Abstract
Purpose
Dry eye disease is a common condition associated with age-related meibomian gland dysfunction (ARMGD). We have previously shown that ARMGD occurs in old mice, similar to that observed in human patients with MGD. To begin to understand the mechanism underlying ARMGD, we generated transcriptome profiles of eyelids excised from young and old mice of both sexes.
Methods
Male and female C57BL/6 mice were euthanized at ages of 3 months or 2 years and their lower eyelids removed, the conjunctival epithelium scrapped off, and the tarsal plate, containing the meibomian glands, dissected from the overlying muscle and lid epidermis. RNA was isolated, enriched, and transcribed into cDNA and processed to generate four non-stranded libraries with distinct bar codes on each adaptor. The libraries were then sequenced and mapped to the mm10 reference genome, and expression results were gathered as reads per length of transcript in kilobases per million mapped reads (RPKM) values. Differential gene expression analyses were performed using CyberT.
Results
Approximately 55 million reads were generated from each library. Expression data indicated that about 15,000 genes were expressed in these tissues. Of the genes that showed more than twofold significant differences in either young or old tissue, 698 were identified as differentially expressed. According to the Gene Ontology (GO) analysis, the cellular, developmental, and metabolic processes were found to be highly represented with Wnt function noted to be altered in the aging mouse.
Conclusions
The RNA sequencing data identified several signaling pathways, including fibroblast growth factor (FGF) and Wnt that were altered in the meibomian glands of aging mice.
Introduction
Meibomian gland dysfunction (MGD) severely impacts vision and lifestyle, and the prevalence of MGD increases with age. MGD typically involves glandular atrophy or a decrease in meibum lipid quality [1,2], although little is known regarding the associated gene expression changes or the mechanism underlying age-related MGD (ARMGD).
The meibomian glands are holocrine, lipid-excreting glands embedded in the collagenous tarsal plate of the eyelid. They are primarily responsible for producing meibum lipids that form the outermost layer of the tear film, preventing tear evaporation, and providing a smooth optical surface. With ARMGD, aqueous tears evaporate faster from the ocular surface leading to decreased tear break-up time, increased tear osmolarity, and the release of inflammatory cytokines [3].
Past clinical histologic studies of human meibomian glands have suggested that hyper-keratinization may play a role in the development of MGD [4], and various animal models have shown that meibomian glands are susceptible to hyper-keratinization and blockage of the gland orifice [5,6,7]. Furthermore, gene expression analysis using microarrays of aged human eyelid tissues with MGD has also identified increased expression of the epidermal keratins 1 and 10 [8]. However, protein analysis of secretions from human patients with MGD has failed to detect keratins 1 and 10 from meibomian gland excreta [9].
Recent studies have implicated cell senescence, gland atrophy, and increased protein-to-lipid ratios of meibum in mouse and human ARMGD [10-13]. Changes in quiescent meibomian gland progenitor cells may be an important underlying factor responsible for the development of gland atrophy with age, although this has yet to be studied because these cells have only recently been identified [14]. The ARMGD mouse model and human subjects also show a change in peroxisome proliferator-activated receptor gamma (PPAR-γ) localization that is associated with age, along with anterior displacement of Marx’s line (mucocutaneous junction) at the eyelid margin, gland atrophy, and ductal plugging of the gland orifice without hyper-keratinization [11,13,15]. Although all of these factors may combine to cause evaporative stress to the ocular surface, the effects of age on the regulatory mechanisms controlling meibomian gland differentiation and function remain unknown.
In this study, we used RNA sequencing to analyze the differential expression of genes in the transcriptome of eyelids from old (2 years) compared to young (3 months) C57BL/6 mice. We discovered that 51 genes were exclusively expressed in male and 50 genes were exclusively expressed in female eyelid tissues. Seventy-seven genes were more highly expressed in young male and female mice. Four hundred and eighteen genes were more expressed in 2-year-old tissue. Gene Ontology (GO) ontology revealed that genes associated with cellular, developmental, and metabolic processes were prevalent, and fibroblast growth factor (FGF) and Wnt functions were significantly altered in the aging mouse. In particular, expression of a member of the Dkk (Dickkopf) family of Wnt signaling antagonists, DKKL1, was found to be decreased in the mouse model of ARMGD. Here, we verify the expression of DKKL1 in human tissue sections and immortalized meibocyte cultures.
Methods
Specimen preparation
A total of ten C57Bl/6 mice aged 3 months (three male and three female) and 2 years (three male and one female) were euthanized with CO2 asphyxiation and cervical dislocation before the lower and upper eyelids were harvested. The overlying conjunctiva was scraped off the eyelids, and the tarsal plates containing the meibomian glands dissected from the overlying muscle and eyelid epidermis and immediately frozen under liquid nitrogen. Animal experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) protocol at University of California, Irvine and adhered to the ARVO Statement on the Use of Animals in Vision Research at all times.
All mice were obtained from our aging C57Bl/6 mouse colony that was exposed to the same environmental room conditions from birth. Furthermore, the young and old mice had the same date of birth. Unfortunately, fewer female mice in the aged cohort survived to 2 years compared to the male mice, reducing the number available for this study. As the goal of this project was to identify only age-related gene expression patterns, eyelid tissue was pooled from individual mice to reduce variation and increase RNA quantity.
RNA isolation and library construction
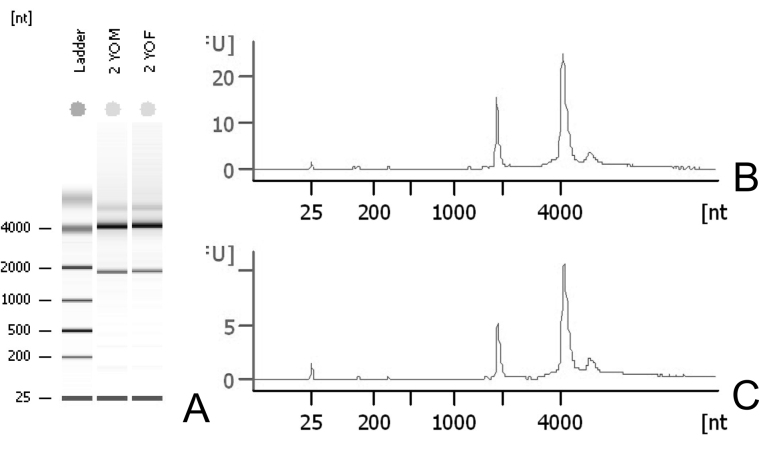
Tissues from each age and sex group were pooled and crushed after liquid nitrogen immersion. The resulting frozen powder was suspended in RLT buffer (Qiagen, Valencia, CA), and RNA was isolated over RNeasy columns (Qiagen). RNA quality and quantity were assessed with the aid of a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), and the results for the 2-year-old groups are shown in Figure 1A. The results obtained from the young mice were indistinguishable (not shown), and none of the samples showed evidence of degradation. As part of the analysis, the Ribosome Integrity Number (RIN) is reported, and in each case, the value was greater than 9 for each (scaled from 1 to 10, with 10 the maximum value).
Figure 1.
Quality results of RNA isolated from mouse eyelids. A: High-quality 18S and 28S RNA was isolated from 2-year-old male and female C57BL/6 mice according to the Bioanalyzer plots of the 2-year-old male (B) and female (C) mouse eyelid meibomian glands. The average quality of sequence information was 32.6, and the Ribosome Integrity Number (RIN) values exceeded 9 on a scale of 1–10.
Total RNA (1 μg) from each pool was enriched for poly A containing RNA using poly dT magnetic beads, and the enriched fraction was chemically fragmented using a TruSeq RNA Sample Prep kit v2 (Illumina, San Diego, CA). The RNA fragments were then transcribed into cDNA using random primers and reverse transcriptase. After second strand synthesis using DNA polymerase and RNase H, the cDNA fragments were blunt ended and ligated to adapters. The adaptor-ligated fragments were then subjected to 11 PCR cycles using primers complementary to the adaptor sequences. PCR amplification protocol: 98 °C for 30 s then 11 cycles of 98 °C for 10 s; 60 °C for 30 s; 72 °C for 30 s and then finished with 72 °C for 5 min and held at 10 °C. This resulted in a non-stranded library for each RNA pool with distinct bar code adapters to discriminate the tissue source (young versus old, male versus female). The libraries were then sequenced on an Illumina HiSeq 2500 using version 3 chemistry within a single column to generate approximately 50 million reads from each library.
The resulting sequences were then assessed for quality using the RobiNA software package. As reported in the supplemental files, the base call quality was routinely high and gave, on average, 100 bps of sequence information per sequence. Additionally, each sequence gave similarly high-quality sequence information with an average quality of 32.6 (Figure 1B,C). Similar results were obtained for the remaining three libraries (not shown).
Gene expression data analysis
FASTQ sequence files were evaluated for quality using the high-performance computing cluster at the University of California, Irvine using RobiNA software [16]. Sequences trimmed of adaptor sequences were mapped to the mm10 reference genome downloaded from UCSC Genome Bioinformatics website (https://genome.ucsc.edu). Using the Tuxedo package available from Bioconductor, sequences were aligned to the mm10 reference genome using the TopHat module [17]. The Cufflinks module was then used to assemble the transcripts and compare or merge them with the other three libraries [18]. Expression results were gathered as reads per length of transcript in kilobases per million mapped reads (RPKM) values for each gene. The genes with RPKM values equal to or greater than 0.125 were considered to be expressed genes (BioMedicalCentral/1471-2164/13/82; Appendix 1). Differential gene expression analyses were then performed using CyberT software.
Genes suggested to be differentially expressed with a Bonferroni p value of less than 10−5 and at least a twofold difference in expression (Appendix 2) were then searched to identify the genes that appeared differentially expressed in young and old mice regardless of sex (those that appear common across groups). Genes common to male and female mice were then evaluated for pathway analyses using the tools available at PantherDB [19].
Immunohistochemistry
Human tissue was collected in accordance with the Institutional Review Board (IRB) at the University of California, Irvine. Mouse tissue collected from the upper eyelids was embedded in Tissue-Tek OCT (optimum cutting temperature; Sakura Finetek USA, Inc., Torrance, CA) freezing medium, frozen in liquid nitrogen, and sectioned at 10 µm using a Leica CM1850 Cryotome (Leica, Wetzlar, Germany).
Frozen sections were hydrated with PBS (1X; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 (dibasic), 1.8 mM KH2PO4 (monobasic), pH 7.4) and then placed in freezing acetone (−20 °C) for 3 min. Tissue sections were then blocked with 1% bovine serum albumin (BSA) in PBS for 30 min at 37 °C and then incubated with rabbit polyclonal anti-DKKL1 at a 1/100 dilution (#ABN 193; EMP Millipore, Darmstadt, Germany) for 1 h at 37 °C, washed with PBS, and reacted with Alexa Fluor 546 secondary antibodies (1/500) against rabbit immunoglobulin (IgG; Invitrogen, Carlsbad, CA) for 1 h at 37 °C. All samples were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; 300 nM) in PBS for 15 min. For negative controls, sections were incubated with secondary antibodies alone. For fluorescent imaging of the sections, a Leica DMI6000B equipped with an automated stage and programmed with MetaMorph software (Molecular Devices, Sunnyvale, CA) was used.
Immortalized mouse meibomian gland cells were cultured in keratinocyte growth media (Lonza, Walkersville, MD) at 37 °C and 5.0% CO2 and treated with (50 µM) rosiglitazone for 7 days before fixation with 2% paraformaldehyde and immunostaining with anti-DKKL1 at a dilution of 1/100 in 5% goat serum and 1X PBS. Untreated cells and cells treated with 50 µM rosiglitazone were immunostained for DKKL1 and imaged for antibody staining (secondary antibody: AlexaFluor 546, 1/500) and lipid vacuoles using bright-field imaging with a Leica fluorescence microscope.
Results
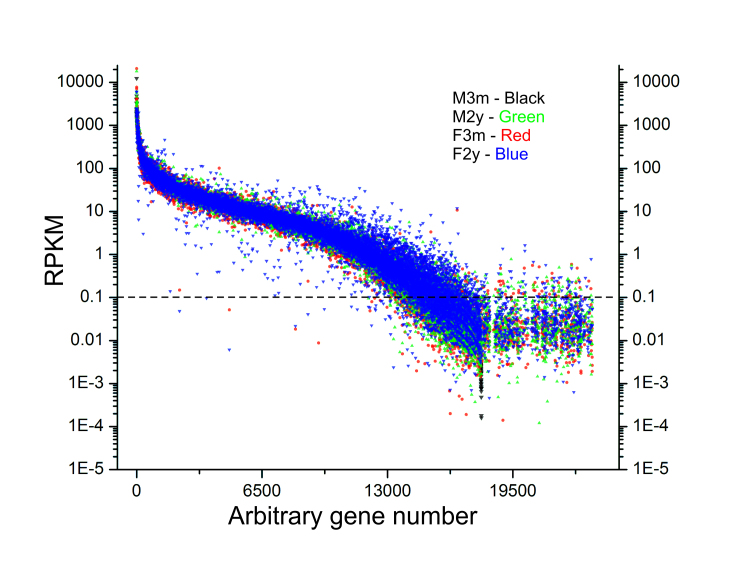
Approximately 15,000 genes were identified with RPKM values greater than or equal to 0.125, and at least ten reads were considered as expressed in each sample (Appendix 1, Figure 2). The most highly represented gene in each library was identified as glutathione S-transferase omega 1 (Gsto1) with an RPKM value approximately threefold higher than any other identified gene. Not unexpected for tissues with a large epithelial component, multiple keratin genes were all highly represented in all libraries; however, no significant differences were seen in the expression of the epidermal cytokeratins 1 and 10 or the cornified envelope proteins, Sprr1a or Sprr2a, which are necessary for keratinization. Using the Venn drawing tool, the majority of expressed genes (14,282) were seen expressed in each source of tissue (Figure 3). Comparing genes that appeared restricted by sex, regardless of age, 51 genes were selectively expressed in male mice while 50 genes were selectively expressed in female mice. Young animals, regardless of sex, selectively expressed 65 genes, while the aged animals selectively expressed 201 genes. These sets are presented in Appendix 2.
Figure 2.
Gene expression levels of 3-month-old and 2-year-old male and female mouse eyelids. Expression results for each gene were gathered from RNA sequencing data as reads per length of transcript in kilobases per million mapped reads (RPKM). Genes with RPKM values equal to or greater than 0.125 were considered to be expressed.
Figure 3.
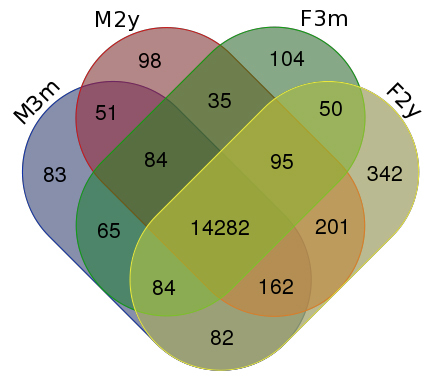
Venn diagram of selective gene expression in 3-month- and 2-year-old male and female mouse eyelids. A total of 14,282 genes were found to be expressed in all specimens, whereas 51 genes were selectively expressed in male mice while 50 genes were selectively expressed in female mice. Young animals, either male or female, selectively expressed 65 genes, while aged mice selectively expressed 201 genes.
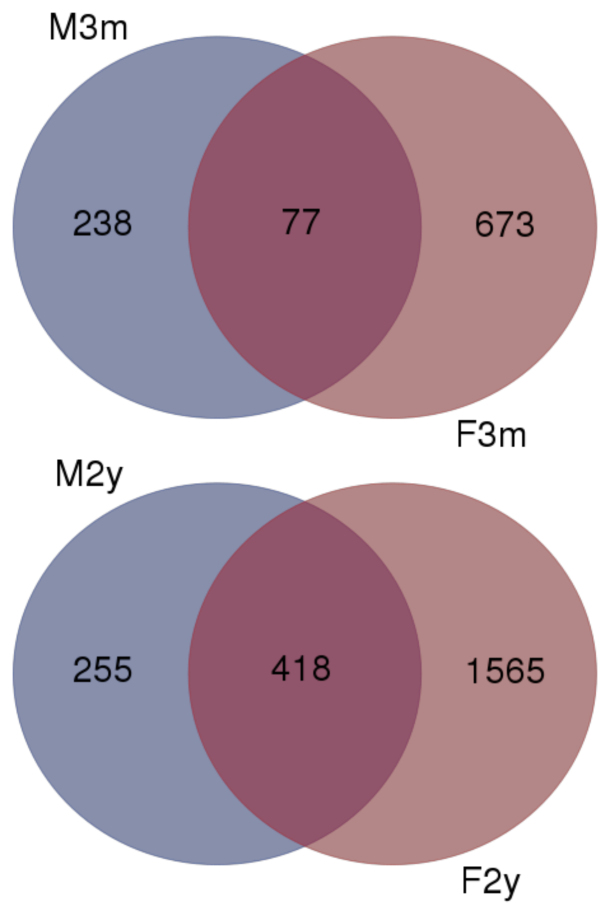
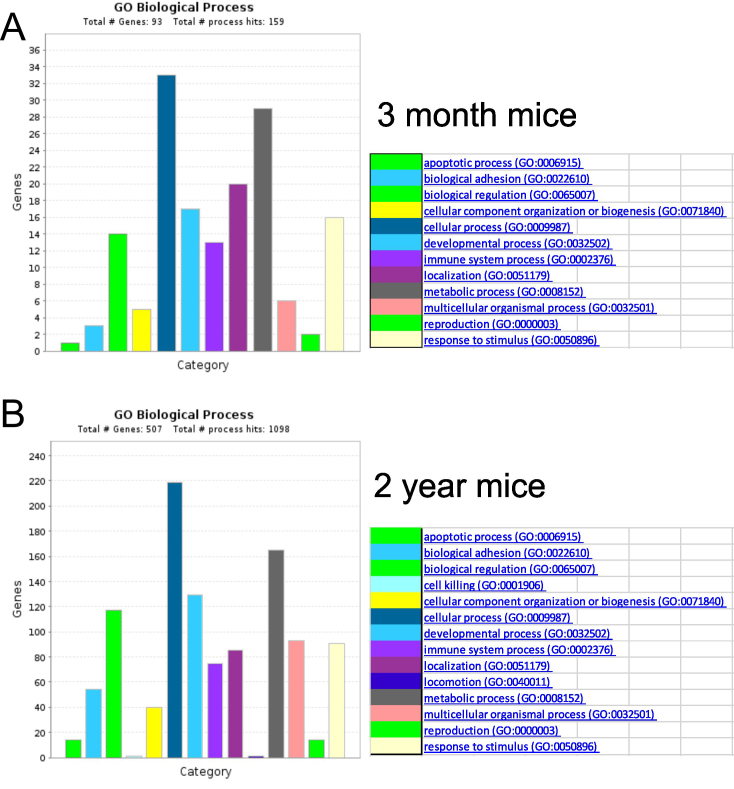
Although comparison of gene lists may contribute information concerning selective expression, it is difficult to assess the absence of any apparent gene in these data sets. Therefore, differential gene expression analyses were performed using CyberT (Appendix 3, Figure 4). To limit the number of candidate genes to be considered possibly differential, the results were restricted to only genes that showed at least a twofold difference and a Bonferroni corrected p value of less than 10−5 (Appendix 2) between the data sets. These analyses identified approximately 3,725 genes as apparently differentially expressed. From these data sets, a further restriction was placed such that only genes common to the male and female mice data sets were considered. As shown in Figure 4, 77 genes were more highly expressed in young male and female mice. Conversely, 418 genes were more prominently expressed in the male and female 2-year-old tissue. Combining these differentially expressed candidate genes with genes that appeared to be selectively expressed in young and old tissues (Figure 3), 567 candidate genes were identified in the 2-year-old mice, while 131 candidates were identified in the 3-month-old mice (Appendix 4). Pathway analysis of these common genes (Figure 5) emphasized that the biologic processes most affected by age are reflected in pathways that involve cellular and metabolic processes. These candidate genes were also assessed for statistical overrepresentation of GO slim biologic pathways within the data. As shown in Figure 5A, in the evaluation of the genes overexpressed in young mice, two pathways were statistically enriched and represent genes involved with gland development and cellular defense. In the evaluation of genes overexpressed in old mice in a similar manner, several pathways were statistically overrepresented (Figure 5B). As seen, the number of pathways and the number of genes represented within these pathways were markedly greater than those seen enriched in the young mice. Interestingly, developmental, differentiation, response to stimulus, cell signaling, and cell regulation pathways are represented, suggesting that older mice have alterations in a multiplicity of processes, distinct from young mice.
Figure 4.
Differential gene expression analysis of 3-month- and 2-year-old mice eyelids performed by CyberT. A total of 3,725 genes were considered differentially expressed by showing at least a twofold difference in the reads per length of transcript in kilobases per million mapped reads (RPKM) values and a Bonferroni corrected p value of less than 10−5 between the data sets. Seventy-seven genes were more highly expressed in male and female 3-month-old mice, whereas 418 genes were found to be expressed higher in the male and female 2-year-old tissue.
Figure 5.
Pathway analysis of gene expression data generated from RNA sequencing. Pathway analysis shows that the biologic processes most affected by aging are cellular and metabolic processes. In young mice, genes involved with meibomian gland development and cellular defense were found to be statistically enriched whereas, in older mice, the number of pathways and genes involving differentiation, cell signaling, and cell regulation pathways were significantly altered.
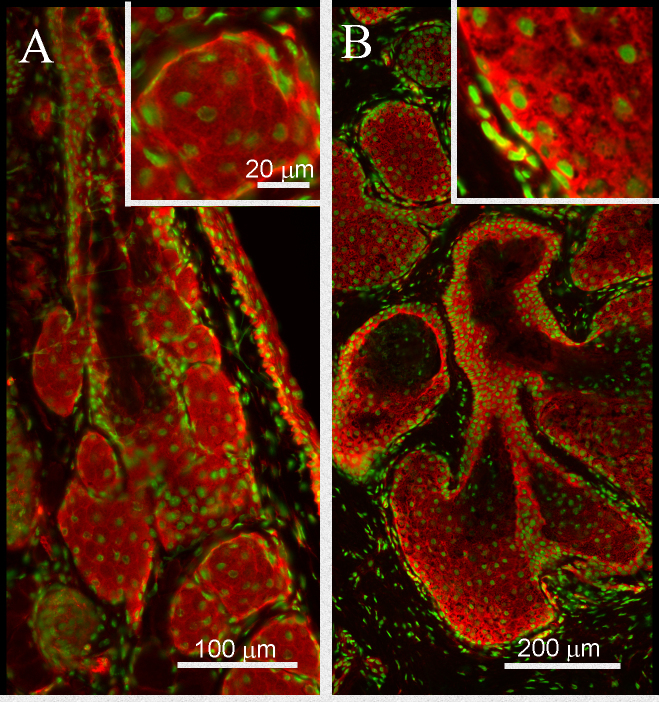
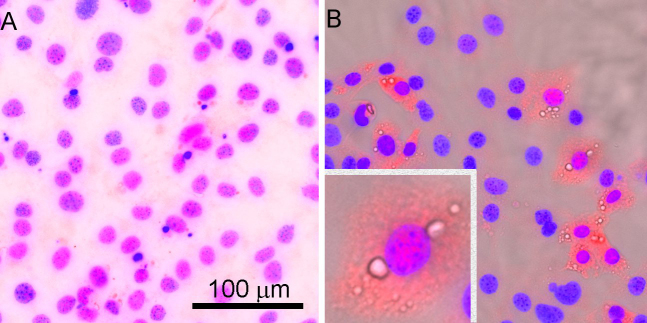
One gene overexpressed in young mice, DKKL1, encodes for a Dickkopf-like protein also referred to as Soggy (SGY-1). To evaluate whether DKKL1 was expressed, immunostaining of mouse and human Meibomian glands was performed. As shown in Figure 6, the mouse (Figure 6A) and human (Figure 6B) meibomian gland ducts and acini show positive staining for DKKL1. At higher magnification, the distribution appeared to be primarily granular within the cytoplasm with some enrichment at the plasma membrane. To explore the possible relationship of DKKL1 with PPAR-γ expression, cultures of immortalized mouse meibocytes were stained for DKKL1 in the presence or absence of rosiglitazone, a PPAR-γ agonist. As shown in Figure 7, cultures grown without added agonist display little if any DKKL1 staining (panel A). The addition of the agonist, which is known to stimulate and induce PPAR-γ, led to a marked increase in staining. In addition, DKKL1-positive cells appeared to be differentiating as lipid-containing vacuoles were present in the cells.
Figure 6.
Immunohistochemistry of mouse and human meibomian gland tissue cryosections. Ten-micron cryosections of optimum cutting temperature (OCT)-embedded (A) mouse and (B) human eyelid tissue was stained with anti-DKKL1 (red) at a 1/100 dilution and Alexa Fluor 546 secondary antibody at a 1/500 dilution while 4′,6-diamidino-2-phenylindole (DAPI) was used to counter-stain the cell nuclei (green).
Figure 7.
Immunocytochemistry of cultured immortalized mouse meibocytes. Mouse meibocytes were cultured in keratinocyte growth media and immunostained with anti-DKKL1 (1/100) after fixation and (A) no treatment or (B) treatment with 50 µM rosiglitazone, a peroxisome proliferator-activated receptor gamma (PPAR-γ) agonist.
Discussion
Using RNA sequencing, we quantified gene expression in the eyelids of young male and female mice compared to old mice with ARMGD. This approach has enabled us to identify candidate genes altered with aging, which may be involved in the mechanisms underlying ARMGD.
Fifty-one genes were found to be selectively expressed in male mice while 50 genes were selectively expressed in female mice. Y chromosome genes were lacking in the female library while the X-inactive specific transcript (Xist), for example, was lacking in the male libraries. Young animals, regardless of sex, selectively expressed 65 genes, while the aged animals selectively expressed 201 genes. When the differentially expressed genes were combined with the selectively expressed genes in young and old tissues, 567 candidate genes were identified in the 2-year-old mice, while 131 candidates were identified in the 3-month-old mice. Pathway analysis of these common genes emphasized that the biologic processes most affected by age are pathways that involve cellular and metabolic processes.
We found no significant difference in the expression of the epidermal cytokeratins 1 and 10 between young and old mice, supporting the hypothesis that hyper-keratinization is not the cause of ARMGD and that other mechanisms are behind this age-related disease. Moreover, no increase was observed in the expression of the cornified envelope protein Sprr1a, which is integral to the process of keratinization and cornification. Without an increase in Sprr1a, the involvement of hyper-keratinization in ARMGD in the aged mice used in his study is unlikely. This evidence further supports the hypothesis that the atrophic model of ARMGD may involve progenitor cell senescence through genetic changes caused by aging.
From the expression data, DKKL1 was found overexpressed in young mice eyelids, and its presence in mouse and human meibomian glands was confirmed with immunohistochemistry. Cultures of immortalized mouse meibocytes were stained for DKKL1, in the presence or absence of a PPAR-γ agonist (50 µM rosiglitazone), and DKKL1 expression increased in cell actively synthesizing lipid. PPAR-γ is integral to meibocyte lipogenesis and the synthesis of meibum that forms part of the tear film. The association of PPAR-γ with DKKL1 expression suggests that DKKL1 may play an important role in meibocyte differentiation and meibum synthesis. This may be important for sebaceous gland function and sebum synthesis in the skin as well as the meibum production of the eyelid meibomian glands.
DKKL1, overexpressed in young mice, was intriguing in that its expression in adult tissues was thought to be limited to testicular tissue [20], yet DKKL1 is expressed in young male and female eyelid tissue. DKKL1 is a homolog to the Dickkopf (DKK1–4) family of proteins that shows unique homology to DKK3 [21]. DKKs are rich in cysteines and typically act as negative regulators of Wnt signaling as they antagonize Wnt/β-catenin signaling through inhibition of Wnt coreceptors such as LRP5 and LRP6. DKKs also show a high affinity for the transmembrane proteins Kremen1 and 2, which are also known to regulate Wnt signaling pathways [22]. Unlike the other DKK family members, DKKL1 does not regulate Wnt/β-catenin canonical signaling [23]. From our work, it is clear that further investigation into the expression of DKKL1 in the holocrine meibomian glands is required to precisely understand DKKL1’s role in lipogenesis.
In conclusion, RNA sequencing technology has precisely quantified the transcriptome and gene expression in young and old mice of both sexes. This huge database of information has provided us with a wealth of targets to analyze as potential factors responsible for ARMGD. Of particular interest is DKKL1, which is significantly downregulated in old mice. This decreased expression in the mouse model of ARMGD suggests that it may serve an important function that is altered with age. Further study of the candidate genes revealed in this study should help unravel the mechanisms behind ARMGD and may offer potential targets, such as DKKL1, for therapy to treat MGD.
Acknowledgments
Supported by the Research to Prevent Blindness Inc. unrestricted grant and NEI R01EY021510.
Appendix 1. Expressed genes in 3 month and 2 year old male and female mouse eyelids
To access the data, click or select the words “Appendix 1.”
Appendix 2. Selectively expressed genes in young versus old and male versus female mouse eyelids
To access the data, click or select the words “Appendix 2.”
Appendix 3. CyberT results of differential gene expression analysis
To access the data, click or select the words “Appendix 3.”
Appendix 4. Candidate genes for 3 month and 2 year old mouse eyelids
To access the data, click or select the words “Appendix 4.”
References
- 1.Lemp MA, Crews LA, Bron AJ, Foulks GN, Sullivan BD. Distribution of aqueous-deficient and evaporative dry eye in a clinic-based patient cohort: a retrospective study. Cornea. 2012;31:472–8. doi: 10.1097/ICO.0b013e318225415a. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22378109&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 2.Horwath-Winter J, Berghold A, Schmut O, Floegel I, Solhdju V, Bodner E, Schwantzer G, Haller-Schober EM. Evaluation of the clinical course of dry eye syndrome. Arch Ophthalmol. 2003;121:1364–8. doi: 10.1001/archopht.121.10.1364. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=14557170&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 3.Mathers W. Evaporation from the ocular surface. Exp Eye Res. 2004;78:389–94. doi: 10.1016/s0014-4835(03)00199-4. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15106917&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 4.Gutgesell VJ, Stern GA, Hood CI. Histopathology of meibomian gland dysfunction. Am J Ophthalmol. 1982;94:383–7. doi: 10.1016/0002-9394(82)90365-8. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=7124880&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 5.Jester JV, Nicolaides N, Kiss-Palvolgyi I, Smith RE. Meibomian gland dysfunction. II. The role of keratinization in a rabbit model of MGD. Invest Ophthalmol Vis Sci. 1989;30:936–45. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=2470694&dopt=Abstract [PubMed] [Google Scholar]
- 6.Jester JV, Rajagopalan S, Rodrigues M. Meibomian gland changes in the rhino (hrrhhrrh) mouse. Invest Ophthalmol Vis Sci. 1988;29:1190–4. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=2458328&dopt=Abstract [PubMed] [Google Scholar]
- 7.Ohnishi Y, Kohno T. Polychlorinated biphenyls poisoning in monkey eye. Invest Ophthalmol Vis Sci. 1979;18:981–4. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=113362&dopt=Abstract [PubMed] [Google Scholar]
- 8.Liu S, Richards SM, Lo K, Hatton M, Fay A, Sullivan DA. Changes in gene expression in human meibomian gland dysfunction. Invest Ophthalmol Vis Sci. 2011;52:2727–40. doi: 10.1167/iovs.10-6482. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21372006&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong BL, Hodson SA, Wigham T, Miller F, Larke JR. Evidence for keratin proteins in normal and abnormal human meibomian fluids. Curr Eye Res. 1991;10:1113–9. doi: 10.3109/02713689109024128. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=1724955&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 10.Suhalim JL, Parfitt GJ, Xie Y, De Paiva CS, Pflugfelder SC, Shah TN, Potma EO, Brown DJ, Jester JV. Effect of desiccating stress on mouse meibomian gland function. Ocul Surf. 2014;12:59–68. doi: 10.1016/j.jtos.2013.08.002. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24439047&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parfitt GJ, Xie Y, Geyfman M, Brown DJ, Jester JV. Absence of ductal hyper-keratinization in mouse age-related meibomian gland dysfunction (ARMGD). Aging (Albany, NY) 2013;5:825–34. doi: 10.18632/aging.100615. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24259272&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jester BE, Nien CJ, Winkler M, Brown DJ, Jester JV. Volumetric reconstruction of the mouse meibomian gland using high-resolution nonlinear optical imaging. Anat Rec (Hoboken) 2011;294:185–92. doi: 10.1002/ar.21305. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21234992&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nien CJ, Paugh JR, Massei S, Wahlert AJ, Kao WW, Jester JV. Age-related changes in the meibomian gland. Exp Eye Res. 2009;89:1021–7. doi: 10.1016/j.exer.2009.08.013. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=19733559&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parfitt GJ, Geyfman M, Xie Y, Jester JV. Characterization of quiescent epithelial cells in mouse meibomian glands and hair follicle/sebaceous glands by immunofluorescence tomography. J Invest Dermatol. 2015;135:1175–7. doi: 10.1038/jid.2014.484. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25398054&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nien CJ, Massei S, Lin G, Nabavi C, Tao J, Brown DJ, Paugh JR, Jester JV. Effects of age and dysfunction on human meibomian glands. Arch Ophthalmol. 2011;129:462–9. doi: 10.1001/archophthalmol.2011.69. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21482872&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–7. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=19289445&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20436464&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–86. doi: 10.1093/nar/gks1118. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23193289&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Q, Wu X, Chen C, Diao R, Lai Y, Huang J, Chen J, Yu Z, Gui Y, Tang A, Cai Z. Developmental expression and function of DKKL1/Dkkl1 in humans and mice. Reproductive biology and endocrinology. RB&E. 2012;10:51. doi: 10.1186/1477-7827-10-51. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22817830&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krupnik VE, Sharp JD, Jiang C, Robison K, Chickering TW, Amaravadi L, Brown DE, Guyot D, Mays G, Leiby K, Chang B, Duong T, Goodearl AD, Gearing DP, Sokol SY, McCarthy SA. Functional and structural diversity of the human Dickkopf gene family. Gene. 1999;238:301–13. doi: 10.1016/s0378-1119(99)00365-0. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10570958&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 22.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–81. doi: 10.1038/sj.onc.1210054. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17143291&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 23.Dakhova O, O’Day D, Kinet N, Yucer N, Wiese M, Shetty G, Ducy P. Dickkopf-like1 regulates postpubertal spermatocyte apoptosis and testosterone production. Endocrinology. 2009;150:404–12. doi: 10.1210/en.2008-0673. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18818293&dopt=Abstract [DOI] [PubMed] [Google Scholar]