Abstract
The past decade of cancer research has ushered in a comprehensive understanding of the way that the sequence of the genome can be coopted during the process of tumorigenesis. However, only recently has the epigenome, and in particular the three-dimensional topology of chromatin, been implicated in cancer progression. Here we review recent findings of how the cancer genome is regulated and dysregulated to effect changes in 3D genome topology. We discuss the impact of the spatial organization of the genome on the frequency of tumorigenic chromosomal translocations and the effects of disruption of the proteins responsible for the establishment of chromatin loops. Alteration of the three-dimensional cancer genome is a rapidly emerging hallmark of multiple cancer subtypes.
Keywords: cancer, chromatin, epigenetics, 3D architecture, CTCF, cohesin
INTRODUCTION
Cancer is largely associated with the sequential acquisition of mutations in a single lineage of cells that ultimately leads to unrestrained proliferation. The foundations of cancer biology were laid through the discovery of oncogenes and tumor suppressors with canonical roles in proliferation and cell cycle control. Decades of research have elucidated the major drivers and the genetic mechanisms responsible for tumorigenesis, identifying point mutations and small-scale alterations that directly affect individual proteins in a one-dimensional fashion. The contribution of the epigenome to this process has become more apparent with the discovery of mutations in genes known to regulate DNA methylation and histone modification. These mutations are largely considered to affect gene expression in a two-dimensional fashion through the modulation of transcription factor recruitment. However, the precise mechanisms by which these alterations of the epigenome contribute to cancer progression has remained elusive. Only recently has the three-dimensional context of the genome been identified as a major player in the development and progression of cancer [1–4].
The genomes of higher eukaryotes are packaged into exquisitely organized hierarchical structures. Linear DNA is wrapped around histone proteins forming the 10 nm nucleosomal fiber which is subsequently folded in three-dimensions to create loops of DNA that form discrete neighborhoods of genes at the sub-megabase level [5,6]. These neighborhoods are formed through the action of multiple proteins including CCCTC-binding factor (CTCF) and the cohesin complex [7,8]. Groups of gene neighborhoods are further organized into large, isolated, megabase structures termed topologically associating domains (TADs) [9–13]. Each of these layers of organization have pronounced effects on gene expression and the control of cell identity and cell fate. The mechanisms by which these three-dimensional interactions are manipulated and coopted in the context of cancer are the subject of this review (Figure 1).
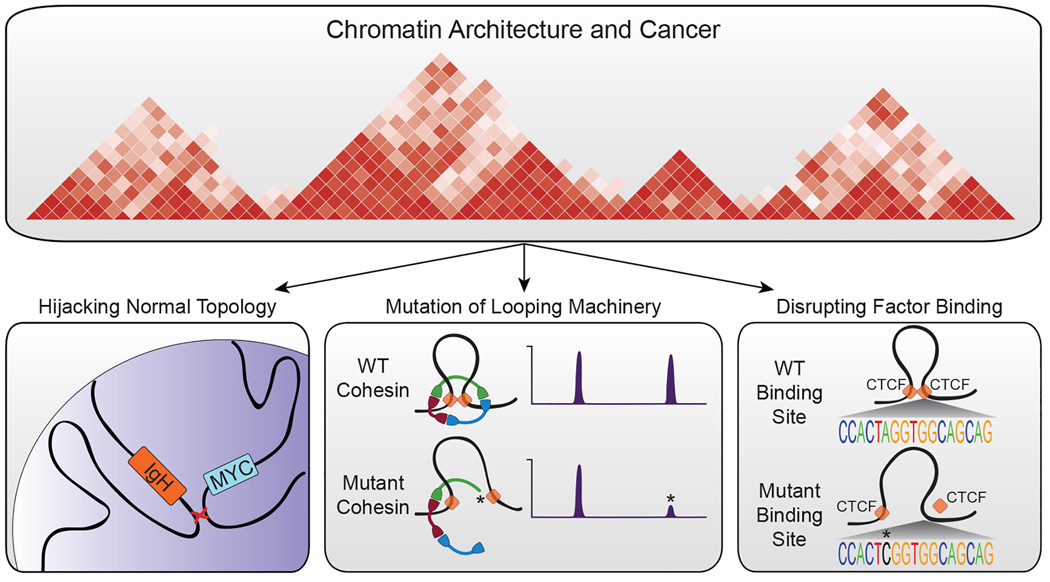
Figure 1. Contribution of chromatin architecture to our understanding of cancer.
Chromatin architecture has been implicated in the pathogenesis of cancer through multiple lines of evidence. The normal topology of the genome has been shown to predispose certain cell types to the acquisition of certain chromosomal translocations such as the MYC/IgH translocations associated with certain types of lymphoma (left panel). The genetic mutation of components involved in chromatin looping, including the cohesin complex (red/green/blue ring) and CTCF (orange squares), has also been observed in many cancer types (center panel). These mutations (illustrated by *) likely cause quantitative changes in factor binding, illustrated here by a change in ChIP-seq signal. Lastly, disruption of regulatory regions that serve as the anchors for looping machinery has been identified in cancer and multiple other diseases (right panel).
Hijacking topology: the contribution of normal DNA architecture to carcinogenesis
In the absence of alteration of the topological structure of the three-dimensional genome, recent work has demonstrated that the normal organization of the genome predisposes certain cell types to the acquisition of specific cancerous lesions. Chromosomal rearrangements, such as translocations, require the formation and incorrect resolution of DNA double strand breaks. Higher order chromatin structure has been shown to play a role in the formation of specific translocations through the spatial coordination of otherwise unrelated DNA sequences. Through the development of sequencing technologies to capture translocation junctions in B lymphocytes, multiple groups have shown that translocations between pairs of DSBs occurring on the same chromosome are strongly preferred over interchromosomal events [1,2]. The constrained physical proximity of intrachromosomal interactions implies that spatial organization of chromosomes influences the translocation process. In line with this hypothesis, modeling of somatic copy-number alterations and genome-wide chromosome conformation capture suggests that the distribution of chromosomal alterations is spatially related to three-dimensional genomic architecture [14]. These results were confirmed by direct comparisons of translocation frequency and spatial proximity of interchromosomal interactions [15,16]. Cumulatively, these studies show that the three-dimensional proximity between two loci is directly proportional to the likelihood of translocation. When combined with a requirement for positive selection in cancer cells, these observations explain the frequency of recurrent translocations such as BCR-ABL and MYC-IGH.
Mutation or genetic alteration of genome organization components
In addition to serving as a template for large-scale chromosomal aberrations, the three-dimensional genomic architecture is often perturbed in cancer through genetic alteration of the proteins involved in the establishment and maintenance of chromatin interactions. In particular, the cohesin complex, a multimeric ring structure involved in mediating looping interactions, has been found to be mutated in a wide variety of cancers. In addition to participating in three-dimensional looping, the cohesin complex is also involved in sister chromatid segregation during mitosis. Given the well-established role for aneuploidy in cancer, it has been hypothesized that mutation of the cohesin complex would contribute to carcinogenesis through the mis-segregation of chromosomes [17,18]. While this may be true in some cases, recent work has made it clear that genetic disruption of the cohesin complex plays a much more subtle role in cancer formation[19–22].
The first evidence that genetic mutation of the cohesin complex may not result in aneuploidy came from the genetic characterization of acute myeloid leukemia (AML), an aggressive malignancy of the bone marrow. AML is one of the most genetically stable adult cancers with minimal aneuploidy and an average of 10–15 coding mutations per patient [23,24]. Cohesin complex mutations were first described in AML in 2012, occurring in approximately 13% of patients [25–27]. These mutations occur in all four members of the cohesin complex (STAG2, SMC3, SMC1A, and RAD21) and are typically missense or truncating mutations. The spectrum of mutations observed implies a loss of function mechanism which is consistent with the finding that cohesin mutated leukemia cells have reduced levels of chromatin-bound cohesin components [3]. Recent work has demonstrated that mutations in the cohesin complex impair hematopoietic progenitor differentiation [19–21], suggesting a clear mechanism by which the cohesin complex may play a role in cancer progression in the absence of aneuploidy.
In addition to AML, the cohesin complex has been found to be mutated in multiple other types of cancer. Most prominently, mutations in STAG2 occur in 20–30% of urothelial bladder carcinoma and are not associated with aneuploidy [22,28–30]. Mutations in the cohesin complex have also been found in glioblastoma [31], medulloblastoma [32], breast cancer [33], pancreatic ductal adenocarcinoma [34], and Ewing sarcoma [35].
Beyond the cohesin complex, recurrent mutations have been observed in CTCF and other cohesin-interacting proteins. The first report of CTCF missense mutations in cancer identified multiple zinc finger domain mutations in breast, prostate, and Wilms’ tumors [36]. These missense mutations each selectively altered CTCF binding to a subset of target sites but did not completely abrogate DNA binding by CTCF. These results imply that selective alteration of chromatin architecture, perhaps in a cell type-specific manner, can play a causative role in cancer development. Notably, point mutations and copy number loss of CTCF are commonly observed in breast cancer [37], prostate cancer [37], and endometrial cancer [38], implicating a haploinsufficient phenotype for CTCF. Indeed, mouse models of CTCF haploinsufficiency indicate a strong predisposition to cancer with 80% of Ctcf heterozygous knockout mice succumbing to cancer by 100 weeks of age compared to only 40% of wildtype littermates. This 50% reduction in Ctcf gene dosage has profound effects on DNA methylation, suggesting a role for CTCF in maintaining the stability of global cytosine methylation [39].
Despite the abundance of cancerous mutations detected in the genes known to regulate chromatin topology, the precise mechanism of action of these mutations remains elusive. Many studies have addressed the consequence of loss of cohesin or CTCF via knockdown or knockout in post-mitotic cells showing widespread disruption of long-range interactions and concomitant changes in the expression of nearby genes [40–44]. However, cancer-associated mutations in these proteins are often heterozygous with the mutated allele expressed, indicating that a reduction in wildtype protein levels by knockdown or knockout may not phenocopy a heterozygous mutation. Future work investigating the consequences of cohesin complex or CTCF mutation on three-dimensional chromatin architecture of cancer cells will provide key insights into the mechanism of action of these mutations. It remains unclear how these mutations confer a carcinogenic phenotype and whether all mutations in genes regulating the three-dimensional genome have the same mechanistic effect.
Genetic and epigenetic dysregulation of chromatin architecture
In the absence of direct disruption of chromatin organizers, genetic or epigenetic dysregulation of the non-coding genome can have profound effects on chromatin architecture. In particular, changes in the sequence or epigenetic milieu of transcription factor binding sites can lead to alterations in chromatin interactions which have broad-reaching effects on gene expression and cellular identity. Greater than 95% of genome-wide association study-identified SNPs are located in intergenic regions and more than 75% associated with DNase I-hypersensitive sites, indicating a strong link to regulatory elements [45]. Additional studies have linked these disease-associated polymorphisms in non-genic regions with regulatory elements involved in chromatin organization and looping [46,47]. Similarly, recent work studying colorectal cancer (CRC) has shown that certain genetic subtypes of CRC are characterized by a predominance of mutations at CTCF binding sites [48]. This enrichment for mutations at CTCF binding sites was only observed in the context of simultaneous cohesin binding, implicating a specific subset of CTCF binding sites in the pathogenesis of CRC. Moreover, these CTCF binding site mutations were highly associated with AT>GC mutations, previously identified as a unique mutational signature in cancer [49], and are enriched at specific positions in the CTCF consensus sequence. Importantly, as few as two SNPs in a CTCF binding site lead to complete abrogation of CTCF binding [50]. Across multiple patient samples, CTCF binding site mutations display a unimodal distribution whereby a small number of patients account for a majority of the mutations in CTCF binding sites. On a more global scale, CTCF binding site mutations are coupled to late replication timing domains and previous studies have shown that these CTCF/cohesin binding sites are not replicated by the leading strand DNA polymerase Pol ε but by another uncharacterized polymerase [44]. Taken together, these results imply that a subset of CRC patients may have global defects in the repair of mutations in CTCF binding sites. Whether these mutations are causative of or merely correlated with cancer progression remains to be shown. Importantly, this mutational signature was not unique to CRC and was observed in multiple other cancer types, suggesting a more universal role for dysregulation of CTCF binding in the pathogenesis of cancer.
CTCF binding patterns can also be influenced by epigenetic modification of its binding sites. DNA methylation of the CTCF consensus binding sequence has been shown to control cell type-specific CTCF binding [51,52], indicating that CTCF occupancy can be readily modulated by reversible epigenetic alterations. As disruption of DNA methylation is a hallmark of multiple types of cancer [53–55], it is possible that changes in DNA methylation directly or indirectly affect CTCF binding. One study of IDH mutant glioma has linked hypermethylation of CTCF binding sites to dissolution of important domain boundaries and aberrant expression of powerful oncogenes [4]. Future, more in-depth studies of CTCF binding in cancer subtypes with altered DNA methylation will help to answer these questions and elucidate other potential mechanisms by which chromatin architecture is disrupted in cancer.
How could changes in chromatin architecture mechanistically lead to cancer?
While recent years of research have enumerated multiple examples of how chromatin architecture is dysregulated in cancer, no studies have provided direct mechanistic insight into how this dysregulation is translated into phenotypes associated with cancer. Multiple models can be posited but no direct evidence exists to distinguish these possibilities (Figure 2).
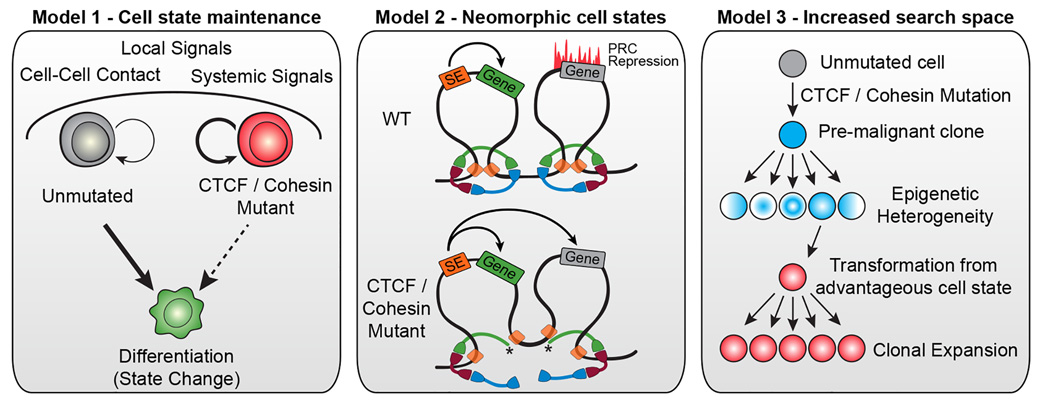
Figure 2. Models for the mechanistic underpinnings of how disruption of chromatin architecture can contribute to cancer progression.
Disruption of the chromatin architecture machinery may abrogate a cell’s ability to establish new chromatin loops and thus inhibit it from changing cellular state (left panel). Alternatively, changes in chromatin domains caused by dysregulation of looping proteins (red/green/blue rings represent the cohesin complex; orange squares represent CTCF) could lead to neomorphic transcriptional states which could be tumorigenic (center panel). In this example, truncation of one member of the cohesin complex (*) leads to dissolution of stabilized loops and inappropriate interaction of a super-enhancer with a previously silent gene. Lastly, pseudo-random cell states could be created through the inactivation of chromatin architecture machinery leading to an increased epigenetic search space. Some of these epigenetic configurations may be more advantageous or may facilitate transformation (right).
Model 1 - Dysregulation of chromatin architecture prevents cell state changes
Mutation of components of the chromatin organization machinery such as CTCF and cohesin may not create new functional states but may, instead, prevent cells from changing states. In the context of cancer, acquisition of a CTCF or cohesin mutation in a stem cell would prevent that cell from differentiation which would increase the likelihood of acquiring additional mutations and potentially bestow a self-renewal phenotype to progeny cells. Indirect evidence supporting this model exists in AML [19–21] whereby mutations in the cohesin complex members lead to defects in differentiation. Mechanistically, this model may be possible through the action of pioneer factors. In the absence of proper CTCF or cohesin function, global chromatin accessibility decreases [44], implying that the only transcription factors capable of binding DNA may be pioneer factors that can bind condensed chromatin. In the context of a stem cell that acquires a mutation in CTCF or cohesin, the expressed pioneer factors would control a stem cell state and would perpetuate that state in the absence of additional changes to the chromatin architecture.
Model 2 - Inappropriate 3D looping and insulation alter the cis-regulation of key genes and create neomorphic cell states
Disruption of the components maintaining chromatin architecture may lead to novel combinations of expressed and repressed genes and contribute to cancer development through the generation of neomorphic cell states. In depth studies of the binding sites and interactions of CTCF and cohesin in embryonic stem cells has shown that super enhancer-associated genes with important functions for cell identity exist in insulated neighborhoods created by looping interactions between two CTCF/cohesin binding sites [56]. Additionally, repressed lineage-specifying developmental regulators are also found in separate insulated neighborhoods. The integrity of these activational and repressive insulated neighborhoods is critical for the proper expression and repression of nearby genes that exist on the outskirts of these neighborhoods. Disruption of the neighborhood boundaries by mutation of CTCF binding sites or by dysregulation of cohesin or CTCF through mutational inactivation would lead to inappropriate expression and/or repression of key developmental genes [56] which could create unnatural cell states that have the potential to cause cancer. One example supporting this model has been demonstrated in glioblastoma whereby hypermethylation of CTCF binding sites leads to reduced CTCF binding at specific domain boundaries. In this study, the loss of one such boundary enables a constitutive enhancer to interact aberrantly with the PDGFRA gene, a prominent oncogene in glioma [4].
Model 3 - Disruption of chromatin architecture increases the epigenetic search space probed by cells and increases the likelihood of developing cancer
The loss of proper DNA repair leads to the acquisition of many more mutations, thus increasing the mutational search space of a given cell [57]. Similarly, disruption of the chromatin architecture of a cell may have pseudo-random effects on gene expression and repression. New chromatin loops may be established and old loops destroyed due to the random nature of whether a functional or non-functional CTCF or cohesin protein is recruited to a given chromosomal location. This increases the epigenetic search space of these cells as they probe pseudo-random chromatin configurations until acquiring an evolutionarily advantageous cell state that can be secured through positive selection. In this way, dysregulation of chromatin architecture may contribute to cancer by increasing epigenetic variability.
Ultimately, these models attribute the consequences of topological alterations to changes in gene expression. It has been well established that changes in gene expression can lead to increased proliferation and decreased differentiation, two key hallmarks of cancer [58]. Moreover, a causative link between changes in chromatin topology and changes in gene expression has been established through multiple lines of evidence [4,59].
DISCUSSION
The intricate interplay between chromatin architecture and cell identity has been extensively explored in the context of healthy cells. However, the involvement of three-dimensional chromatin organization in the pathogenesis of cancer has only recently been acknowledged. Work elucidating the interplay of spatial proximity and frequency of chromosomal translocations has enhanced our understanding of how the natural organization of chromatin can be co-opted to generate recurrent translocations directly responsible for cancer progression. Moreover, high-throughput sequencing efforts have identified mutations in the genes encoding for components of the chromatin organization machinery, such as CTCF and cohesin, as well as mutations in the sites bound by these factors. These genetic studies have shown that dysregulation of chromatin architecture may be a central hallmark of tumorigenesis in multiple cancer types. Nevertheless, it is still unclear precisely how, for example, a mutation in the cohesin complex affects chromatin organization and contributes to the pathogenesis of cancer. While it is known that programmed changes in genome topology occur during the normal process of differentiation [5,12,60,61], little is known about how genome organization changes in the setting of cancer. Recent work has shown that tumor cells exhibit a similar overall genomic architecture to their normal cell counterparts with TAD and sub-TAD compartments; however, characteristic and important local differences exist [4,13,62,63]. These minor differences may hold the key to understanding the epigenetics of cancer. As genome wide techniques for assaying chromatin conformation become more feasible and widely applied to the study of primary patient cancers, the answers to these questions will become clearer.
Acknowledgments
We would like to thank Brook Barajas and Adam Rubin for thoughtful discussion and critical review of this work. M.R.C. acknowledges funding from the National Institutes of Health training grant R25CA180993. V.G.C. acknowledges support from U.S. Public Health Service Award R01GM035463 from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. Translocation-Capture Sequencing Reveals the Extent and Nature of Chromosomal Rearrangements in B Lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho Y-J, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. Genome-wide Translocation Sequencing Reveals Mechanisms of Chromosome Breaks and Rearrangements in B Cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kon A, Shih L-Y, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 4. Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2015 doi: 10.1038/nature16490. Using integrative epigenomic profiling of IDH wild type and IDH mutant glioma samples, Flavahan et al. demonstrate that hypermethylation of specific CTCF sequences leads to loss of CTCF at a domain boundary, allowing for aberrant constitutive expression of PDGFRA, a known glioma oncogene. This is the first study to demonstrate a direct connection between a well-studied cancer mutation and disruption of chromatin architecture in a primary patient sample.
- 5. Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong C-T, Hookway TA, Guo C, Sun Y, et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. Through the study of embryonic stem cell differentiation, Phillips-Cremins et al. demonstrate the fundamental principles governing the reorganization of the three-dimensional genome during an active cellular process. They provide strong evidence suggesting that cell-type-specific chromatin organization occurs at the submegabase scale and characterize the various architectural proteins that establish the hierarchical genomic organization in humans.
- 6.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips JE, Corces VG. CTCF: Master Weaver of the Genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misteli T. Beyond the Sequence: Cellular Organization of Genome Function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 9.Hou C, Li L, Qin ZS, Corces VG. Gene Density, Transcription, and Insulators Contribute to the Partition of the Drosophila Genome into Physical Domains. Mol. Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the Xinactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman-aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;33292:289–294. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fudenberg G, Getz G, Meyerson M, Mirny L. High-order chromatin architecture determines the landscape of chromosomal alterations in cancer. Nat. Biotechnol. 2011;29:1109–1113. doi: 10.1038/nbt.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, McCord RP, Ho Y-J, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial Organization of the Mouse Genome and Its Role in Recurrent Chromosomal Translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engreitz JM, Agarwala V, Mirny LA. Three-Dimensional Genome Architecture Influences Partner Selection for Chromosomal Translocations in Human Disease. PLoS One. 2012;7:e44196. doi: 10.1371/journal.pone.0044196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barber TD, McManus K, Yuen KWY, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl. Acad. SciUSA. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solomon Da, Kim T, Diaz-Martinez La, Fair J, Elkahloun AG, Harris BT, Toretsky Ja, Rosenberg Sa, Shukla N, Ladanyi M, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mazumdar C, Shen Y, Xavy S, Zhao F, Reinisch A, Li R, Corces MR, Flynn RA, Buenrostro JD, Chan SM, et al. Leukemia-Associated Cohesin Mutants Dominantly Enforce Stem Cell Programs and Impair Human Hematopoietic Progenitor Differentiation. Cell Stem Cell. 2015 doi: 10.1016/j.stem.2015.09.017. In this study, Mazumdar et al. provide one of the first in-depth phenotypic characterizations of a cancerous mutation in a protein involved in chromatin organization. The authors demonstrate that mutation of various components of the cohesin complex in acute myeloid leukemia leads to defects in differentiation, linking chromatin architecture to a carcinogenic phenotype for the first time.
- 20.Viny AD, Ott CJ, Spitzer B, Rivas M, Meydan C, Papalexi E, Yelin D, Shank K, Reyes J, Chiu A, et al. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J. Exp. Med. 2015;212:1819–1832. doi: 10.1084/jem.20151317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, Wang K, Kayembe C, Rocha PP, Raviram R, Gong Y, et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J. Exp. Med. 2015;212:1833–1850. doi: 10.1084/jem.20151323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balbás-Martínez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Márquez M, Vazquez M, Lapi E, Castro-Giner F, Beltran S, Bayés M, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat. Genet. 2013;45:1464–1469. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.TCGA Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence MS, Stojanov P, Polak P, Kryukov G V, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts S a, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding L, Ley TJ, Larson DE, Miller C a, Koboldt DC, Welch JS, Ritchey JK, Young M a, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. The Origin and Evolution of Mutations in Acute Myeloid Leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, Majeti R. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 2012;4:1–10. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon Da, Kim J-S, Bondaruk J, Shariat SF, Wang Z-F, Elkahloun AG, Ozawa T, Gerard J, Zhuang D, Zhang S, et al. Frequent truncating mutations of STAG2 in bladder cancer. Nat. Genet. 2013;45:1428–1430. doi: 10.1038/ng.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, Dean M, Huang Y, Jia W, Zhou Q, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat. Genet. 2013;45:1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor CF, Platt FM, Hurst CD, Thygesen HH, Knowles MA. Frequent inactivating mutations of STAG2 in bladder cancer are associated with low tumour grade and stage and inversely related to chromosomal copy number changes. Hum. Mol. Genet. 2014;23:1964–1974. doi: 10.1093/hmg/ddt589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brennan CWW, Verhaak RGWGW, McKenna A, Campos B, Noushmehr H, Salama SRR, Zheng S, Chakravarty D, Sanborn JZZ, Berman SHH, et al. The Somatic Genomic Landscape of Glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones DTW, Jäger N, Kool M, Zichner T, Hutter B, Sultan M, Cho Y-J, Pugh TJ, Hovestadt V, Stütz AM, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Zainal SN, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evers L, Perez-Mancera PA, Lenkiewicz E, Tang N, Aust D, Knösel T, Rümmele P, Holley T, Kassner M, Aziz M, et al. STAG2 is a clinically relevant tumor suppressor in pancreatic ductal adenocarcinoma. Genome Med. 2014;6:9. doi: 10.1186/gm526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, Patidar R, Hurd L, Chen L, Shern JF, et al. The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent STAG2 Mutation. PLoS Genet. 2014;10:e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filippova GN, Qi C-F, Ulmer JE, Moore JM, Ward MD, Hu YJ, Loukinov DI, Pugacheva EM, Klenova EM, Grundy PE, et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter its DNA-binding specificity. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- 37.Filippova GN, Lindblom A, Meincke LJ, Klenova EM, Neiman PE, Collins SJ, Doggett N a, Lobanenkov VV. A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosom. Cancer. 1998;22:26–36. [PubMed] [Google Scholar]
- 38.Walker C, Miranda M, O’Hern M, McElroy J, Coombes K, Bundschuh R, Cohn D, Mutch D, Goodfellow P. Patterns of CTCF and ZFHX3 Mutation and Associated Outcomes in Endometrial Cancer. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kemp CJ, Moore JM, Moser R, Bernard B, Teater M, Smith LE, Rabaia Na, Gurley KE, Guinney J, Busch SE, et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014;7:1020–1029. doi: 10.1016/j.celrep.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando Da, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuin J, Dixon JR, van der Reijden MIJA, Ye Z, Kolovos P, Brouwer RWW, van de Corput MPC, van de Werken HJG, Knoch TA, van IJcken WFJ, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sofueva S, Yaffe E, Chan W-C, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, et al. Transcription Factor Binding in Human Cells Occurs in Dense Clusters Formed around Cohesin Anchor Sites. Cell. 2013;154:801–813. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 45.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels Pa, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 47. Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, Martin AR, Greenside PG, Srivas R, Phanstiel DH, Pekowska A, et al. Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell. 2015;162:1–15. doi: 10.1016/j.cell.2015.07.048. By integrating ChIP-seq data for three histone marks with Hi-C and ChIA-PET-based chromatin contact maps from lymphoblastoid cell lines, the authors discover a large number of histone quantitative trait loci. These loci are enriched for common sequence variants associated with autoimmune diseases, enabling the prediction of target genes of disease-associated variants from GWAS studies. Such techniques could be applied to cancer data in the future.
- 48. Katainen R, Dave K, Pitkänen E, Palin K, Kivioja T, Välimäki N, Gylfe AE, Ristolainen H, Hänninen Ua, Cajuso T, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet. 2015;47:818–821. doi: 10.1038/ng.3335. Katainen et al. profile 213 primary human colorectal cancer samples and identify frequent point mutations at CTCF/cohesin-binding sites (as determined by ChIP-seq). This represents the first study identifying CTCF/cohesin-binding sites as a major mutational hotspot in the noncoding cancer genome.
- 49.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio Sa, JR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maurano MT, Wang H, John S, Canfield T, Lee K, Stamatoyannopoulos Ja. Role of DNA Methylation in Modulating Transcription Factor Occupancy. Cell Rep. 2015;12:1–12. doi: 10.1016/j.celrep.2015.07.024. Maurano et al. investigate the degree to which DNA methylation affects transcription factor binding. The authors demonstrate that 98% of unoccupied, methylated CTCF recognition sequences remain unoccupied after loss of DNA methylation at that site. This provides the first compelling genome-wide evidence demonstrating that DNA methylation alone is not the primary gatekeeper of transcription factor binding.
- 53.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ley TJ, Ding L, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, et al. DNMT3A Mutations in Acute Myeloid Leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons W, Jones S, Zhang X, Lin JC-H, Leary R, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia G, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of Cell Identity Genes Occurs in Insulated Neighborhoods in Mammalian Chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. Through the use of cohesin ChIA-PET in embryonic stem cells, the authors demonstrate that key cell identity genes and their enhancers are contained within insulated neighborhoods flanked by interacting CTCF sites. These results indicate that disruption of these insulated neighborhoods would have profound effects on local gene expression and serve as the basis for Model 2 of this review.
- 57.Loeb LA. A Mutator Phenotype in Cancer. Cancer Res. 2001;61:3230–3239. [PubMed] [Google Scholar]
- 58.Hanahan D, Weinberg RA. Hallmarks of Cancer : The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Li L, Lyu X, Hou C, Takenaka N, Nguyen HQ, Ong C-T, Cubeñas-Potts C, Hu M, Lei EP, Bosco G, et al. Widespread Rearrangement of 3D Chromatin Organization Underlies Polycomb-Mediated Stress-Induced Silencing. Mol. Cell. 2015 doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. Through the study of embryonic stem cells and four separate embryonic stem cell-derived lineages, Dixon et al. demonstrate that 36% of the active and inactive chromosomal compartments change during differentiation. This work demonstrates that chromatin architecture is rapidly and coordinately changed during normal differentiation.
- 61.Fraser J, Ferrai C, Chiariello AM, Schueler M, Rito T, Laudanno G, Barbieri M, Moore BL, Kraemer DC, Aitken S, et al. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol Syst Biol. 2015;11:1–14. doi: 10.15252/msb.20156492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rickman DS, Soong TD, Moss B, Mosquera JM, Dlabal J, Terry S, MacDonald TY, Tripodi J, Bunting K, Najfeld V, et al. Oncogene-mediated alterations in chromatin conformation. Proc. Natl. Acad. Sci. 2012;109:9083–9088. doi: 10.1073/pnas.1112570109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rousseau M, Ferraiuolo MA, Crutchley JL, Wang XQ, Miura H, Blanchette M, Dostie J. Classifying leukemia types with chromatin conformation data. Genome Biol. 2014;15:R60. doi: 10.1186/gb-2014-15-4-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]