Abstract
Purpose
To describe the clinical and genetic findings in a patient with autosomal recessive bestrophinopathy (ARB) and his healthy parents.
Methods
The patient and his healthy non-consanguineous parents underwent detailed ophthalmic evaluations including electro-oculography (EOG), spectral-domain optical coherence tomography (SD-OCT), and fundus autofluorescence (FAF) imaging. Mutation analysis of the BEST1 gene was performed by Sanger sequencing.
Results
The FAF images showed multiple spots of increased autofluorescence, and the sites of these spots corresponded to the yellowish deposits detected by ophthalmoscopy. SD-OCT showed cystoid macular changes and a shallow serous macular detachment. The Arden ratio of the EOG was markedly reduced to 1.1 in both eyes. Genetic analysis of the proband detected two sequence variants of the BEST1 gene in the heterozygous state: a novel variant c.717delG, p.V239VfsX2 and an already described c.763C>T, p.R255W variant associated with Best vitelliform macular dystrophy and ARB. The proband’s father carried the c.717delG, p.V239VfsX2 variant in the heterozygous state, and the mother carried the c.763C>T, p.R255W variant in the heterozygous state. The parents who were heterozygous for the BEST1 variants had normal visual acuity, EOG, SD-OCT, and FAF images.
Conclusions
In a truncating BEST1 mutation, the phenotype associated with ARB is most likely due to a marked decrease in the expression of BEST1 promoted by the nonsense-mediated decay surveillance mechanism, and it may depend on the position of the premature termination of the codon created.
Keywords: Autosomal recessive bestrophinopathy, BEST1, Fundus autofluorescence, Electro-oculography (EOG)
Introduction
BEST1 (VMD2) is a gene located on chromosome 11 (11q12.3) that encodes for the 585 amino acid transmembrane protein bestrophin 1 which is located on the basolateral aspect of retinal pigment epithelial (RPE) cells [1, 2]. Although the functional role of bestrophin-1 within the RPE has not been determined definitively, it has been postulated to function as a Ca2+-activated Cl− channel [3], a regulator of voltage-gated Ca2+ channels [4], and a HCO3− channel [5]. Mutations in BEST1 therefore affect the RPE metabolism and consequently the outer retinal function with which the RPE is intimately associated.
Mutations of the BEST1 gene have been associated with different ocular phenotypes [6]. The first disease shown to be caused by BEST1 sequence variants was Best vitelliform macular dystrophy (BVMD) [1], a retinal disease characterized by bilateral yellowish yolk-colored lesion in the macula. BEST1 mutations are also associated with several other eye diseases including adult-onset vitelliform macular dystrophy (AOVMD) [7], autosomal dominant vitreo-retinochoroidopathy (ADVIRC) [8], retinitis pigmentosa [9], and the microcornea, retinal dystrophy, cataract, and posterior staphyloma (MRCS) syndrome [10].
Autosomal recessive bestrophinopathy (ARB), first described in detail in 2008, is another member of the phenotypic spectrum associated with mutations in the BEST1 gene [11]. The characteristics of this disorder include a progressive reduction in central vision, absence of the electro-oculographic (EOG) light rise, and reduced full-field electroretinograms (ERGs). None of the patients have the vitelliform lesions typical of Best disease, but have a diffuse irregularity of the reflex from the RPE including dispersed punctate flecks [11]. All of the patients have an accumulation of fluid within and/or beneath the neurosensory retina in the macular area [11].
ARB has been reported to be due to either compound heterozygous or homozygous BEST1 gene mutations [6, 11]. A recent manuscript described an ocular phenotype similar to ARB associated with a single heterozygous mutation of the BEST1 gene [12].
Several mutations associated with ARB have been reported to be involved in causing dominant Best disease when they were present in the heterozygous state [6, 7, 13–15]. The clinical phenotype of some patients with recessive bestrophinopathy is distinct from that seen in Best disease, while in others it is similar to the typical phenotype observed in autosomal dominant vitelliform dystrophy [16, 17]. Identification of additional families with recessive bestrophinopathy and detailed characterization of the clinical phenotypes of homozygous and heterozygous individuals will assist in establishing the phenotype–genotype correlations in patients with BEST1-associated diseases.
Methods
The protocol of this study conformed to the tenets of the Declaration of Helsinki and was approved by the Institutional Review Board of the Nippon Medical School. A signed written informed consent was obtained from the patient and his parents after the nature and possible consequences of the study were explained.
Blood samples were collected from the patient and his parents, and genomic DNA was isolated from the peripheral white blood cells using a blood DNA isolation kit (NucleoSpin Blood XL; Macherey–Nagel, Germany). The DNA was used as the template to amplify the BEST1 gene. The coding regions and flanking introns of the BEST1 gene were amplified by polymerase chain reaction (PCR) using primers synthesized by Greiner Bio-One (Tokyo, Japan). The PCR products were purified (ExoSAP-IT; USB Corp., USA) and were used as the template for sequencing. Both strands were sequenced on an automated sequencer (Bio Matrix Research; Chiba, Japan).
The ophthalmological examinations included measurements of the best-corrected visual acuity (BCVA), refractive error and axial length, slit-lamp biomicroscopy, ophthalmoscopy, fundus photography, fundus autofluorescence (FAF) imaging, fluorescein angiography (FA), SD-OCT, full-field electroretinography (ERG), multifocal ERGs (mfERGs), and electro-oculography (EOG). The EOGs and ERGs were recorded using an extended testing protocol conforming to the International Society for Clinical Electrophysiology of Vision standards. The ERGs were elicited and recorded with a LED built-in electrode (LE2000, Tomey, Japan). The mfERGs were recorded using a commercial mfERG system (VERIS Science; Electro-Diagnostic Imaging, Inc. Redwood City, CA, USA). The FAF images were acquired with the TRC-NW8Fplus retinal camera (TOPCON, Tokyo, Japan), and the SD-OCT images were acquired with a Cirrus HD-OCT (Carl Zeiss Meditec).
Results
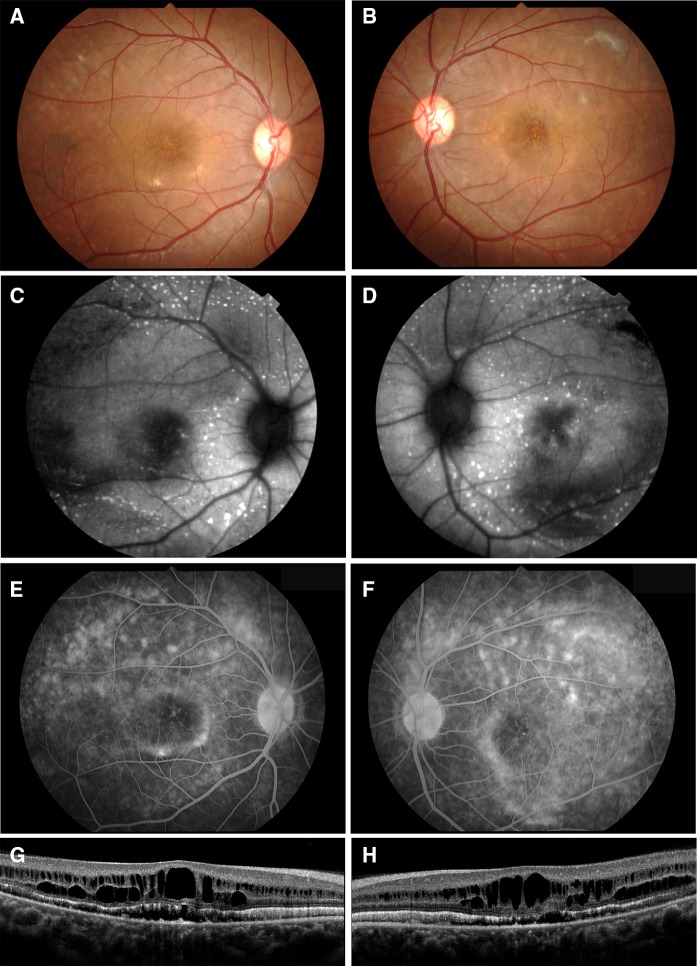
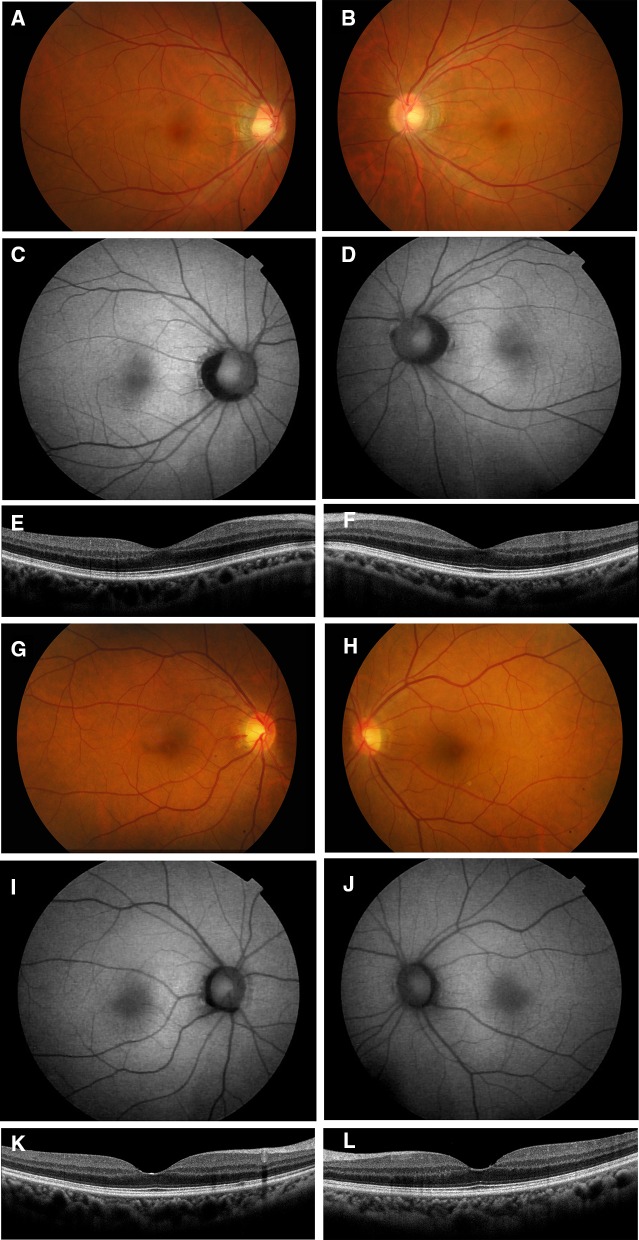
The patient was a 25-year-old man whose decimal best-corrected visual acuity (BCVA) was 0.9 in the right eye and 0.3 in the left eye. His refraction was S + 0.5 C-1.25 at 180 in the right eye and S + 0.5 C-2.0 at 175 in the left eye. Axial length was 23.71 mm in the right eye and 23.85 mm in the left eye. The intraocular pressure and anterior ocular segments were within normal limits in both eyes. Fundus examinations revealed a cystoid macular lesion and multiple yellowish deposits throughout the posterior pole of both eyes (Fig. 1). The vitelliform lesions that are typical of Best disease were not observed (Fig. 1). FAF imaging showed multiple hyper-autofluorescent spots in the peripheral retina of both eyes, and the site of the spots corresponded with the yellowish deposits observed by ophthalmoscopy (Fig. 1). FAF imaging also detected a hypo-autofluorescent lesion in the macula of both eyes (Fig. 1). FA showed widespread patchy hyper-fluorescence (Fig. 1). The SD-OCT images showed cystoid changes in the macula and shallow serous retinal detachments in both eyes. There was a thickening and hyper-reflectivity at the areas corresponding to ellipsoid and interdigitation zones of the photoreceptors in the SD-OCT images (Fig. 1).
Fig. 1.
Fundus photographs, autofluorescence images, fluorescein angiograms, and SD-OCT images from patient with autosomal recessive bestrophinopathy (ARB) (proband, II-1). Fundus photographs (a, b), autofluorescence images (c, d), fluorescein angiograms (e, f), and SD-OCT images (g, h) are shown. Results from the right eye (a, c, e, g) and left eye (b, d, f, h) are shown. Fundus photograph shows cystoid macular lesions and multiple yellowish deposits throughout the posterior pole of both eyes. FAF images show multiple hyper-autofluorescent regions in the peripheral retina of both eyes. FAF images also show a hypo-autofluorescent lesion in the macular of both eyes. Fluorescein angiograms show widespread patchy hyper-fluorescence. The SD-OCT images show cystoid macular changes and shallow serous retinal detachments in both eyes. There is also a thickening and hyper-reflectivity at the areas corresponding to the ellipsoid and interdigitation zones
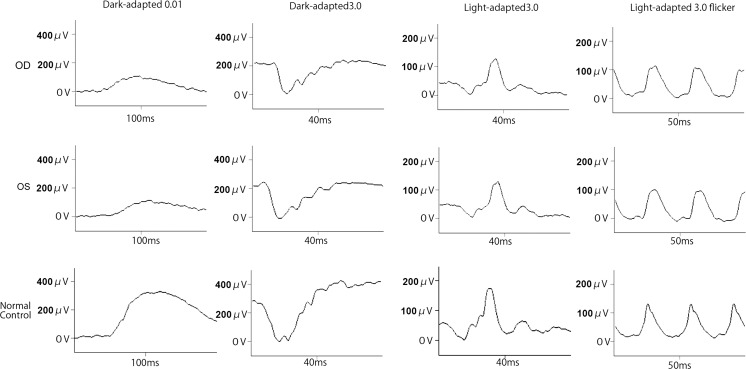
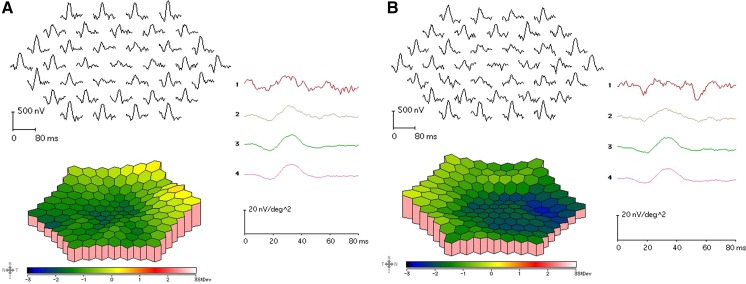
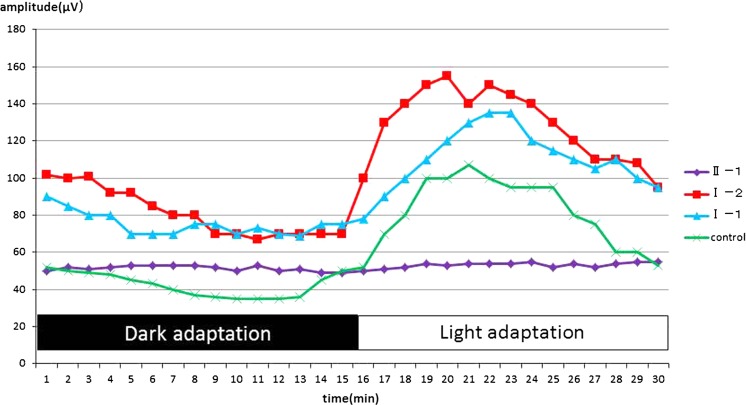
The amplitudes of both the cone and rod full-field ERGs were reduced, and the waveforms were similar in both eyes (Fig. 2). The amplitudes of the mfERGs were reduced in the central and peripheral sectors of both eyes (Fig. 3). The Arden ratio of the EOG was 1.1 in both eyes with a dark trough 15 min after beginning the measurements and a light peak 15 min from the beginning of the light phase (Fig. 4).
Fig. 2.
Full-field electroretinograms (ERGs). Full-field ERGs recorded from the right eye (top) and left eye (middle) of the proband (II-1) are shown. The ERGs recorded from a normal control are also shown (bottom). The dark-adapted 0.01, dark-adapted 3.0, light-adapted 3.0, and light-adapted 3.0 flicker ERGs are shown. The results of all the responses show a slight reduction of the b-wave amplitudes in both eyes
Fig. 3.
Multifocal ERGs. The mfERGs, topographic map, and average densities of the rings of the multifocal ERGs of right eye (a) and left eye (b) of the proband are shown. The amplitudes of the mfERGs in the foveal area are severely reduced in both eyes
Fig. 4.
Electro-oculograms (EOGs). The EOGs of the right eyes of the proband, proband’s parents, and normal control are shown. The Arden ratio of the EOG of the proband is markedly reduced with an absence of the light rise. The EOGs of the parents have a normal Arden ratio and light rise
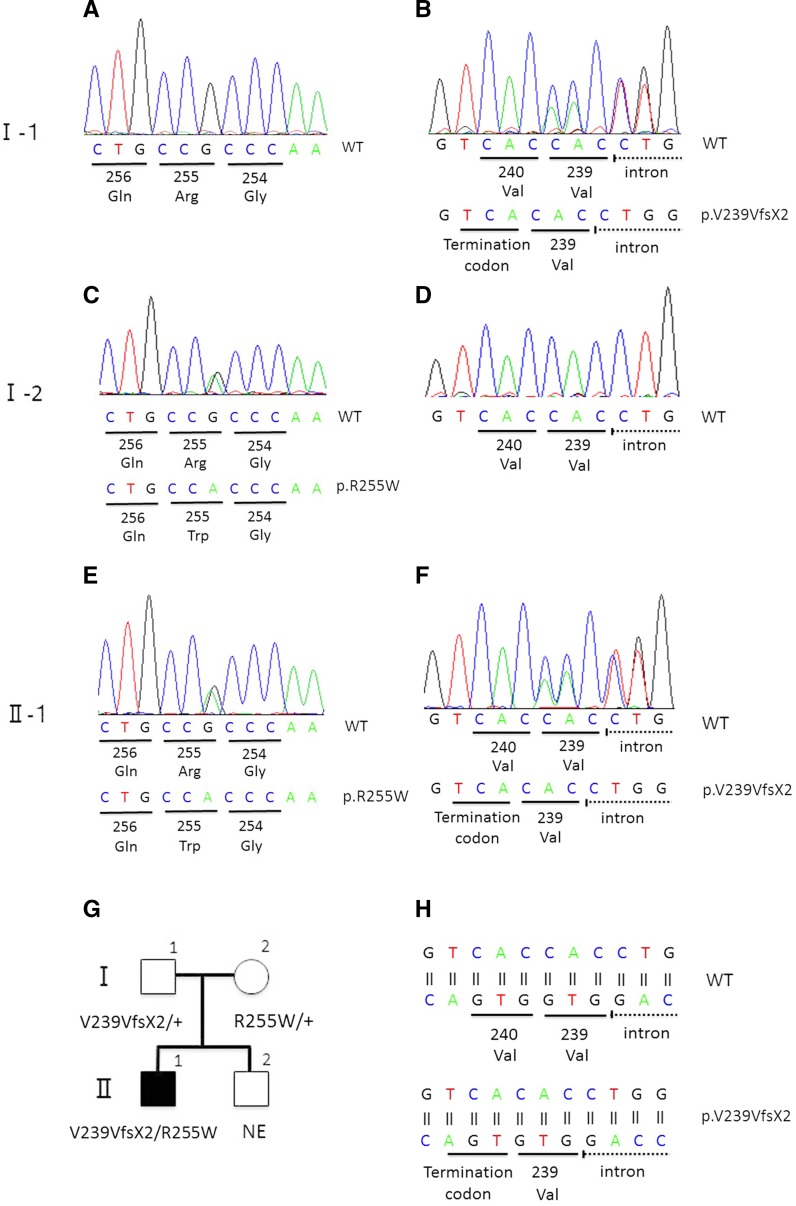
Mutation analysis of the BEST1 gene in the proband showed two heterozygous sequence variants. One was a novel variant, c.717delG, p.V239VfsX2, and the other was a variant previously reported, c.763C>T, p.R255W. Both variants were found in exon 7 (Fig. 5).
Fig. 5.
Molecular genetic findings and a pedigree chart. Sequence chromatograms of the proband’s father (I-1; a, b), mother (I-2; c, d), and the proband (II-1; e, f) are shown. Sequence chromatograms around the amino acid position 255 (a, c, e) and 240 (b, d, e) are shown. Results of reverse strand of the sequence chromatograms are shown (a–f). A single-nucleotide mutation (c.763C>T) results in the substitution of tryptophan for arginine at amino acid position 255 (p.R255W) in the mother and proband (c, e). A deletion mutation (c.717delG) results in the synonymous substitution of valine for valine at amino acid position 239 and a frame shift that leads to a premature termination codon at two amino acid residues downstream from the mutation (p.V239VfsX2) in the father and proband (b, f). Pedigree charts for the segregation analysis are shown (g). Schematic diagram of the deletion mutation (c.717delG) in the proband (bottom) and wild type (top) are shown (h). A frame shift mutation leads to a premature termination codon at two amino acid residues downstream from the mutation
The proband’s father (I-1, 57 years old) and mother (I-2, 57 years old) had normal visual acuity, and their fundus, FAF, and SD-OCT images were also within normal limits (Fig. 6). The EOGs of both parents had a normal light rise with normal Arden ratio in both eyes (Fig. 5). Mutation analyses of the parents identified a c.717delG, p.V239VfsX2 variant in the father and a c.763C>T, p.R255W variant in the mother in the heterozygous state.
Fig. 6.
Fundus photographs, fundus autofluorescence image, and SD-OCT images from the parents of the proband. Fundus photographs (a, b, g, h), autofluorescence (c, d, i, j), and SD-OCT images (e, f, k, l) are shown. Results from the father (a–f) and mother (g–l) are shown. Fundus appearance, FAF, and SD-OCT of the proband’s parents are normal
Discussion
The imaging and functional data obtained on our patient are in good agreement with the findings from previous reports of ARB. The characteristic features of ARB are a clinically recognizable retinal dystrophy with yellowish subretinal lesions scattered in the posterior pole that have marked diffuse fundus autofluorescence abnormalities [18–24]. The SD-OCT findings of previous ARB cases included diffuse intraretinal cystic spaces across both the inner and outer plexiform layers, subretinal fluid with shallow serous retinal detachment, and thickening and hyper-reflectivity of the ellipsoid and interdigitation zones which may represent an elongation of the photoreceptors [21, 24, 25]. The Arden ratio of the EOGs of patients with ARB is reported to be low with an absence of the light rise [11, 19, 23]. The imaging and functional findings in our patient are typical of ARB.
The BEST1 mutation, c.717delG, p.V239VfsX2, has not been reported and not included in the SNP database. The allele frequency of the variant was estimated from two databases; the Human Genetic Variation Database (HGVD; http://www.genome.med.kyoto-u.ac.jp/SnpDB/about.htm) which is specific for the Japanese population, and the ExAC Browser (Beta)(http://exac.broadinstitute.org) database. Both of these databases did not contain the allele frequency of the variant, which indicates that this variant is very rare.
Although most mutations associated with BVMD are missense mutations that do not compromise protein synthesis, the few ARB-causing mutations reported to date are premature truncations or nonsense substitutions that lead to early transcript degradation or non-functional proteins. These are associated with a null phenotype (Table 1). In truncating BEST1 mutations, the null phenotype associated with ARB is attributed to a severe decrease in BEST1 expression promoted by the nonsense-mediated decay (NMD) surveillance mechanism [26]. Recent evidence supports the idea that NMD degradation depends on the position of the premature translation termination codons. Pomares et al. [26] reported that the BEST1 transcripts in a patient who carried the premature stop codon at position 230 are preserved in only 13 % of the case, while the BEST1 transcripts of a patient who carry a premature stop codon in position 349 are preserved in 22 % of the case. Patients who carry the premature stop codon in position 230 have a characteristic ARB phenotype, while patients who carry a premature stop codon in position 349 have ophthalmological features resembling both ARB and BVMD [26]. Thus, the residual amount of aberrant protein can promote a negative effect causing a mixed phenotype of both ARB and BVMD traits. This hypothesis was supported by previous reports of biallelic BEST1 mutations with at least a premature termination codon (Table 1). Although patients 2 and 6 of Table 1 had the BVMD phenotype which is not consistent with the hypothesis, the same second allele mutation (R141H) may be associated with the BVMD phenotype [21, 27]. Our patient with a premature termination codon at position 240 is consistent with the hypothesis that the patient should have an ARB phenotype.
Table 1.
Summary of previously reported biallelic BEST1 mutations with premature termination codon
| Patient number | Allele 1 | Allele 2 | ||||||
|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino acid | Termination position | Nucleotide | Amino acid | Termination position | Fundus appearance | References | |
| 1 | c.15C>A | p.Y5X | 5 | c.430A>G | p.S144G | ARB | Lacassagne et al. [30] | |
| 2 | c.87C>G | p.Y29X | 29 | c.422G>A | p.R141H | BVMD | Schatz et al. [27] | |
| 3 | c.102C>T | p.E35WfsX11 | 45 | c.1470_1471delCA | p.H490QfsX24 | 513 | ARB | Davidson et al. [18] |
| 4 | c.172_173dupCA | p.Q58HfsX4 | 61 | c.584C>T | p.A195V | ARB | Borman et al. [21] | |
| 5 | c.175_176dupCA | p.Q59HfsX3 | 61 | c.175_176dupCA | p.Q59HfsX3 | 61 | ARB | Boon et al. [22] |
| 6 | c.475C>T | p.Q159X | 159 | c.422G>A | p.R141H | BVMD | Borman et al. [21] | |
| 7 | c.519delA | p.K173NfsX2 | 174 | c.860G>A | p.W287X | 287 | ARB | Tian et al. [29] |
| 8 | c.598C>T | p.R200X | 200 | c.598C>T | p.R200X | 200 | ARB | Burgess et al. [11] |
| 9 | c.263_279del17 | p.L88LfsX138 | 225 | c.584C>T | p.A195V | ARB | Gerth et al. [25] | |
| 10 | c.521_522delTG | p.L174EfsX57 | 230 | c.521_522delTG | p.L174EfsX57 | 230 | ARB | Pomares et al. [26] |
| 11 | c.762delG | p.R255GfsX4 | 258 | c.74G>A | p.R25Q | ARB | Boon et al. [22] | |
| 12 | c.1100 + 1G>A | p.V317PfsX33 | 349 | c.1100 + 1G>A | p.V317PfsX33 | 349 | ARB + BVMD | Pomares et al. [26] |
| 13 | c.1066C>T | p.R356X | 356 | c.550C>T | p.P184S | ARB | Borman et al. [21] | |
| 14 | c.1038duC | p.Y347LfsX54 | 400 | c.553A>C | p.H178P | ARB | Borman et al. [21] | |
| 15 | c.1212delC | p.P404PfsX78 | 481 | c.637G>A | p.E213K | ARB | Silva et al. [31] | |
| 16 | c.1415delT | p.L472PfsX10 | 481 | c.1415delT | p.L472PfsX10 | 481 | BVMD | Bitner et al. [16] |
| 17 | c.1470_1471delCA | p.H490QfsX24 | 513 | c.584C>T | p.A195V | BVMD | Kinnick et al. [32] | |
| 18 | c.1669delG | p.E557NfsX52 | 608 | c.934G>A | p.D312N | BVMD | Sodi et al. [33] | |
The other mutation found in this study (R255W) was reported to be present in both a BVMD family in the heterozygous state and two ARB families in the compound heterozygous state [28, 29]. In the BVMD family with the R255W mutation, the parents of the proband were not genetically examined [28]. In the ARB families with the R255W mutation, each parent of the proband was heterozygous carriers of the R255W mutation and they were healthy [29]. Our data do not explain why the mother of our patient who carried the heterozygous R255W mutation did not have BVMD. One possibility is that the mutation exhibits reduced penetrance for the phenotype. The other possibility is that the previously described BVMD patient who had heterozygous c.763C>T, p.R255W mutation may have had an undiscovered second allele mutation such as a large deletion.
In some cases, it is difficult to differentiate ARB from BVMD and to speculate on the prognosis of the disease. Identifying the genetic defect of BEST1 gene and position of the premature termination codon may help in differentiating the ARB from BVMD and predict the prognosis of the disease.
Acknowledgments
We thank Professor Emeritus Duco Hamsaki of the Bascom Palmer Eye Institute, University of Miami School of Medicine, Miami, FL, for discussions and editing our manuscript.
Compliance with ethical standards
Conflict of interest
All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest, or non-financial interest in the subject matter or materials discussed in this manuscript.
Statements of human rights
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Statement of welfare of animals
No animals were involved in the study.
References
- 1.Petrukhin K, Koisti MJ, Bakall B, Li W, Xie G, Marknell T, Sandgren O, Forsman K, Holmgren G, Andreasson S, Vujic M, Bergen AA, McGarty-Dugan V, Figueroa D, Austin CP, Metzker ML, Caskey CT, Wadelius C. Identification of the gene responsible for Best macular dystrophy. Nat Genet. 1998;19:241–247. doi: 10.1038/915. [DOI] [PubMed] [Google Scholar]
- 2.Marmorstein AD, Marmorstein LY, Rayborn M, Wang X, Hollyfield JG, Petrukhin K. Bestrophin, the product of the Best vitelliform macular dystrophy gene (VMD2), localizes to the basolateral plasma membrane of the retinal pigment epithelium. Proc Natl Acad Sci USA. 2000;97:12758–12763. doi: 10.1073/pnas.220402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun H, Tsunenari T, Yau KW, Nathans J. The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc Natl Acad Sci USA. 2002;99:4008–4013. doi: 10.1073/pnas.052692999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenthal R, Bakall B, Kinnick T, Peachey N, Wimmers S, Wadelius C, Marmorstein A, Strauss O. Expression of bestrophin-1, the product of the VMD2 gene, modulates voltage-dependent Ca2+ channels in retinal pigment epithelial cells. FASEB J. 2006;20:178–180. doi: 10.1096/fj.05-4495fje. [DOI] [PubMed] [Google Scholar]
- 5.Qu Z, Hartzell HC. Bestrophin Cl-channels are highly permeable to HCO3. Am J Physiol Cell Physiol. 2008;294:C1371–C1377. doi: 10.1152/ajpcell.00398.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boon CJ, Klevering BJ, Leroy BP, Hoyng CB, Keunen JE, den Hollander AI. The spectrum of ocular phenotypes caused by mutations in the BEST1 gene. Prog Retin Eye Res. 2009;28:187–205. doi: 10.1016/j.preteyeres.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Krämer F, White K, Pauleikhoff D, Gehrig A, Passmore L, Rivera A, Rudolph G, Kellner U, Andrassi M, Lorenz B, Rohrschneider K, Blankenagel A, Jurklies B, Schilling H, Schütt F, Holz FG, Weber BH. Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration. Eur J Hum Genet. 2000;8:286–292. doi: 10.1038/sj.ejhg.5200447. [DOI] [PubMed] [Google Scholar]
- 8.Yardley J, Leroy BP, Hart-Holden N, Lafaut BA, Loeys B, Messiaen LM, Perveen R, Reddy MA, Bhattacharya SS, Traboulsi E, Baralle D, De Laey JJ, Puech B, Kestelyn P, Moore AT, Manson FD, Black GC. Mutations of VMD2 splicing regulators cause nanophthalmos and autosomal dominant vitreo-retinochoroidopathy (ADVIRC) Invest Ophthalmol Vis Sci. 2004;45:3683–3689. doi: 10.1167/iovs.04-0550. [DOI] [PubMed] [Google Scholar]
- 9.Davidson AE, Millar ID, Urquhart JE, Burgess-Mullan R, Shweikh Y, Parry N, O’Sullivan J, Maher GJ, McKibbin M, Downes SM, Lotery AJ, Jacobson SG, Brown PD, Black GC, Manson FD. Missense mutations in a retinal pigment epithelium protein, bestrophin-1, cause retinitis pigmentosa. Am J Hum Genet. 2009;85:581–592. doi: 10.1016/j.ajhg.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaelides M, Urquhart J, Holder GE, Restori M, Kayali N, Manson FD, Black GC. Evidence of genetic heterogeneity in MRCS (microcornea, rod-cone dystrophy, cataract, and posterior staphyloma) syndrome. Am J Ophthalmol. 2006;141:418–420. doi: 10.1016/j.ajo.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Burgess R, Millar ID, Leroy BP, Urquhart JE, Fearon IM, De Baere E, Brown PD, Robson AG, Wright GA, Kestelyn P, Holder GE, Webster AR, Manson FD, Black GC. Biallelic mutation of BEST1 causes a distinct retinopathy in humans. Am J Hum Genet. 2008;82:19–31. doi: 10.1016/j.ajhg.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toto L, Boon CJ, Di Antonio L, Battaglia Parodi M, Mastropasqua R, Antonucci I, Stuppia L, Mastropasqua L (2015) BESTROPHINOPATHY: a spectrum of ocular abnormalities caused by the c.614T>C mutation in the BEST1 gene. Retina. doi:10.1097/IAE.0000000000000950 [DOI] [PubMed]
- 13.Krämer F, Mohr N, Kellner U, Rudolph G, Weber BH. Ten novel mutations in VMD2 associated with Best macular dystrophy (BMD) Hum Mutat. 2003;22:418. doi: 10.1002/humu.9189. [DOI] [PubMed] [Google Scholar]
- 14.Lotery AJ, Munier FL, Fishman GA, Weleber RG, Jacobson SG, Affatigato LM, Nichols BE, Schorderet DF, Sheffield VC, Stone EM. Allelic variation in the VMD2 gene in best disease and age-related macular degeneration. Invest Ophthalmol Vis Sci. 2000;41:1291–1296. [PubMed] [Google Scholar]
- 15.Boon CJ, Klevering BJ, den Hollander AI, Zonneveld MN, Theelen T, Cremers FP, Hoyng CB. Clinical and genetic heterogeneity in multifocal vitelliform dystrophy. Arch Ophthalmol. 2007;125:1100–1106. doi: 10.1001/archopht.125.8.1100. [DOI] [PubMed] [Google Scholar]
- 16.Bitner H, Mizrahi-Meissonnier L, Griefner G, Erdinest I, Sharon D, Banin E. A homozygous frameshift mutation in BEST1 causes the classical form of Best disease in an autosomal recessive mode. Invest Ophthalmol Vis Sci. 2011;52:5332–5338. doi: 10.1167/iovs.11-7174. [DOI] [PubMed] [Google Scholar]
- 17.Burgess R, MacLaren RE, Davidson AE, Urquhart JE, Holder GE, Robson AG, Moore AT, Keefe RO, Black GC, Manson FD. ADVIRC is caused by distinct mutations in BEST1 that alter pre-mRNA splicing. J Med Genet. 2009;46:620–625. doi: 10.1136/jmg.2008.059881. [DOI] [PubMed] [Google Scholar]
- 18.Davidson AE, Sergouniotis PI, Burgess-Mullan R, Hart-Holden N, Low S, Foster PJ, Manson FD, Black GC, Webster AR. A synonymous codon variant in two patients with autosomal recessive bestrophinopathy alters in vitro splicing of BEST1. Mol Vis. 2010;16:2916–2922. [PMC free article] [PubMed] [Google Scholar]
- 19.Guerriero S, Preising MN, Ciccolella N, Causio F, Lorenz B, Fischetto R. Autosomal recessive bestrophinopathy: new observations on the retinal phenotype—clinical and molecular report of an Italian family. Ophthalmologica. 2011;225:228–235. doi: 10.1159/000324472. [DOI] [PubMed] [Google Scholar]
- 20.Piñeiro-Gallego T, Álvarez M, Pereiro I, Campos S, Sharon D, Schatz P, Valverde D. Clinical evaluation of two consanguineous families with homozygous mutations in BEST1. Mol Vis. 2011;17:1607–1617. [PMC free article] [PubMed] [Google Scholar]
- 21.Borman AD, Davidson AE, O’Sullivan J, Thompson DA, Robson AG, De Baere E, Black GC, Webster AR, Holder GE, Leroy BP, Manson FD, Moore AT. Childhood-onset autosomal recessive bestrophinopathy. Arch Ophthalmol. 2011;129:1088–1093. doi: 10.1001/archophthalmol.2011.197. [DOI] [PubMed] [Google Scholar]
- 22.Boon CJ, van den Born LI, Visser L, Keunen JE, Bergen AA, Booij JC, Riemslag FC, Florijn RJ, van Schooneveld MJ. Autosomal recessive bestrophinopathy: differential diagnosis and treatment options. Ophthalmology. 2013;120:809–820. doi: 10.1016/j.ophtha.2012.09.057. [DOI] [PubMed] [Google Scholar]
- 23.Sharon D, Al-Hamdani S, Engelsberg K, Mizrahi-Meissonnier L, Obolensky A, Banin E, Sander B, Jensen H, Larsen M, Schatz P. Ocular phenotype analysis of a family with biallelic mutations in the BEST1 gene. Am J Ophthalmol. 2014;157:697–709. doi: 10.1016/j.ajo.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Fung AT, Yzer S, Goldberg N, Wang H, Nissen M, Giovannini A, Merriam JE, Bukanova EN, Cai C, Yannuzzi LA, Tsang SH, Allikmets R. New best1 mutations in autosomal recessive bestrophinopathy. Retina. 2015;35:773–782. doi: 10.1097/IAE.0000000000000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerth C, Zawadzki RJ, Werner JS, Héon E. Detailed analysis of retinal function and morphology in a patient with autosomal recessive bestrophinopathy (ARB) Doc Ophthalmol. 2009;118:239–246. doi: 10.1007/s10633-008-9154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pomares E, Burés-Jelstrup A, Ruiz-Nogales S, Corcóstegui B, González-Duarte R, Navarro R. Nonsense-mediated decay as the molecular cause for autosomal recessive bestrophinopathy in two unrelated families. Invest Ophthalmol Vis Sci. 2012;53:532–537. doi: 10.1167/iovs.11-7964. [DOI] [PubMed] [Google Scholar]
- 27.Schatz P, Klar J, Andréasson S, Ponjavic V, Dahl N. Variant phenotype of Best vitelliform macular dystrophy associated with compound heterozygous mutations in VMD2. Ophthalmic Genet. 2006;27:51–56. doi: 10.1080/13816810600677990. [DOI] [PubMed] [Google Scholar]
- 28.Wong RL, Hou P, Choy KW, Chiang SW, Tam PO, Li H, Chan WM, Lam DS, Pang CP, Lai TY. Novel and homozygous BEST1 mutations in Chinese patients with Best vitelliform macular dystrophy. Retina. 2010;30:820–827. doi: 10.1097/IAE.0b013e3181c700c1. [DOI] [PubMed] [Google Scholar]
- 29.Tian R, Yang G, Wang J, Chen Y. BEST1 gene mutations in Chinese patients with bestrophinopathy. Mol Vis. 2014;20:1594–1604. [PMC free article] [PubMed] [Google Scholar]
- 30.Lacassagne E, Dhuez A, Rigaudière F, Dansault A, Vêtu C, Bigot K, Vieira V, Puech B, Defoort-Dhellemmes S, Abitbol M. Phenotypic variability in a French family with a novel mutation in the BEST1 gene causing multifocal best vitelliform macular dystrophy. Mol Vis. 2011;17:309–322. [PMC free article] [PubMed] [Google Scholar]
- 31.Silva RA, Berrocal AM, Lam BL, Albini TA (2013) Novel mutation in BEST1 associated with retinoschisis. JAMA Ophthalmol 131:794–798. Erratum in (2013): JAMA Ophthalmol 131:1249 [DOI] [PubMed]
- 32.Kinnick TR, Mullins RF, Dev S, Leys M, Mackey DA, Kay CN, Lam BL, Fishman GA, Traboulsi E, Iezzi R, Stone EM. Autosomal recessive vitelliform macular dystrophy in a large cohort of vitelliform macular dystrophy patients. Retina. 2011;31:581–595. doi: 10.1097/IAE.0b013e318203ee60. [DOI] [PubMed] [Google Scholar]
- 33.Sodi A, Menchini F, Manitto MP, Passerini I, Murro V, Torricelli F, Menchini U. Ocular phenotypes associated with biallelic mutations in BEST1 in Italian patients. Mol Vis. 2011;17:3078–3087. [PMC free article] [PubMed] [Google Scholar]