-actin and LSD1 is shown as loading controls for cytoplasmic and nuclear extracts respectively and densitometric quantification is combined data from two independent experiments represented as mean ± SD. ND: not detected, *p< 0.05, **p< 0.01. See also Figure S5.
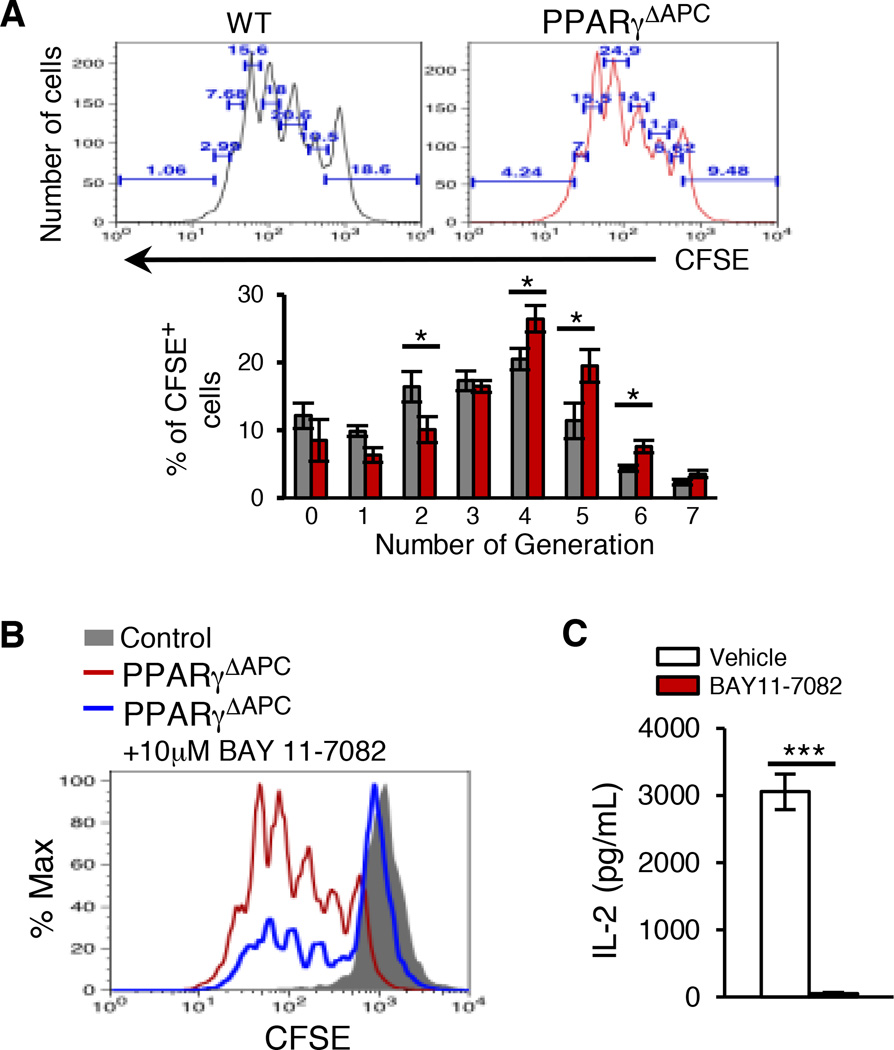
To further investigate a causal relationship between reduced H2O2 and heightened NF-κB activation in the CD11c+ cells, we co-cultured CD11c+ DCs from tolerized CD11c-specific PPARγ-deficient and sufficient mice with CFSE labeled Treg-depleted OT-II CD4+ T cells in the presence of OVA peptide and assessed T cell proliferation. The PPARγ-deficient cells induced better T cell proliferation compared to WT cells as determined by quantitation of %CFSE+ cells in each population of divided cells (Figure 5A). T cell proliferation induced by the PPARγ-deficient CD11c+ cells was drastically inhibited by pre-treatment of the DCs with the specific inhibitor of IκBα phosphorylation, BAY 11–7082 (Figure 5B and Figure S6). A significant decrease in the level of IL-2 was also observed in co-cultures stimulated with DCs pre-treated with BAY-117082 (Figure 5C).
Figure 5. Absence of PPARγ in CD11c+ DCs promotes T cell proliferation via NF-κB activation.
CFSE-labeled Treg-depleted OT-II splenic CD4+ T cells were stimulated with either vehicle or BAY11 −7082 (10 µM) pre-treated CD11c+ DCs sorted from tolerized PPARγΔAPC and WT mice. (A) T cell proliferation was assessed after 72 hr by estimating CFSE dilution using flow cytometry and number of cells per generation was calculated. (B) Overlay plot showing T cell proliferation as CFSE dilution when stimulated in presence of either vehicle or BAY11 −7082 (10µM) pre-treated CD11c+ DCs sorted from tolerized PPARγΔAPC mice. Data shown are representative of three independent experiments. (C) Expression of IL-2 was assessed in the PPARγΔAPC DC-T cell co-culture supernatant by ELISA. Data for panels A and C are shown as mean values ± SEM and composite of three independent experiments. *p< 0.05, ***p< 0.00. See also Figure S6.
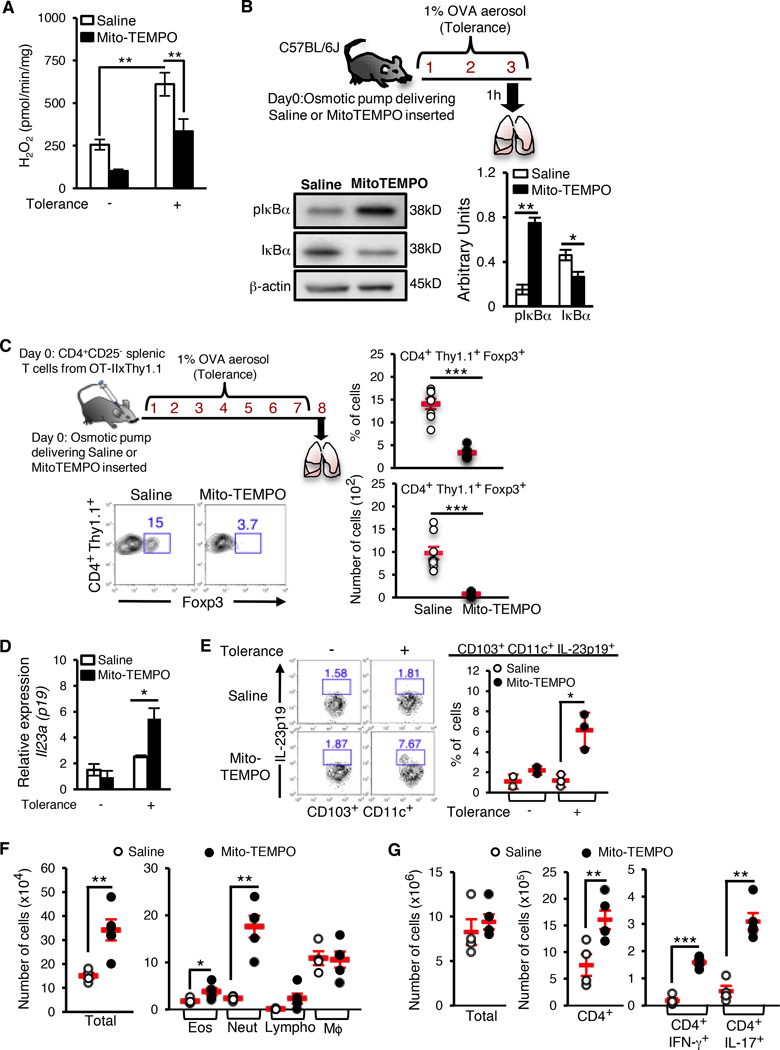
Scavenging mitochondrial ROS promotes NF-κB activation and impairs airway tolerance
To further confirm the specific role of mitochondrial ROS in suppression of NF-κB activation, we made use of the mitochondrially-targeted ROS scavenger Mito-TEMPO (Dikalova et al., 2010; Murphy, 1997). As shown in Figure 6A, use of Mito-TEMPO reduced H2O2 production from the CD11c+ cells isolated from C57/BL6J tolerized mice. Next, we used Mito-TEMPO in vivo to demonstrate the importance of mitochondrial ROS in blocking NF-κB activation and expression of pro-inflammatory cytokine genes during tolerance thereby favoring Treg generation and immune suppression. Mito-TEMPO was instilled into mice using an osmotic pump and exposed the mice to aerosolized OVA. Lung CD11c+ cells isolated from mice that received Mito-TEMPO showed a higher level of pIκBa and a lower level of IκBa as compared to those that received saline (Figure 6B). While the results of these experiments do not exclude a role for other cellular superoxide-generating systems (including NADPH oxidase), in aggregate, our data demonstrate that mitochondrially-derived H2O2 contributes significantly to NF-κB inhibition. Since Foxp3 induction in adoptively transferred naïve CD4+ T cells is severely impaired in CD103 DC-deficient Batf3−/− mice (Khare et al., 2013), we asked whether quenching of mitochondrial ROS by Mito-TEMPO in vivo, which would cause NF-κB activation in all DCs including CD103+ DCs, would similarly affect Foxp3 induction in naïve CD4+ T cells. Treatment with Mito-TEMPO blunted Foxp3 induction in adoptively transferred CD4+CD25- T cells from OT-IIxThy1.1 mice (Figure 6C). Conversely, we detected increased expression of the pro-inflammatory cytokine IL-23p19 (Figure 6D and 6E) in CD103+ DCs isolated from Mito-TEMPO-treated tolerized mice as compared to that in controls. Compared to vehicle-treated controls, tolerized mice that received Mito-TEMPO showed a significant increase in neutrophil infiltration and a smaller increase in eosinophil infiltration in the airways when challenged in the inflammation model (Figure 6F). The accompanying CD4+ T cell response showed a prominent increase in IL-17 expression accompanied by an increase in IFN-γ expression (Figure 6G). Therefore, results obtained with Mito-TEMPO treatment of mice were in close agreement with those derived using PPARγΔAPC mice (Khare et al., 2015) thereby demonstrating that a major mechanism by which PPARγ regulates immune tolerance is by controlling mitochondrial ROS generation.
Figure 6. Absence of PPARγ-dependent mitochondrial ROS generation in CD11c+ cells induces a Th17 immune response.
Osmotic pumps delivering either saline or Mito-TEMPO (0.7 mg/kg/day) were surgically inserted in C57/BL6J mice that were then exposed to inhaled OVA (as indicated). (A) Amplex red assay was used to quantitate H2O2 generation in sorted lung CD11c+ cells. Combined data from two independent experiments expressed as mean ± SEM. (B) Representative western blot and densitometry quantification to detect expression of pIκBα and total IκBα levels in lung CD11c+ cells. (C) 106 CD4+ CD25− splenic T cells from OT-IIxThy1.1 mice were adoptively transferred following osmotic pump implantation. The recipient mice were exposed to inhaled OVA (upper left panel) and Foxp3 induction in the donor T cells in the lungs of the recipient mice was assessed. Representative flow cytometry data from one of two experiments (bottom left) and frequency (top right) and number (bottom right) of CD4+ Thy1.1+ Foxp3+ T cells are shown. Symbols in the graphs represent individual mice (n=6), horizontal lines show the mean values and error bars denote SEM. (D) Expression of Il23a (p19) mRNA in CD103+ CD11c+ lung cells sorted from naïve and tolerized saline- or Mito-TEMPO-treated mice, relative to hprt1. (E) Flow cytometry analysis (left) and frequency (right) of IL-23p19-expressing lung CD103+CD11c+ cells from naïve and tolerized saline- or Mito-TEMPO-treated mice. Symbols in the graphs represent individual mice (n=3), horizontal lines show the mean values and error bars denote SEM. (F–G) Following osmotic pump implantation, mice were tolerized to OVA and tested for induction of tolerance as described under methods. 24 hr after the last treatment, various parameters of inflammation were analyzed. (F) Total cell numbers (left), and eosinophils and neutrophils (right) in the BAL fluid, and (G) Number of total lung cells, CD4+ T cells and IL-17A+ and IFN-γ+ CD4+ T cells in the lungs of saline- or Mito-TEMPO-treated mice were enumerated. Symbols in the graphs represent individual mice (n=5), horizontal lines show the mean values and error bars denote SEM. *p< 0.05, **p< 0.01, ***p< 0.001.
Discussion
Our study highlights a previously unappreciated function of PPARγ in regulating immune tolerance induced by low doses of inhaled antigen by invoking mitochondrial metabolism in lung APCs. We show that PPARγ prevents nuclear translocation of NF-κB limiting binding to target sites in the promoters of pro-inflammatory cytokine genes. Use of a targeted gene array revealed differential expression of enzymes involved in mitochondrial FAO between WT and PPARγ-deficient cells. PPARγ was required for optimal Cpt1a and Cpt1b mRNA expression in CD11c+ cells, Cpt1 proteins being required for FFA translocation to mitochondria. PPARγ also promoted expression of Fads2 in response to inhaled antigen, Fads-2 being shown to catalyze de novo generation of PUFAs. We show that the OCR of WT cells from tolerized mice is inhibited by blockade of Cpt1 and is substantially reduced in the absence of PPARγ. We provide evidence of mitochondrial respiration involving Complex I for increased H2O2 generation in response to inhaled antigen in WT cells, which inhibited IκBα phosphorylation/NF-κB activation in these cells. It is interesting to note that mitochondrial Complex III activity causing increased ROS generation has been associated with IL-2 production and NF-AT activation in T cells (Sena et al., 2013). Loss of PPARγ in the CD11c+ cells led to undetectable SOD activity explaining reduced H2O2 generation in the cells. Scavenging of mitochondrial ROS induced IκBα phosphorylation and promoted expression of cytokine genes, which favored T helper responses and inhibited Treg development with loss of immune tolerance.
Our gene expression data and FFA analyses collectively suggest that an important task of PPARγ is to promote mitochondrial oxidation of PUFAs in lung APCs. It is important to note that in the CD11c+ cells during inflammation, we have detected lower expression of PPARγ (Khare et al., 2015). Thus, although PUFAs are present in CD11c+ cells under conditions of inflammation (data not shown), lower levels of PPARγ translating into lower SOD activity would prevent an increase in H2O2 level in these cells promoting NF-κB activation. The relevance of weak NF-κB activation by H2O2 observed in in vitro studies by treating cell lines with H2O2 or by using antioxidants such as N-acetyl cysteine (NAC) or pyrrolidine dithiocarbamate (PDTC) has been a subject of considerable debate (Oliveira-Marques et al., 2009). The positive effect of H2O2 on NF-κB activation observed in these studies has been largely cell type-dependent and in some studies showed no evidence of IκBα degradation but instead involved tyrosine phosphorylation and dissociation of IκBα from NF-κB (Beraud et al., 1999). Also, reassessment of the effects of NAC and PDTC showed that they attenuated NF-κB activation independent of their antioxidant function. Limited information exists about the role of H2O2 in the immune system. Our data show that PPARγ-deficient CD11c+ cells from antigen-exposed mice have undetectable SOD activity and a lower H2O2 level. A peroxisome proliferator-response element (PPRE) was previously identified in the promoter of the CuZnSOD gene (Yoo et al., 1999). In addition, cardiac PPARγwas shown to be important for the expression of MnSOD (Ding et al., 2007). An increase in mitochondrial H2O2 level mediated by PPARγ-promoted SOD activity would result in increased cytosolic H2O2 level due to free diffusion of H2O2 across the mitochondrial membrane resulting in phosphorylation and ultimately degradation of IκBα. Interestingly, SOD inactivation has been associated with asthma pathophysiology (Comhair et al., 2005). Our study suggests that under ambient levels of exposure to environmental antigens, PPARγ-dependent increase in H2O2 serves a prophylactic function by blocking NF-κB activation and its nuclear translocation thereby preventing its recruitment to cytokine promoters. However, we propose that with the use of next generation TZDs with minimum side-effects, PPARγ can be also utilized therapeutically once inflammation is induced and NF-κB is recruited to target promoters where transrepression functions of PPARγ involving PPARγ SUMOylation can be exploited (Pascual et al., 2005). Taken together, diverse mechanisms can be utilized by PPARγ to exercise its anti-inflammatory effects, which may also depend on the specific ligand engaged by this nuclear receptor.
In summary, our study demonstrates a protective role of mitochondria in CD11c+ cells as a source of beneficial H2O2, the generation of which requires PPARγ. In various capacities, mitochondria are now considered master regulators of danger signaling (Galluzzi et al., 2012). While immune tolerance is desirable to blunt allergic and autoimmune diseases, it is an undesirable outcome in cancer that prevents anti-tumor immune responses. In general, oxidative stress is associated with deleterious consequences including tumorigenesis (Finkel, 2003). High levels of H2O2 are associated with many cancers (Lisanti et al., 2011), which based on the results of the present study, may promote Treg cells that impair anti-cancer immunity. Targeting mitochondria by specific small molecules (Murphy, 1997) to either promote or quench H2O2 production may provide therapeutic benefit to either blunt inflammatory diseases like asthma and various autoimmune diseases or to induce anti-tumor immunity.
Experimental Procedures
Mice
C57BL/6J (00664), PPARγfl/fl (004584) and CD11c-Cre (008068) mice were purchased from The Jackson Laboratory and were housed in the Department of Laboratory Animal Resources (DLAR) at the University of Pittsburgh. PPARγfl/fl and CD11c-Cre mice were bred to generate mice with deletion of PPARγ in CD11c+ cells (CD11c-Cre-PPARγfl/fl) and littermate controls (PPARγfl/fl) (Khare et al., 2015). CD11c-Cre mice were also used as controls and were found to be functionally similar to PPARγfl/fl mice (data not shown). OT-IIxThy1.1 transgenic mice, a gift from Dr. Lauren Cohn at Yale University (New Haven), were bred in the DLAR facility. All mice were housed under pathogen-free conditions and used between 6–12 weeks of age. All protocols involving animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pittsburgh.
Exposure of mice to antigens to induce tolerance and assessment of airway inflammation
The mice were exposed to aerosolized OVA for 3 consecutive days and lung cells were harvested 1hr after the last exposure, as shown in Figure 1A. For comparison of tolerance models, mice were tolerized by exposure to OVA for 10 consecutive days, as described before (Ostroukhova et al., 2004). In experiments where mice were exposed to the allergen Derp 1 of house dust mites, 20 µl of LoTox Natural Derp 1 protein (nDer p 1; Indoor Biotechnologies) resuspended in PBS at a concentration of 1 mg/ml was intranasally administered once for 3 consecutive days. For assessing the role of reactive oxygen species (ROS) in the induction of tolerance versus pro-inflammatory cytokines, mini-osmotic pumps (Alzet model 2001, 1 µL/hr, 7 days OR Alzet model 2002, 0.5µL/hr, 14 days), containing either Mito-TEMPO (0.7 mg/kg/day) or saline were subcutaneously inserted into WT mice under anesthesia. The mice were then subjected to aerosolized OVA for 20 min each day over a 3-day or a 10-day period (McMenamin et al., 1994; Ostroukhova et al., 2004). To test for induction of tolerance, the tolerized mice were subjected to an inflammation model as previously described by us involving intranasal administration of OVA along with the adjuvant cholera toxin (CT) (List Biochemicals) (Khare et al., 2015; Khare et al., 2013; Oriss et al., 2005). mice were challenged with aerosolized OVA daily for the next 7 days. The degree of airway inflammation and/or resistance to antigen challenge (tolerance) was assessed in mice 24 hr following completion of the protocol(s) outlined in the preceding section as described previously (Khare et al., 2013; Krishnamoorthy et al., 2012; Oriss et al., 2005).
Cell isolation and sorting
The lungs of euthanized mice were perfused with sterile PBS, dissected and enzymatically digested as described previously (Khare et al., 2015; Khare et al., 2013; Oriss et al., 2005). Lung cells were then physically dissociated on a gentleMACS Dissociator (Miltenyi Biotec) according to the manufacture’s protocol. Single cell suspensions were obtained by passing the dissociated tissue through a 70 µM cell strainer (BD Falcon). Magnetic separation utilizing anti-CD11c microbeads (Miltenyi Biotec) was used to enrich for CD11c+ cells. The lung CD11c+ cells were sorted into CD11c+ DCs (low auto-fluorescence) to greater than 90% purity using a FACSAria cell sorter (BD Immunocytometry Systems) for DC-T cell co-culture.
Antibodies and flow cytometry
Cell-surface markers were analyzed on several cell types using a variety of reagents. Cells were stained in 1X PBS containing 2% (wt/vol) FBS with anti-CD4-APC or PE-Cy7 (RM4–5; BD Biosciences), anti-CD11c-APC, PercP-Cy5.5, PE-Cy7 (HL3, BD Biosciences), anti-CD103-PE (M290; BD Biosciences), anti-CD11b–PE-Cy7, PE-CF594 (M1/70; BD Biosciences), anti-CD90.1-PE (OX-7; BD Biosciences), anti-MHCII FITC (NIMR4; Southern Biotech), anti-CD45-PE-Cy7 (30F11; Biolegend), anti-Siglec-F-PE, PE-CF594 (E50–2440, BD Biosciences) and anti-F4/80-Alexa fluor 647, PE (BM8, Invitrogen). Intracellular staining was performed according to the manufacturer’s instructions. Intracellular staining with anti-IL-17A-PE (eBio17B7; eBioscience), anti-IFN-γ-FITC (XMG1.2; BD Biosciences) was assessed in cells stimulated for a total of 6 hr with PMA (50 ng/ml; Sigma) and ionomycin (1 µg/ml; Sigma) in the presence of monensin (BD Biosciences) for the final 3 hr of incubation according to the manufacturer’s instructions (BD Biosciences). For intracellular cytokine staining in DCs, lung cells were cultured ex vivo in the presence of monensin alone for 5 hr and stained with anti-IL-23p19-efluor 660 (fc23cpg; ebiosciences) according to the manufacturer’s instructions. Stained cells were examined on a FACSCalibur and FACSAria flow cytometers (BD Immunocytometry Systems) and the data were analyzed using FlowJo software (Tree Star).
Induction of Foxp3+ CD4+ T cells in vivo
Treg-depleted CD4+ T (CD4+CD25−) cells were isolated from OT-IIxThy1.1 spleens as described before (Khare et al., 2015; Krishnamoorthy et al., 2012) and were adoptively transferred intravenously (106 cells per recipient) into CD11c-Cre-PPARγfl/fl (PPARγΔAPC) and PPARγfl/fl (WT) mice respectively. 24 hr following the adoptive transfer of cells, the recipient mice were exposed to OVA aerosols for 10 consecutive days and then were sacrificed one day following the last challenge. PBS-perfused lungs were harvested and single cell suspensions were prepared. In vivo induction of Foxp3 was assessed by staining for donor mouse strain-specific cell-surface marker anti-CD90.1 along with anti-CD4 and anti-Foxp3. CD4+ T cells positive for donor cell surface marker were selected for analysis of Foxp3 expression.
RNA isolation and quantitative or semi-quantitative RT-PCR
Sorted lung CD11c+ cells from CD11c-Cre-PPARγfl/fl and PPARγfl/fl mice were lysed using RLT plus Buffer (Qiagen) and RNA was extracted using an RNeasy Plus Micro kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized using a High Capacity cDNA Reverse Transcription Kit (Life Technologies) and was used as a template for quantitative PCR according to the manufacturer’s instructions (Life Technologies). Primer and probe sets for assaying individual gene expression were purchased from Life Technologies (Taqman Gene Expression Assays). qRT-PCR reactions were carried out using the ABI PRISM 7700 Sequence System (Applied Biosystems) at the Genomics Research Core at the University of Pittsburgh. Results were analyzed using the SDS 2.2.2 software. mRNA expression was calculated using the 2−ΔCt method using hprt1 as internal reference control.
PPAR target PCR array
Lung CD11c+ cells were sorted from naïve or tolerized (OVA-exposed) WT an PPARγΔAPC mice. CD11c+ cells isolated from naïve WT mice were treated with either vehicle or 10 µM Rosiglitazone (RSG) in vitro for 4 hr at 37°C. Total RNA was isolated as described above and subjected to cDNA synthesis using RT² First Strand Kit (Qiagen). Gene expression from four biological replicates was analyzed using RT² Profiler™ PCR Array Mouse PPAR Targets (Cat # PAMM-149Z, Qiagen) according to manufacturer’s instructions. Data analysis was carried out using web-based analysis software available on Qiagen’s website. Heat-map was generated using HeatMapImage module available at http://genepattern.broadinstitute.org/.
CFSE Proliferation Assay
Treg-depleted splenic CD4+ T cells from OT-II mice were labeled with CFSE (Vybrant CFDA-SE cell tracer kit; Invitrogen) as described previously (Hui, PNAS, 2008). Briefly, CD4+ CD25− T cells were labeled with 5 µM CFSE for 10 min at 37°C and co-cultured in presence of 1µg/mL OVA-peptide with CD11c+ DCs sorted from tolerized PPARγΔAPC and WT mice that were pre-treated for 1 hr at 37°C with either vehicle or 10µM BAY11 −7082 (EMD Milipore) at a DC:T cell ratio of 1:10. CD4+ T cell proliferation was assessed after 72 hr using flow cytometry. IL-2 level in culture supernatants was assessed by ELISA.
GC/MS analysis for free fatty acids
CD11c+ and CD11c− cells isolated from the lungs of naïve and tolerized WT an PPARγΔAPC mice were frozen at −80°C under inert condition and sent to Lipid Maps, Lipidomics Core (University of California, San Diego) for assaying concentrations of free fatty acid using GC/MS (Quehenberger et al., 2011). For the analysis of free fatty acids, the biological material was supplemented with a cocktail of deuterated internal fatty acid standards and extracted twice with 0.05 N methanolic HCl/isooctane (1:3, v/v) and the combined isooctane layers were evaporated to dryness. The extracted free fatty acids were derivatized with pentafluorobenzyl bromide and the fatty acid esters were analyzed by GC/MS on an Agilent 6890N gas chromatograph equipped with an Agilent 5973 mass selective detector (Agilent, Santa Clara, CA). Fatty acid quantitation was achieved by the stable isotope dilution method using concentration curves generated from fatty acid quantitative standards using identical conditions. The raw data obtained was then subjected to analysis and the unknowns compared to the standard curves generated and represented as pmol/106 cells.
Western Blot analysis
Total cell lysates and nuclear and cytoplasmic extracts were prepared from sorted and/or treated CD11c+ and CD11c− cells as described in text using lysis buffer (Cell signaling) and nuclear extraction kit (Panomics), respectively. Lysates (15 µg) were separated on 10% SDS-PAGE gels and then transferred onto PVDF membranes (Millipore). Nonspecific binding was blocked by incubation in blocking buffer (5% nonfat milk in TBST), followed by sequential incubations with the primary antibody and appropriate horseradish peroxidase-conjugated secondary antibody (Thermo Scientific), each diluted in blocking buffer. Immunoreactive protein was detected using enhanced chemiluminescence (ECL) reagents (Thermo Scientific). Antibodies against κBα (Santa Cruz biotech), phospho-IκBα, NF-κB subunit p65, c-rel, LSD1 (Cell Signaling), phospho IKKα/β, IKKα/β (Cell Signaling), MnSOD (EMD Millipore), Cu-ZnSOD (Enzo lifesciences) and β-actin (Cell signaling) were used for western blot analysis. Densitometric analysis was performed using the ImageJ software.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were done according to the manufacturer’s protocol (Pierce) with slight modifications. In brief, CD11c+ cells were sorted from the lungs of tolerize (3 day or 10 day models) WT and PPARγΔAPC mice were fixed with 1% formaldehyde, quenched using 1M glycine, lysed and chromatin digested using ChIP grade Micrococcal Nuclease for 15 minutes at 37°C. Each pull-down was performed using 0.3×106 cells with anti-PPARγ (Abcam) using Protein A/G beads. Beads were washed with high salt and lithium chloride-containing buffers. Chelex-100 Resin was added directly to the beads, the beads heated to 95°C for 10 minutes and incubated at 55°C with Proteinase K. Samples were heated again and supernatants collected were used for PCR with the following ChIP primers: IL-6 (−60bp) (forward 5’-CCCACCCTCCAACAAAGATT-3’ and reverse 5’-TGAGCTACAGACATCCCCAGT-3’), p19 (−153bp) (forward 5’-GGAACTGAAGCGGCATACCT-3’ and reverse 5’-AAGGTCCCTGCACTGTAAGG-3’), p19 (−123bp) (forward 5’-AGGTGATCTAAGAAGGTGGCT-3’ and reverse 5’-TTCGAAGTCTGGTTCCCGTG-3’). The primers and the respective binding sites were designed and validated using Cold Spring softwares NCBI Primer Blast and Match-1.0.
Chymotrypsin- and Trypsin-like protease activity
Lung CD11c+ cells were sorted from 3 day tolerized WT and PPARγΔAPC mice and chymotrypsin-like and trypsin-like activities associated with the proteasome complex was measured from 10,000 cells/well using a homogeneous, luminescent Proteasome-Glo™ Cell-Based Assay (Promega) according to the manufacturer’s instructions. The plates were read on Veritas microplate luminometer (Turner Biosystems).
Superoxide dismutase (SOD) activity
Extracts from lung CD11c+ cells sorted from tolerized WT and PPARγΔAPC mice were prepared and assayed for SOD activity using a colorimetric Superoxide Dismutase activity kit (Cayman Chemicals) according to the manufacturer’s instructions.
Detection of Reactive oxygen species (ROS)
Intracellular H2O2 level in CD11c+ cells from naïve in the presence or absence of RSG (1 h at 37°C), tolerized WT and PPARγΔAPC mice (1 hr after the last exposure to inhaled allergen) in the presence or absence of rotenone, L-NAME or antimycin A (Sigma) and from WT mice treated with either saline or Mito-TEMPO, administered through mini-osmotic pumps (naïve and OVA-tolerized) was assessed by Amplex Red assay. For intracellular detection of H2O2, lung CD11c+ cells isolated from tolerized WT and PPARγΔAPC mice were incubated in the presence or absence of 10 µM PF6-AM (Adooq Bioscience) with or without antigen (10 µg/ml OVA (or 1 µg/ml nDer p1) for 40 min at 37 C. After washing to remove OVA and excess PF6-AM, cells were fixed with 4% paraformaldehyde (PFA) at 4° C for 15 mins. For positive control, PF6-AM-loaded cells were exposed to 100 µM H2O2 for 15 mins before fixing with PFA. To ascertain specificity of PF6-AM for H2O2, 10 µM Catalase was added to cells along with exogenous H2O2. Fixed cells were cytospun onto clean glass slides and processed for imaging. Thresholds were initially set for both WT and PPARγ-deficient cells to minimize non-specific background signal due to autofluorescence. The images were captured using Olympus Fluoview 1000 confocal microscope with a 20X objective and quantification was performed using Nikon NIS-Elements software based on uniformly set intensity threshold and normalized per nucleus.
Complex I and III assay activity
Complex I and III assays were performed in CD11c+ cells lysed by three cycles of freeze/thaw as previously described (Shiva et al., 2007). Briefly, Complex I activity was measured by spectrophotometrically monitoring the rotenone-sensitive oxidation of NADH at 340nm. Complex III activity was assessed by spectrophotometrically monitoring the reduction of cytochrome c (550nm) in the presence of ubiquinol.
DM NQ treatment
Lung CD11c+ cells were sorted from tolerized WT and PPARγΔAPC mice and were treated ex vivo with 200 µM DMNQ (2,3-Dimethoxy-1,4-naphthoquinone; Enzo life sciences) for 1 hr at 37°C. Effect on intracellular H2O2 and IκB levels were assessed as described above.
Measurement of oxygen consumption rate
Oxygen consumption rate (OCR) of CD11c+ cells isolated from tolerized WT an PPARγΔAPC mice was measured at the basal level. One batch of cells in each group was treated with 200 µM Etomoxir (Eto) and thereafter with 1 µM oligomycin (Oligo), 1.5 µM FCCP, and 0.1 µM rotenone plus 1 µM antimycin (Ant A+Rot) using the XF-96 Seahorse system. Cells were plated in triplicate. Seahorse assays were performed in unbuffered DMEM media supplemented with 2.5 mM glucose and 1 mM glutamine.
Mitochondrial DNA estimation assay
To determine the ratio of mitochondrial DNA to nuclear DNA in CD11c+ cell from naïve and tolerized WT and PPARγΔAPC mice, 2−ΔΔCt was calculated using difference in Ct values of nucleus-encoded h19 gene and mitochondria-encoded nd1 for each individual sample.
Assessment of mitochondrial morphology and volume
Formalin-fixed CD11c+ cells from naïve and tolerized (OVA and Derp1) WT and PPARγΔAPC mice were stained using anti-Tom20 (Santa Cruz, SC-11416) to label mitochondria. Cells were co-stained with phalloidin-cy3 (to stain F-actin,; Life Technologies, R415) and DAPI (to stain nuclei; Southern Biotech). Confocal Z-stacks were collected using a 60X (1.43NA) optic on a Nikon A1 equipped with GASP detectors and NIS Elements software (Nikon Inc). The confocal datasets were deconvolved using the 3D Landweber capabilities of Nikon Elements (Nikon Inc) and then imported into Imaris (version) software (Bitplane) for surface rendering and calculation of mitochondrial volumes. Mitochondrial fragmentation was assessed using the sphericity parameter (defined as the ratio of the surface area of the given object to the surface area of a sphere with the same volume as the given object).
Statistical analyses
Student’s unpaired two-tailed t-test was used for comparisons between two groups. One-way ANOVA with Tukey’s post hoc test was used for comparisons between multiple groups, when appropriate. Differences between groups were considered significant when p<0.05. All statistical analyses were performed using Prism 5 software (GraphPad Prism software).
Supplementary Material
Acknowledgments
We thank Dr. Oswald Quehenberger at LIPID MAPS, University of California San Diego, for his expert help and advice in the analysis of free fatty acids in lung cells, and Morgan Jessup for quantitation of H2O2 by imaging. This work was supported by US National Institutes of Health grants AI048927 and AI106684 (to A.R.), HL113956 (to A.R. and P.R.), and HL114453, AI100012 and HL122307 (to P.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
A.K. designed and performed experiments, analyzed data and wrote the manuscript. M.R. designed and performed PCR target array, mitochondrial bioenergetics assays, mtDNA estimation, ELISA and analyzed data, K.C. designed and performed immunoblotting experiments, ChIP assays and analyzed data, S.D. performed confocal microscopy and analyzed data, C.C carried out OCR measurements and analyzed data, C.K. performed amplex red assays and analyzed data, K.Q. performed confocal microscopy and analyzed data, C.S-C. and S.W. assisted with assessment of mitochondrial morphology, C.M. assisted with experiments involving osmotic pump installations and animal surgeries, and analyzed data. A.K. and T.B.O performed flow sorting, R.Huff performed animal surgeries. R.H. analyzed lung histology, P.R. designed experiments and analyzed data. S.S designed ROS-related experiments and analyzed data. A.R. conceived the study, designed experiments, analyzed data and wrote the manuscript.
The authors declare no competing financial interests.
References
- Basu S, Rajakaruna S, Dickinson BC, Chang CJ, Menko AS. Endogenous hydrogen peroxide production in the epithelium of the developing embryonic lens. Mol Vis. 2014;20:458–467. [PMC free article] [PubMed] [Google Scholar]
- Belvisi MG, Hele DJ, Birrell MA. Peroxisome proliferator-activated receptor gamma agonists as therapy for chronic airway inflammation. Eur J Pharmacol. 2006;533:101–109. doi: 10.1016/j.ejphar.2005.12.048. [DOI] [PubMed] [Google Scholar]
- Beraud C, Henzel WJ, Baeuerle PA. Involvement of regulatory and catalytic subunits of phosphoinositide 3-kinase in NF-kappaB activation. Proc Natl Acad Sci U S A. 1999;96:429–434. doi: 10.1073/pnas.96.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD. The sites and topology of mitochondrial superoxide production. Exp Gerontol. 2010;45:466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner GR. Superoxide dismutase in redox biology: the roles of superoxide and hydrogen peroxide. Anticancer Agents Med Chem. 2011;11:341–346. doi: 10.2174/187152011795677544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cates EC, Fattouh R, Wattie J, Inman MD, Goncharova S, Coyle AJ, Gutierrez-Ramos JC, Jordana M. Intranasal exposure of mice to house dust mite elicits allergic airway inflammation via a GM-CSF-mediated mechanism. J. Immunol. 2004;173:6384–6392. doi: 10.4049/jimmunol.173.10.6384. [DOI] [PubMed] [Google Scholar]
- Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- Comhair SA, Xu W, Ghosh S, Thunnissen FB, Almasan A, Calhoun WJ, Janocha AJ, Zheng L, Hazen SL, Erzurum SC. Superoxide dismutase inactivation in pathophysiology of asthmatic airway remodeling and reactivity. Am J Pathol. 2005;166:663–674. doi: 10.1016/S0002-9440(10)62288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curotto de Lafaille MA, Kutchukhidze N, Shen S, Ding Y, Yee H, Lafaille JJ. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29:114–126. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- da Silva AF, Mariotti FR, Maximo V, Campello S. Mitochondria dynamism: of shape, transport and cell migration. Cell Mol Life Sci. 2014;71:2313–2324. doi: 10.1007/s00018-014-1557-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson BC, Peltier J, Stone D, Schaffer DV, Chang CJ. Nox2 redox signaling maintains essential cell populations in the brain. Nat Chem Biol. 2011;7:106–112. doi: 10.1038/nchembio.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikalova AE, Bikineyeva AT, Budzyn K, Nazarewicz RR, McCann L, Lewis W, Harrison DG, Dikalov SI. Therapeutic targeting of mitochondrial superoxide in hypertension. Circ Res. 2010;107:106–116. doi: 10.1161/CIRCRESAHA.109.214601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Fu M, Qin Q, Lewis W, Kim HW, Fukai T, Bacanamwo M, Chen YE, Schneider MD, Mangelsdorf DJ, et al. Cardiac peroxisome proliferator-activated receptor gamma is essential in protecting cardiomyocytes from oxidative damage. Cardiovasc Res. 2007;76:269–279. doi: 10.1016/j.cardiores.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- Forman HJ, Kennedy JA. Role of superoxide radical in mitochondrial dehydrogenase reactions. Biochem Biophys Res Commun. 1974;60:1044–1050. doi: 10.1016/0006-291x(74)90418-5. [DOI] [PubMed] [Google Scholar]
- Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol. 2012;13:780–788. doi: 10.1038/nrm3479. [DOI] [PubMed] [Google Scholar]
- Guillou H, Zadravec D, Martin PG, Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog Lipid Res. 2010;49:186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Guth AM, Janssen WJ, Bosio CM, Crouch EC, Henson PM, Dow SW. Lung environment determines unique phenotype of alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2009;296:L936–L946. doi: 10.1152/ajplung.90625.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- James AM, Collins Y, Logan A, Murphy MP. Mitochondrial oxidative stress and the metabolic syndrome. Trends Endocrinol Metab. 2012;23:429–434. doi: 10.1016/j.tem.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Kalyanaraman B, Darley-Usmar V, Davies KJ, Dennery PA, Forman HJ, Grisham MB, Mann GE, Moore K, Roberts LJ, 2nd, Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med. 2012;52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Katano-Toki A, Satoh T, Tomaru T, Yoshino S, Ishizuka T, Ishii S, Ozawa A, Shibusawa N, Tsuchiya T, Saito T, et al. THRAP3 interacts with HELZ2 and plays a novel role in adipocyte differentiation. Mol Endocrinol. 2013;27:769–780. doi: 10.1210/me.2012-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare A, Chakraborty K, Raundhal M, Ray P, Ray A. Cutting Edge:Dual Function of PPARγ in CD11c+ Cells Ensures Immune Tolerance in the Airways. J. Immunol. 2015;195:431–435. doi: 10.4049/jimmunol.1500474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare A, Krishnamoorthy N, Oriss TB, Fei M, Ray P, Ray A. Cutting edge: inhaled antigen upregulates retinaldehyde dehydrogenase in lung CD103+ but not plasmacytoid dendritic cells to induce Foxp3 de novo in CD4+ T cells and promote airway tolerance. J. Immunol. 2013;191:25–29. doi: 10.4049/jimmunol.1300193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn SH, Wouters EF, Vos N, Janssen-Heininger YM. Cytokine-induced activation of nuclear factor-kappa B is inhibited by hydrogen peroxide through oxidative inactivation of IkappaB kinase. J Biol Chem. 2001;276:35693–35700. doi: 10.1074/jbc.M104321200. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy N, Khare A, Oriss TB, Raundhal M, Morse C, Yarlagadda M, Wenzel SE, Moore ML, Peebles RS, Ray A, Ray P. Early infection with respiratory syncytial virus impairs regulatory T cell function and increases susceptibility to allergic asthma. Nat. Med. 2012;18:1525–1530. doi: 10.1038/nm.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht BN, Hammad H. Lung dendritic cells in respiratory viral infection and asthma: from protection to immunopathology. Annu. Rev. Immunol. 2012;30:243–270. doi: 10.1146/annurev-immunol-020711-075021. [DOI] [PubMed] [Google Scholar]
- Lin VS, Dickinson BC, Chang CJ. Boronate-based fluorescent probes: imaging hydrogen peroxide in living systems. Methods Enzymol. 2013;526:19–43. doi: 10.1016/B978-0-12-405883-5.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti MP, Martinez-Outschoorn UE, Lin Z, Pavlides S, Whitaker-Menezes D, Pestell RG, Howell A, Sotgia F. Hydrogen peroxide fuels aging, inflammation, cancer metabolism and metastasis: the seed and soil also needs “fertilizer”. Cell Cycle. 2011;10:2440–2449. doi: 10.4161/cc.10.15.16870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi A, Grasso P, Moreno M, de Lange P, Silvestri E, Lanni A, Goglia F. Interrelated influence of superoxides and free fatty acids over mitochondrial uncoupling in skeletal muscle. Biochim Biophys Acta. 2008;1777:826–833. doi: 10.1016/j.bbabio.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Loschen G, Azzi A, Richter C, Flohe L. Superoxide radicals as precursors of mitochondrial hydrogen peroxide. FEBS Lett. 1974;42:68–72. doi: 10.1016/0014-5793(74)80281-4. [DOI] [PubMed] [Google Scholar]
- McMenamin C, Pimm C, McKersey M, Holt PG. Regulation of IgE responses to inhaled antigen in mice by antigen- specific gamma delta T cells. Science. 1994;265:1869–1871. doi: 10.1126/science.7916481. [DOI] [PubMed] [Google Scholar]
- Murphy MP. Selective targeting of bioactive compounds to mitochondria. Trends Biotechnol. 1997;15:326–330. doi: 10.1016/S0167-7799(97)01068-8. [DOI] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nethery D, Callahan LA, Stofan D, Mattera R, DiMarco A, Supinski G. PLA(2) dependence of diaphragm mitochondrial formation of reactive oxygen species. J Appl Physiol (1985) 2000;89:72–80. doi: 10.1152/jappl.2000.89.1.72. [DOI] [PubMed] [Google Scholar]
- Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, Chawla A. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Marques V, Marinho HS, Cyrne L, Antunes F. Role of hydrogen peroxide in NF-kappaB activation: from inducer to modulator. Antioxid Redox Signal. 2009;11:2223–2243. doi: 10.1089/ars.2009.2601. [DOI] [PubMed] [Google Scholar]
- Oriss TB, Ostroukhova M, Seguin-Devaux C, Dixon-McCarthy B, Stolz DB, Watkins SC, Pillemer B, Ray P, Ray A. Dynamics of Dendritic Cell Phenotype and Interactions with CD4+ T Cells in Airway Inflammation and Tolerance. J. Immunol. 2005;174:854–863. doi: 10.4049/jimmunol.174.2.854. [DOI] [PubMed] [Google Scholar]
- Ostroukhova M, Seguin-Devaux C, Oriss TB, Dixon-McCarthy B, Yang L, Ameredes BT, Corcoran TE, Ray A. Tolerance induced by inhaled antigen involves CD4(+) T cells expressing membrane-bound TGF-beta and FOXP3. J. Clin. Invest. 2004;114:28–38. doi: 10.1172/JCI20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry JD, Pointon AV, Lutz U, Teichert F, Charlwood JK, Chan PH, Athersuch TJ, Taylor EL, Singh R, Luo J, et al. Pivotal role for two electron reduction in 2,3-dimethoxy-1,4-naphthoquinone and 2-methyl-1,4-naphthoquinone metabolism and kinetics in vivo that prevents liver redox stress. Chem Res Toxicol. 2009;22:717–725. doi: 10.1021/tx800472z. [DOI] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat Rev Immunol. 2009;9:778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- Poole DC, Sexton WL, Farkas GA, Powers SK, Reid MB. Diaphragm structure and function in health and disease. Med Sci Sports Exerc. 1997;29:738–754. doi: 10.1097/00005768-199706000-00003. [DOI] [PubMed] [Google Scholar]
- Quehenberger O, Armando AM, Dennis EA. High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim Biophys Acta. 2011;1811:648–656. doi: 10.1016/j.bbalip.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Nobs SP, Kurrer M, Rehrauer H, Thiele C, Kopf M. Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol. 2014;15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiva S, Sack MN, Greer JJ, Duranski M, Ringwood LA, Burwell L, Wang X, MacArthur PH, Shoja A, Raghavachari N, et al. Nitrite augments tolerance to ischemia/reperfusion injury via the modulation of mitochondrial electron transfer. J Exp Med. 2007;204:2089–2102. doi: 10.1084/jem.20070198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM. Decisions about dendritic cells: past, present, and future. Annu. Rev. Immunol. 2012;30:1–22. doi: 10.1146/annurev-immunol-100311-102839. [DOI] [PubMed] [Google Scholar]
- Striz I, Wang YM, Svarcova I, Trnka L, Sorg C, Costabel U. The phenotype of alveolar macrophages and its correlation with immune cells in bronchoalveolar lavage. Eur Respir J. 1993;6:1287–1294. [PubMed] [Google Scholar]
- Tschopp J. Mitochondria: Sovereign of inflammation. Eur J Immunol. 2011;41:1196–1202. doi: 10.1002/eji.201141436. [DOI] [PubMed] [Google Scholar]
- Wahli W, Michalik L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol Metab. 2012;23:351–363. doi: 10.1016/j.tem.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Wakil SJ, Abu-Elheiga LA. Fatty acid metabolism: target for metabolic syndrome. J Lipid Res. 2009;(50 Suppl):S138–S143. doi: 10.1194/jlr.R800079-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf HP, Brenner KV. The effect of etomoxir on glucose turnover and recycling with [1–14C], [3–3H]-glucose tracer in pigs. Horm Metab Res. 1988;20:204–207. doi: 10.1055/s-2007-1010794. [DOI] [PubMed] [Google Scholar]
- Xia Y, Dawson VL, Dawson TM, Snyder SH, Zweier JL. Nitric oxide synthase generates superoxide and nitric oxide in arginine-depleted cells leading to peroxynitrite-mediated cellular injury. Proc Natl Acad Sci U S A. 1996;93:6770–6774. doi: 10.1073/pnas.93.13.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Chang MS, Rho HM. Induction of the rat Cu/Zn superoxide dismutase gene through the peroxisome proliferator-responsive element by arachidonic acid. Gene. 1999;234:87–91. doi: 10.1016/s0378-1119(99)00176-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.