Abstract
Few single nucleotide polymorphisms (SNPs) associated with the risk of renal cell carcinoma (RCC) have been identified, yet genetic predisposition contributes significantly to this malignancy. We previously showed that follistatin-like 1 (FSTL1) was significantly down-regulated in clear cell RCC (ccRCC), in particular metastatic ccRCC. In the present study, we systemically investigated the associations of the 6 SNPs within FSTL1-coding genomic region with RCC risk and postoperative prognosis. Age- and gender-matched case-control study (417 vs 855) indicated that rs1259293 variant genotype CC was significantly associated with an increased risk of RCC, with an odds ratio of 2.004 (95% confidence internal [CI] = 1.190–3.375). Multivariate Cox regression analysis in 309 of 417 cases showed that rs1259293 genotype (CC vs TT + CT) independently predicted an unfavorable prognosis, with a hazard ratio of 2.531 (95% CI = 1.052–6.086). Expression of FSTL1 was significantly higher in adjacent renal tissues than in tumors, and significantly higher in the tissues with rs1259293 TT genotype than in those with rs1259293 TC+CC genotypes. rs1259293 C allele might generate a CTCF binding site that blocks trans-activation of FSTL1 expression. Our results indicate that rs1259293 is associated with an increased risk and unfavorable postoperative prognosis of RCC, possibly by down-regulating FSTL1 expression in renal tissues.
Renal cell carcinoma (RCC) is the major cancer type in kidney and accounts for about 3% of all human malignancies, with a male-to-female ratio of approximately 2:11,2. It derives from renal tubular epithelial cells. Clear cell RCC (ccRCC) accounts for 80% of RCC. A recent twin study carried out in nordic countries has demonstrated that genetic factors contribute greatly to the occurrence of RCC3. Genome-wide association studies (GWAS) in western populations have shown that two loci mapped to endothelial PAS domain protein 1 (EPAS1) gene, encoding hypoxia inducible factor (HIF)-2α, on 2p21 (rs11894252 and rs7579899), a locus on 11q13.3 (rs7105934), a locus mapped to SCARB1, encoding the scavenger receptor class B, member 1, on 12q24.31 (rs4765623), a locus at a transcriptional enhancer of cyclin D1-encoding (CCND1) gene (rs7105934) at 11q13.3, two loci (rs718314 and rs1049380) in the inositol 1,4,5-triphosphate receptor, type 2 (ITPR2) gene on 12p11.23, and a locus (rs35252396) located at 8q24.21were significantly associated with RCC susceptibility4,5,6,7. However, these GWAS-identified RCC-risk single nucleotide polymorphisms (SNPs) identified in western populations are rarely replicated in Chinese populations8,9. Interestingly, the RCC-risk polymorphisms in genes important for reductive metabolism of chemical exposure in western populations are also not replicated in Chinese10,11,12,13. The genetic polymorphisms and their biological effects on signaling proteins that promote RCC development are largely unknown in Chinese population.
In our previous study investigating global gene expression profiling in RCC cells with different metastatic potentials, we showed that follistatin-like 1 (FSTL1) was significantly down-regulated in metastatic ccRCC compared to primary ccRCC cells; furthermore, the mRNA levels of FSTL1 were also significantly lower in ccRCC tissues than in adjacent renal tissue14. FSTL1, located on human chromosome 3, can stimulate cell cycle entry and division of pre-existing cardiomyocytes, thus improving cardiac function15. FSTL1 functions in cardio-renal communication. The lack of FSTL1 production by myocytes promotes glomerular and tubulointerstitial damage in the kidney16. FSTL1 is locally expressed in the loop of Henle in the kidney, and play a role in protecting the kidney from acute nephrotoxic injury via mediating interleukin-1β suppression17. The role of FSTL1 in cancer is complex and controversial. In cancer metastatic to bone, FSTL1 can mediate cancer cell invasion18. In ovarian and endometrial cancers, FSTL1 functions as a tumor suppressor through pro-apoptotic activities19. However, to our knowledge, the role of SNPs that affect FSTL1 expression within the context of malignancy had not been reported.
In the present study, a locus mapped to FSTL1 was not only proven to be associated with RCC risk, but also predicted postoperative prognosis of RCC. The risk genotype of rs1259293 was significantly correlated to reduced FSTL1 expression in adjacent renal tissues. Our results indicate that variant genotype of rs1259293 facilitates development of RCC by down-regulating FSTL1.
Results
Characteristics of study population and SNPs
A total of 417 RCC patients including 368 ccRCC cases and 855 healthy controls were enrolled in this study. Age and gender were matched between 417 cases and 855 controls (Table 1). Of the 417 RCC patients enrolled, 108 including 94 ccRCC patients were lost to follow-up after surgery. There were no statistically significant differences in age, gender, and tumor stage between the 309 patients enrolled and the 108 patients excluded in prognosis analysis (data not shown).
Table 1. Demographic and pathological characteristics of the study subjects.
| Characteristic | Case-control study |
Survival analysis | ||
|---|---|---|---|---|
| RCC cases (%) N = 417 | Controls (%) N = 855 | P | RCC cases (%) N = 309 | |
| Age (years) | ||||
| Mean ± SD | 56.82 ± 12.85 | 58.44 ± 15.24 | 0.543 | 56.88 ± 13.06 |
| ≤60 | 258 (61.87) | 544 (63.63) | – | 190 (61.49) |
| >60 | 159 (38.13) | 311 (36.37) | – | 119 (38.51) |
| Gender | ||||
| Male | 281 (67.39) | 609 (71.23) | 0.161 | 216 (69.90) |
| Female | 136 (32.61) | 246 (28.77) | – | 93 (30.10) |
| Histology | ||||
| Clear cell | 368 (88.25) | – | – | 274 (88.67) |
| Papillary | 13 (3.12) | – | – | 11 (3.56) |
| Chromophobe | 10 (2.40) | – | – | 8 (2.59) |
| Unclassified | 26 (6.24) | – | – | 16 (5.18) |
| AJCC stage | ||||
| I–II | 373 (89.45) | – | – | 266 (86.08) |
| III–IV | 44 (10.55) | – | – | 43 (13.92) |
Abbreviation: AJCC = American Joint Committee on Cancer; RCC = renal cell carcinoma.
From the HapMap projects (www.hapmap.org), FSTL1 coding region was located at 121595817-121652515 of chromosome 3. On the basis of the information in the HapMap project Chinese Han population database, we selected candidate tagged FSTL1-related SNPs at an r2 threshold of 0.80 and minor allele frequency (MAF) of no less than 20% using the Haploview 4.2 program. Six tagged SNPs (rs1105219, rs1259293, rs1402372, rs2673704, rs11708686, and rs1259339) were selected (Supplementary Fig. 1). Of the 6 SNPs, rs11708686 was located in the 3′UTR and the other 5 SNPs were in the intron regions.
Association of FSTL1-related SNPs with the risk of RCC
The case-control study was designed to investigate the association of FSTL1-related SNPs with the risk of RCC. Six SNPs (rs1105219, rs1259293, rs1259339, rs1402372, rs2673704, and rs11708686) were genotyped in 417 cases and 855 healthy controls. All the six SNP candidates were conformed to Hardy-Weinberg equilibrium (HWE) in healthy controls (P > 0.05). Table 2 presents the genotype distributions of the 6 SNPs in healthy controls and RCC patients including ccRCC patients. Compared to healthy controls, the variant genotype CC of rs1259293 in the intron 2 of FSTL1 coding region was significantly associated with an increased risk of RCC, with an odds ratio (OR) of 2.004 and 95% confidence interval (CI) of 1.190–3.375 (P = 0.009). Similarly, the CC genotype of rs1259293 was also significantly associated with an increased risk of ccRCC (OR = 2.014, 95% CI = 1.171–3.463, P = 0.011). The other 5 SNPs (rs11708686, rs1105219, rs1259339, rs1402372, and rs2673704) were not significantly related to the risk of RCC or ccRCC.
Table 2. The associations of FSTL1 polymorphisms with the risk of RCC and ccRCC.
| Genotype | RCC cases No (%) | ccRCC cases No (%) | Controls No (%) | Adjusted OR (95% CI)a | P-valuea | Adjusted OR (95% CI)b | P-valueb |
|---|---|---|---|---|---|---|---|
| rs11708686 (RCC n = 412, ccRCC n = 364, Control n = 836) H-W P = 0.415 | |||||||
| GG | 70 (16.99) | 64 (17.58) | 154 (18.42) | 1.000 (Reference) | – | – | – |
| AG | 203 (49.27) | 178 (48.90) | 423 (50.60) | 1.069 (0.769–1.486) | 0.691 | 1.027 (0.730–1.445) | 0.877 |
| AA | 139 (33.74) | 122 (33.52) | 259 (30.98) | 1.181 (0.830–1.682) | 0.355 | 1.136 (0.788–1.638) | 0.494 |
| AG+AA | 342 (83.01) | 300 (82.42) | 682 (81.58) | 1.115 (0.816–1.524) | 0.493 | 1.067 (0.773–1.474) | 0.692 |
| G allele | 343 (41.63) | 306 (42.00) | 731 (43.72) | 1.000 (Reference) | – | – | – |
| A allele | 481 (58.37) | 422 (58.00) | 941 (56.28) | 1.095 (0.925–1.298) | 0.291 | 1.077 (0.903–1.286) | 0.408 |
| rs1105219 (RCC n = 397, ccRCC n = 352, Control n = 829) H-W P = 0.262 | |||||||
| AA | 87 (21.91) | 77 (21.88) | 168 (20.27) | 1.000 (Reference) | – | – | – |
| AG | 205 (51.64) | 183 (51.99) | 428 (51.63) | 0.933 (0.685–1.270) | 0.658 | 0.935 (0.679–1.289) | 0.682 |
| GG | 105 (26.45) | 92 (26.14) | 233 (28.11) | 0.860 (0.607–1.218) | 0.395 | 0.854 (0.594–1.229) | 0.395 |
| AG+GG | 310 (78.09) | 275 (78.13) | 661 (79.73) | 0.906 (0.676–1.214) | 0.508 | 0.906 (0.669–1.229) | 0.527 |
| A allele | 379 (47.73) | 337 (47.87) | 764 (46.08) | 1.000 (Reference) | – | – | – |
| G allele | 415 (52.27) | 367 (52.13) | 894 (53.92) | 0.930 (0.785–1.103) | 0.405 | 0.927 (0.777–1.106) | 0.400 |
| rs2673704 (RCC n = 412, ccRCC n = 362, Control n = 851) H-W P = 0.170 | |||||||
| TT | 21 (5.10) | 18 (4.97) | 34 (4.00) | 1.000 (Reference) | – | – | – |
| CT | 135 (32. 77) | 124 (34.25) | 241 (28.32) | 0.915 (0.509–1.644) | 0.765 | 0.982 (0.531–1.815) | 0.953 |
| CC | 256 (62.14) | 220 (60.77) | 576 (67.69) | 0.705 (0.400–1.243) | 0.228 | 0.710 (0.392–1.288) | 0.260 |
| CT + CC | 391 (94.90) | 344 (95.03) | 817 (96.00) | 0.770 (0.440–1.347) | 0.360 | 0.795 (0.442–1.429) | 0.443 |
| T allele | 177 (21.48) | 160 (22.10) | 309 (18.16) | 1.000 (Reference) | – | – | – |
| C allele | 647 (78.52) | 564 (77.90) | 1393 (81.84) | 0.810 (0.658–0.997) | 0.046 | 0.783 (0.632–0.971) | 0.026 |
| rs1259293 (RCC n = 417, ccRCC n = 368, Control n = 855) H-W P = 0.124 | |||||||
| TT | 247 (59.23) | 212 (57.61) | 515 (60.23) | 1.000 (Reference) | – | – | – |
| CT | 140 (33.57) | 130 (35.33) | 307 (35.91) | 0.946 (0.736–1.217) | 0.666 | 1.023 (0.788–1.327) | 0.864 |
| CC | 30 (7.19) | 26 (7.07) | 33 (3.86) | 2.004 (1.190–3.375) | 0.009 | 2.014 (1.171–3.463) | 0.011 |
| CT + CC | 170 (40.77) | 156 (42.39) | 340 (39.77) | 1.042 (0.820–1.323) | 0.737 | 1.112 (0.868–1.426) | 0.400 |
| T allele | 634 (76.02) | 554 (75.27) | 1337 (78.19) | 1.000 (Reference) | – | – | – |
| C allele | 200 (23.98) | 182 (24.73) | 373 (21.81) | 1.134 (0.932–1.380) | 0.209 | 1.179 (0.962–1.445) | 0.112 |
| rs1402372 (RCC n = 386, ccRCC n = 344, Control n = 835) H-W P = 0.744 | |||||||
| CC | 58 (15.03) | 52 (15.12) | 114 (13.65) | 1.000 (Reference) | – | – | – |
| AC | 172 (44.56) | 157 (45.64) | 395 (47.31) | 0.868 (0.603–1.251) | 0.449 | 0.889 (0.609–1.298) | 0.543 |
| AA | 156 (40.41) | 135 (39.24) | 326 (39.04) | 0.963 (0.664–1.397) | 0.844 | 0.928 (0.630–1.366) | 0.704 |
| AC+AA | 328 (84.97) | 292 (84.88) | 721 (86.35) | 0.909 (0.645–1.282) | 0.586 | 0.901 (0.631–1.287) | 0.567 |
| C allele | 288 (37.31) | 261 (37.94) | 623 (37.31) | 1.000 (Reference) | – | – | – |
| A allele | 484 (62.69) | 427 (62.06) | 1047 (62.69) | 1.004 (0.842–1.199) | 0.961 | 0.979 (0.815–1.176) | 0.820 |
| rs1259339 (RCC n = 386, ccRCC n = 340, Control n = 789) H-W P = 0.610 | |||||||
| CC | 21 (5.44) | 18 (5.29) | 34 (4.31) | 1.000 (Reference) | – | – | – |
| CT | 131 (33.94) | 109 (32.06) | 271 (34.35) | 0.788 (0.440–1.412) | 0.423 | 0.762 (0.412–1.407) | 0.385 |
| TT | 234 (60.62) | 213 (62.65) | 484 (61.34) | 0.794 (0.450–1.403) | 0.427 | 0.842 (0.464–1.529) | 0.573 |
| CT + TT | 365 (94.56) | 322 (94.71) | 755 (95.69) | 0.792 (0.452–1.386) | 0.413 | 0.813 (0.452–1.463) | 0.489 |
| C allele | 173 (22.41) | 145 (21.32) | 339 (21.48) | 1.000 (Reference) | – | – | – |
| A allele | 599 (77.59) | 535 (78.68) | 1239 (78.52) | 0.950 (0.771–1.170) | 0.627 | 1.010 (0.810–1.258) | 0.932 |
aRCC patients vs healthy controls.
bccRCC patients vs healthy controls.
Abbreviations: FSTL1 = follistatin-like 1; RCC = renal cell carcinoma; ccRCC = clear cell renal cell carcinoma; CI = confidence interval.
OR = odds ratio; H-W = Hardy-Weinberg.
FSTL1-related SNPs predicted postoperative prognosis in RCC
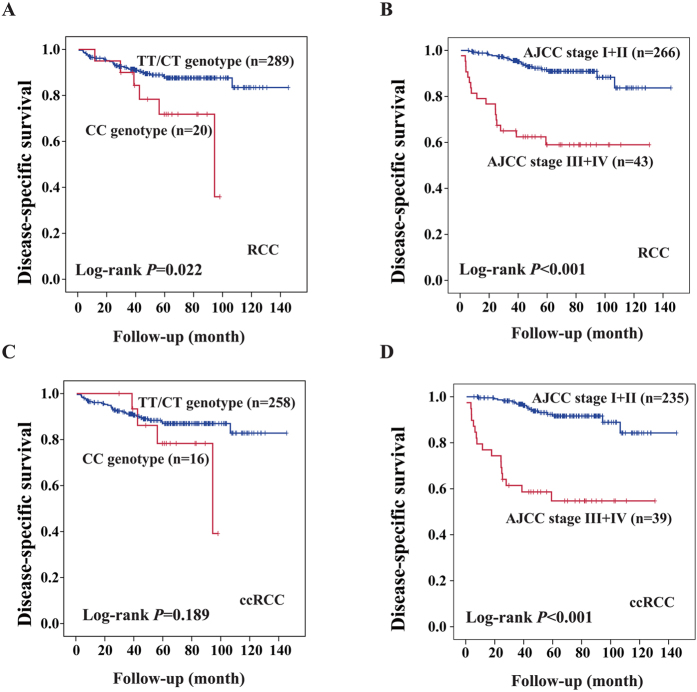
Of the 417 RCC patients, 309 RCC patients including 274 ccRCC patients were successfully followed up after surgery. Up to the last follow-up date on January 30, 2015, 38 patients including 34 ccRCC patients died of RCC following relapse. The association of each SNP with disease-specific survival (DSS) of RCC was evaluated using Kaplan-Meier analysis. It was found that the CC genotype of rs1259293 significantly predicted an unfavorable postoperative prognosis in RCC (Log-rank test, P = 0.022, Fig. 1A) and that none of other 5 SNPs were statistically associated with postoperative prognosis in RCC (data not shown). The CC genotype of rs1259293 did not significantly predict prognosis in ccRCC patients (Log-rank test, P = 0.189, Fig. 1C), however, the average survival durations of ccRCC patients with TT/CT genotypes and CC genotype of rs1259293 were 128.517 ± 3.074 and 85.455 ± 5.681 months, respectively. Advanced AJCC stages (III-IV vs I-II) also predicted an unfavorable postoperative prognosis in RCC (Log-rank test, P < 0.001, Fig. 1B); this effect was replicated in ccRCC (Log-rank test, P < 0.001, Fig. 1D). No significant differences in age, gender, and AJCC stage were found between RCC patients and ccRCC patients with TT/CT genotypes or CC genotypes. Multivariate Cox regression analysis including age, gender, AJCC stage, and rs1259293 genotype showed that AJCC stage (III- IV vs I-II) and rs1259293 genotype (CC vs TT + CT) conferred an unfavorable postoperative prognosis in RCC independently (Table 3).
Figure 1. Kaplan-Meier analysis and Log-rank test showed the effect of the factors significantly associated with the risk of renal cell carcinoma (RCC) on predicting postoperative prognosis in RCC.
(A) Comparison of the CC genotype and the TC and TT genotypes of rs1259293 in predicting disease-specific survival (DSS) in RCC. (B) Comparison of advanced AJCC stages (III+IV) and early stages (I+II) in predicting DSS in RCC. (C) Comparison of the CC genotype and the TC and TT genotypes of rs1259293 in predicting DSS in clear cell RCC (ccRCC). (D) Comparison of advanced AJCC stages (III+IV) and early stages (I+II) in predicting DSS in ccRCC.
Table 3. Factors significantly predicted disease-specific survival in multivariate Cox proportional hazards model in 309 RCC patients.
| Variables | HR (95% CI) | P |
|---|---|---|
| Age | 1.031 (1.004, 1.058) | 0.024 |
| Gender (male vs female) | 1.308 (0.641, 2.670) | 0.461 |
| AJCC stage (III–IV vs I–II) | 5.907 (3.045, 11.460) | <0.001 |
| rs1259293 genotype (CC vs TT + CT) | 2.531 (1.052, 6.086) | 0.038 |
Abbreviation: AJCC = American Joint Committee on Cancer; FSTL1 = follistatin-like 1; HR = hazard ratio; RCC = renal cell carcinoma.
Association of rs1259293 genotypes with FSTL1 expression in renal tissues
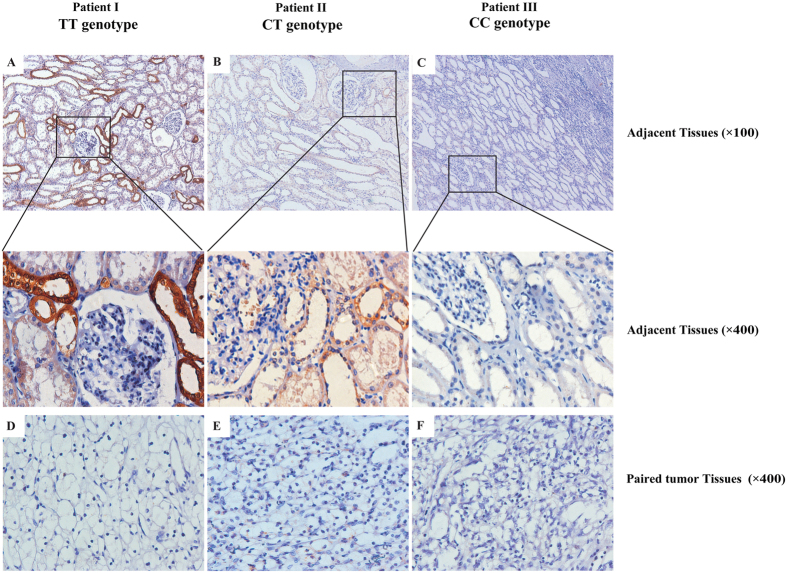
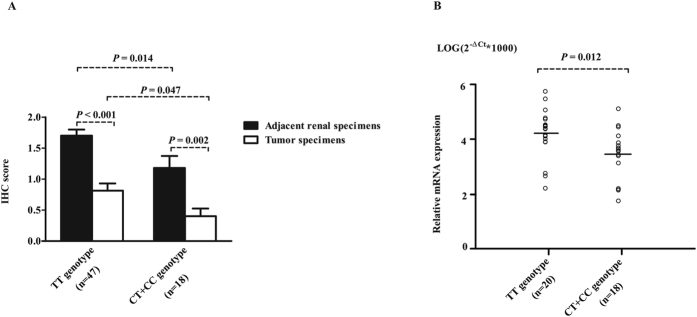
To determine whether rs1259293 genotype was related to the expression level of FSTL1 in the kidney, we examined FSTL1 expression in formalin-fixed paraffin-embedded (FFPE) specimens of ccRCC and the paired adjacent pathologically normal renal tissues from 65 patients using immunohistochemistry (IHC). In adjacent pathological normal renal tissues, FSTL1 immunostaining was selectively positive in the cytoplasm of the loop of Henle near distal convoluted tubules; moreover, the expression of FSTL1 was higher in renal tissues with rs1259293 TT genotype than in those with rs1259293 CT genotype and FSTL1 was almost negative in those with the CC genotype (Fig. 2). Spearman co-efficient test showed that rs1259293 TT genotypes (vs CT + CC genotype) was significantly correlated to higher IHC score of FSTL1 expression in adjacent renal tissues (P = 0.041) and but not significantly correlated to FSTL1 expression in tumors (Table 4). Furthermore, rs1259293 TT genotypes (vs CT + CC genotype) was significantly correlated to higher FSTL1 expression if the IHC scores of ccRCC and the paired adjacent normal renal specimens were combined as a value of a patient (P = 0.007). In RCC patients with rs1259293 TT genotype, IHC score of FSTL1 was significantly higher in adjacent normal renal tissues than in tumor tissues. As sample size of RCC patients with rs1259293 CC genotype was small (n = 5), we combined RCC patients with rs1259293 CC genotype and those with rs1259293 TC genotype. IHC score of FSTL1 was significantly higher in adjacent normal renal tissues than in tumor tissues in the RCC patients with rs1259293 CT + CC genotypes. The IHC score was also significantly higher in adjacent normal renal tissues of patients with rs1259293 TT genotype than in those with rs1259293 CT + CC genotypes; the same was true in tumor tissues. The expression level of FSTL1 mRNA was significantly higher in adjacent renal tissues with TT genotype than in those with CT + CC genotypes by detecting the corresponding FFPE specimens, which was accordance with the IHC results. These data were presented in Fig. 3.
Figure 2. Expression pattern of FSTL1 in adjacent normal renal tissues and paired ccRCC tissues using immunohistochemistry.
(A) Representative FSTL1 immunostaining in adjacent normal renal tissues of the patient with rs1259293 TT genotype. (B) Representative FSTL1 immunostaining in adjacent normal renal tissues of the patient with rs1259293 CT genotype. (C) Representative FSTL1 immunostaining in adjacent normal renal tissues of the patient with rs1259293 CC genotype. (D) Representative FSTL1 immunostaining in paired tumor tissues of the patient with rs1259293 TT genotype. (E) Representative FSTL1 immunostaining in paired tumor tissues of the patient with rs1259293 CT genotype. (F) Representative FSTL1 immunostaining in paired tumor tissues of the patient with rs1259293 CC genotype.
Table 4. The correlation between rs1259293 genotypes and FSTL1 expression in ccRCC tissues and paired adjacent normal tissues from 65 ccRCC patients using spearman association test.
| Paired samples | rs1259293 genotypes | IHC scores of FSTL1 No (%) |
P | rs | |
|---|---|---|---|---|---|
| −/+ | ++/+++ | ||||
| Adjacent normal tissues | TT | 18 (27.7) | 29 (44.6) | 0.041 | −0.255 |
| CT + CC | 12 (18.5) | 6 (9.2) | – | – | |
| ccRCC tissues | TT | 38 (58.5) | 9 (13.8) | 0.179 | −0.169 |
| CT + CC | 17 (26.2) | 1 (1.5) | – | – | |
| Adjacent normal tissues and ccRCC tissues | TT | 10 (15.4) | 37 (56.9) | 0.007 | −0.332 |
| CT + CC | 10 (15.4) | 8 (12.3) | – | – | |
Abbreviations: FSTL1 = follistatin-like 1; IHC = Immunohistochemistry; ccRCC = clear cell renal cell carcinoma.
Figure 3. Comparison of FSTL1 expression level in adjacent renal tissues and paired tumor tissues of the ccRCC patients.
(A) IHC scores of FSTL1 immunostaining in tumors and adjacent renal tissues of patients with rs1259293 TT genotype (n = 47) and those with rs1259293 TC/CC genotypes (n = 18). (B) The levels of FSTL1 mRNA in adjacent renal tissues of 20 newly enrolled patients from the 47 patients with rs1259293 TT genotype and all the 18 patients with rs1259293 TC/CC genotypes. IHC, immunohistochemistry.
Discussion
In this study, we showed that a genetic polymorphism at rs1259293, a locus that has never been linked to any disease so far, predisposed RCC risk. Compared to the major genotype TT, the variant genotype CC of rs1259293 was significantly associated with an increased risk of RCC; furthermore, the CC genotype (vs TT + CT) of rs1259293 predicted an unfavorable postoperative prognosis in RCC independently. Thus, the abundance of rs1259293 CC genotype can predispose the susceptibility and unfavorable prognoses of RCC. rs1259293 CC genotype predisposed the susceptibility of ccRCC; however, it did not significantly predict postoperative prognosis in ccRCC. This result is mainly due to small sample size and the long-term survival nature of ccRCC. In the 309 RCC patients involved in the survival study, 20 died of this malignancy; whereas 16 of 274 ccRCC patients died of this malignancy. Thus, 20.0% of deaths (4/20) were taken off from the subsequent prognostic analysis in ccRCC patients, which undoubtedly affected the statistical power. The average survival durations of ccRCC patients with TT/CT genotypes and CC genotype of rs1259293 were 128.517 ± 3.074 and 85.455 ± 5.681 months, respectively. No significant differences in age, gender, and AJCC stage were found between RCC and ccRCC patients with TT/CT genotypes or CC genotypes. The difference in the postoperative prognosis should be mainly contributed by rs1259293 genotypes. Although rs1259293 CC genotype did not predict an unfavorable postoperative prognosis in ccRCC statistically, the trend was quite apparent (Fig. 1C). Thus, we believe that rs1259293 is an important genetic risk factor of RCC on the basis of our results in a case-control study Chinese RCC patients because it is also an independent prognostic factor for RCC in the cohort study. Allelic frequencies of SNPs differ among populations with different racial backgrounds. We checked allelic frequencies of rs1259293 in African American, Caucasian American, and Chinese Han population in the HapMap project (www.hapmap.org). It was found that the frequency of CC genotype at rs1259293 was 5.3% in African American, 53% in Caucasian American, and 6.7% in Chinese Han population. Interestingly, the incidence of RCC was 12.5/105 in male Caucasian American and 6.7/105 in female Caucasian American; whereas the incidence of RCC was 5.5/105 in Chinese men and 2.7/105 in Chinese women20. Higher CC frequency at rs1259293 is associated with higher RCC incidence in Caucasian than in Chinese, suggesting that the CC genotype at rs1259293 plays a critical role in renal carcinogenesis. However, this relationship might be not evident between African American and Chinese, because the incidence of RCC was 15.2/105 in male African American and 7.3/105 in female African American20. This is possibly because other strong factors such as chronic kidney disease overwhelm the effect of rs1259293 CC genotype. We also found that urolithiasis independently increased the risk of RCC in Chinese13. Having a history of chronic kidney disease is associated with an almost 3-fold increased risk of RCC and this association is strongest among black people (OR = 10.4 [95% CI = 6.0–17.9])21. Nevertheless, the association of rs1259293 with RCC risk and RCC prognosis should be validated in populations with different racial background.
To elucidate the mechanisms by which the rs1259293 genotype predisposed the susceptibility and predicted postoperative prognosis in RCC, we investigated the association of the rs1259293 genotype with FSTL1 expression in tumors and adjacent renal tissues. We found that the IHC score of FSTL1 expression in adjacent renal tissues reduced consecutively from the cortex with rs1259293 TT genotype, those with rs1259293 TC genotype, and those with rs1259293 CC genotype (Fig. 2). The C allele at rs1259293 was proven to be significantly correlated to low IHC score of FSTL1 in adjacent normal renal tissues (Table 4). Expression of FSTL1 was significantly higher in adjacent normal renal tissues than in paired tumor tissues and significantly higher in the tumor or paired adjacent renal tissues of RCC patients with rs1259293 TT genotype than in those with rs1259293 CT + CC genotypes at the protein level (Fig. 3A). Interestingly, the level of FSTL1 mRNA was also significantly higher in adjacent renal tissues with rs1259293 TT genotype than in those with rs1259293 TC+CC genotypes (Fig. 3B). These results indicate that FSTL1 might be a tumor suppressor in RCC while rs1259293 C allele suppresses the transcription of FSTL1 gene. Furthermore, we searched ensemble database at http://asia.ensembl.org/Homo_sapiens/Variation/ and found that rs1259293 C allele, rather than the T allele, generated a CCCTC-binding factor (CTCF)-binding site. CTCF that can bind many enhancer-blocking elements is the only known major insulator-binding protein in the vertebrates and plays important roles in the barrier activity of insulators22. Reduced CTCF binding is associated with loss of insulation between topological domains and aberrant gene activation23. A genetic variant rs60507107 in the binding site of CTCF was found to be associated with an increased risk of lung cancer24. Enhanced binding of CTCF to the sequence with the C allele of rs1259293 may serve as an insulator that blocks active trans-activation of FSTL1 promoter and/or enhancer, thus reducing FSTL1 expression. Based on this straightforward mechanistic rationale and the results of our study, it is reasonable to speculate that FSTL1 is a tumor suppressor in RCC, and rs1259293 CC genotypes contribute to low FSTL1 expression, which therefore predicts a poor prognosis. The role of FSTL1 in RCC and its associations with RCC risk factors such as hypertension, obesity, and diabetes13,25,26 and RCC protective factors such as the use of statins or vitamin C27,28 merit extensive investigation.
In conclusion, the present study systemically investigated the associations of the 6 FSTL1-related SNPs with RCC risk and postoperative prognosis, and identified a new locus rs1259293 whose variant genotype significantly increased RCC risk and predicted an unfavorable postoperative prognosis. Expression of FSTL1 was significantly higher in adjacent normal renal tissues than in paired tumor tissues and significantly higher in the tumor or paired adjacent renal tissues of RCC patients with rs1259293 TT genotype than in those with rs1259293 CT + CC genotype. rs1259293 C allele may generate a CTCF-binding site increasing the binding of CTCF as insulator that blocks active trans-activation of FSTL1 enhancer, thus repressing the expression of FSTL1. Further large-scale, well-designed, different racial population-based studies are warranted to elucidate the impact of rs1259293 on RCC risk and postoperative prognosis.
Methods
Study population
Peripheral blood samples, tumor tissues and paired adjacent renal tissues were collected from the patients who received curative nephrectomy and were pathologically confirmed RCC at the 1st affiliated hospital of Second Military Medical University from Dec 1998 to Nov 2011. The histology for each case was re-confirmed by at least two pathologists. Healthy controls were recruited from Healthy Examination Center of the 1st affiliated hospital for individuals receiving routine physical examinations between May 2006 and November 2011. All healthy controls had no medical history of genetic diseases, chronic renal diseases or cancer. Demographic information was collected using standard questionnaire by checking their medical records. All participants were Han Chinese. This study was approved by the institutional review board of Second Military Medical University. The methods were carried out in accordance with the approved guidelines. The study subjects provided written informed consents.
Selection of SNPs
Online Haploview 4.2 software was applied to retrieve the FSTL1-related SNPs from Chinese Han population database in HapMap project (http://hapmap.ncbi.nlm.nih.gov/). Candidate tagged SNPs were selected based on following restrictions: (1) they were located at chromsome 3, 121595000–121653000 region; (2) each had minor allele frequency (MAF) of >20% in Chinese Han population, rs1259339 with MAF of 19% was manually added; (3) each had an r2 of >0.80. Thus, 6 tagged SNPs (rs1105219, rs1259293, rs1402372, rs2673704, rs11708686, and rs1259339) were selected.
Case-control study
The minimum sample size of case group was 376, which was determined by the formula of 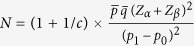 (α = 0.05, β = 0.10, c = 2, p0 = 0.2, OR = 1.6,
(α = 0.05, β = 0.10, c = 2, p0 = 0.2, OR = 1.6, 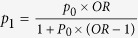 ,
, 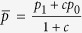 ,
, 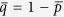 ). Thus, 417 RCC cases and 855 healthy controls met the need of the minimum sample size in this study. Cases and controls were age- and gender-matched on frequency. Of 417 RCC cases, 368 were diagnosed as ccRCC. Genomic DNA was isolated from peripheral blood of cases and healthy controls, and then subjected for genotyping using the fluorescent-probe real-time quantitative PCR assay as previously described29,30. The sequences of the primers and probes are listed in Supplementary Table 1. For quality control purposes, two blank controls were included in each 96-well plate, and more than 5% of samples were randomly selected for duplication, yielding a 100% concordance.
). Thus, 417 RCC cases and 855 healthy controls met the need of the minimum sample size in this study. Cases and controls were age- and gender-matched on frequency. Of 417 RCC cases, 368 were diagnosed as ccRCC. Genomic DNA was isolated from peripheral blood of cases and healthy controls, and then subjected for genotyping using the fluorescent-probe real-time quantitative PCR assay as previously described29,30. The sequences of the primers and probes are listed in Supplementary Table 1. For quality control purposes, two blank controls were included in each 96-well plate, and more than 5% of samples were randomly selected for duplication, yielding a 100% concordance.
Cohort study
All of the cases genotyped in case-control study were invited to participate in cohort study. The patients were followed up by phone or face-to-face interview every 6 months according to our standard epidemiologic procedure. We excluded the RCC patients who lost contact information and refused to adhere to the follow-up study requirements. The last follow up date was Jan 30, 2015, with a median follow-up duration of 56.56 months (interquartile range: 38.06–81.38 months). Death from RCC relapse was defined as an event. Patients alive at the last follow-up and died of conditions unrelated to RCC were censored. DSS was measured in months from the date of receiving surgery to the date that patient died of ccRCC.
IHC
Full sections of FFPE specimens of ccRCC and the paired adjacent normal renal tissues were processed using standard techniques. Antigen retrieval was conducted with in 0.01M Tris-EDTA buffer (pH = 8.0) for 25 min at 100 °C in an electric cooker. Sections were blocked and incubated overnight with anti-FSTL1 (C-term) (1:50 dilution; Abgent, AP10534b, San Diego, CA) overnight at 4 °C. FSTL1 was detected using DAB staining system. Staining evaluation was performed independently by three investigators (Tan XJ, Liu Y, and Yu YW) who were blind to the clinicopathological characteristics and outcome of the patients as previously described30. Briefly, an immunoreactive score was ranked by negative (−), slightly positive (+), moderately positive (++) and strongly positive (+++) according to the extent and intensity of staining. Furthermore, we accessed each pathological site of the adjacent normal renal tissues independently, and then summed up as the score of adjacent normal renal tissues. There was a close agreement on immunoreactive scores (90%) between two investigators. In cases of disagreement, consensus was obtained after discussion.
quantitative RT-PCR
The total of 38 FFPE specimens of adjacent renal tissues (20 with TT genotype; 18 with CT + CC genotypes) were involved in this assay. Total RNAs were isolated using RNeasy FFPE kit (Qiagen, 73504, Stockach, Germany) and reverse transcribed to cDNA, and subjected for quantitative RT-PCR as previously described30. The primers of FSTL1 were sense AAATGCAGCTCCCTGTCCAA and reverse ACTCTTGCCCTCCTCCCATAG. The primers of GAPDH were sense TGACTTCAACAGCGACACCCA and reverse CACCCTGTTGCTGTAGCCAAA. The relative normalized quantity of FSTL1 expression was calculated as previously described31.
Statistical analysis
HWE was examined by using online analytical tools (http://ihg.gsf.de). Demographic characteristics between cases and controls were analyzed using Chi-square test. Differences in continuous variables were tested by Student t test. Unconditional logistic regression model was conducted to calculate odds ratios (ORs) and their 95% confidence internals (CIs) of the association between the SNPs and RCC risk, adjusting for age and gender. Non-parametric analyses of Spearman correlation test was used to assess the correlation of rs1259293 genotypes to FSTL1 expression. For postoperative prognosis analysis, DSSs and their 95% CIs were estimated by the Kaplan-Meier method. The log-rank test was applied to compare DSS between groups. All statistical tests were two-sided and conducted using Statistical Program for Social Sciences (SPSS 16.0, Chicago, IL, USA). A P-value of <0.05 was considered as statistically significant.
Additional Information
How to cite this article: Liu, Y. et al. A genetic polymorphism affects the risk and prognosis of renal cell carcinoma: association with follistatin-like protein 1 expression. Sci. Rep. 6, 26689; doi: 10.1038/srep26689 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Key Basic Research Program (973 program) (2015CB554000 to GC) and the National Natural Science Foundation of China (81520108021 and 91529305 to G.C., 81101928 to X.T.).
Footnotes
The authors declare no competing financial interests.
Author Contributions Conceived and designed the experiments: G.C., X.T. and T.C.T. Supervised the study: G.C. Drafted and revised the manuscript: G.C. Performed the experiments: Y.L., X.T., Y.Y., Y.D. C.N., X.H. and Z.L. Participated in the sample collection and follow-up study: Y.L., X.H., X.T., J.H., D.S., W.L. and J.Y. Conducted statistical analysis: Y.L., X.H., X.T., H.Z. and G.C. Administration: G.C. All authors reviewed the manuscript.
References
- Ferlay J. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127, 2893–2917 (2010). [DOI] [PubMed] [Google Scholar]
- Han X. et al. Incidence and survival analysis of renal cell carcinoma patients among permanent residents in Yangpu district of Shanghai during 2002–2012. Acad J Sec Mil Med Univ 35, 8–13 (2014). [PubMed] [Google Scholar]
- Mucci L. A. et al. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA 315, 68–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue M. P. et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet 43, 60–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodel J. et al. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat Genet 44, 420–425, S421–422 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. et al. A genome-wide association study identifies a novel susceptibility locus for renal cell carcinoma on 12p11.23. Hum Mol Genet 21, 456–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J. et al. A common variant at 8q24.21 is associated with renal cell cancer. Nat Commun 4, 2776 (2013). [DOI] [PubMed] [Google Scholar]
- Cao Q. et al. Chromosome 11q13.3 variant modifies renal cell cancer risk in a Chinese population. Mutagenesis 27, 345–350 (2012). [DOI] [PubMed] [Google Scholar]
- Su T. et al. A GWAS-identified susceptibility locus on chromosome 11q13.3 and its putative molecular target for prediction of postoperative prognosis of human renal cell carcinoma. Oncol Lett 6, 421–426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. E. et al. Occupational trichloroethylene exposure and renal carcinoma risk: evidence of genetic susceptibility by reductive metabolism gene variants. Cancer Res 70, 6527–6536 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Shi H., Hou Q., Mo Z. & Xie X. GSTM1 and GSTT1 polymorphisms contribute to renal cell carcinoma risk: evidence from an updated meta-analysis. Sci Rep 5, 17971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F. D., Ma P., Sui C. G., Tian X. & Jiang Y. H. Association between cytochrome P450 1A1 (CYP1A1) gene polymorphisms and the risk of renal cell carcinoma: a meta-analysis. Sci Rep 5, 8108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang G. et al. Risk factor for clear cell renal cell carcinoma in Chinese population: a case-control study. Cancer Epidemiol 36, 177–182 (2012). [DOI] [PubMed] [Google Scholar]
- Tan X. et al. Global analysis of metastasis-associated gene expression in primary cultures from clinical specimens of clear-cell renal-cell carcinoma. Int J Cancer 123, 1080–1088 (2008). [DOI] [PubMed] [Google Scholar]
- Wei K. et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature 525, 479–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa S. et al. Cardiac myocyte-derived follistatin-like 1 prevents renal injury in a subtotal nephrectomy model. J Am Soc Nephrol 26, 636–646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D. C. et al. Follistatin-like 1 regulates renal IL-1β expression in cisplatin nephrotoxicity. Am J Physiol Renal Physiol 299, F1320–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo-Saito C., Fuwa T., Murakami K. & Kawakami Y. Targeting FSTL1 prevents tumor bone metastasis and consequent immune dysfunction. Cancer Res 73, 6185–6193 (2013). [DOI] [PubMed] [Google Scholar]
- Chan Q. K. et al. Tumor suppressor effect of follistatin-like 1 in ovarian and endometrial carcinogenesis: a differential expression and functional analysis. Carcinogenesis 30, 114–121 (2009). [DOI] [PubMed] [Google Scholar]
- Znaor A., Lortet-Tieulent J., Laversanne M., Jemal A. & Bray F. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol 67, 519–530 (2015). [DOI] [PubMed] [Google Scholar]
- Hofmann J. N. et al. Chronic kidney disease and risk of renal cell carcinoma: differences by race. Epidemiology 26, 59–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S. et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19, 24–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J. et al. Systematical analyses of variants in CTCF-binding sites identified a novel lung cancer susceptibility locus among Chinese population. Sci Rep 5, 7833 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weikert S. & Ljungberg B. Contemporary epidemiology of renal cell carcinoma: perspectives of primary prevention. World J Urol 28, 247–252 (2010). [DOI] [PubMed] [Google Scholar]
- Deckers I. A. et al. Polymorphisms in genes of the renin-angiotensin-aldosterone system and renal cell cancer risk: interplay with hypertension and intakes of sodium, potassium and fluid. Int J Cancer 136, 1104–1116 (2015). [DOI] [PubMed] [Google Scholar]
- Bravi F. et al. Food groups and renal cell carcinoma: a case-control study from Italy. Int J Cancer 120, 681–685 (2007). [DOI] [PubMed] [Google Scholar]
- Jia L., Jia Q., Shang Y., Dong X. & Li L. Vitamin C intake and risk of renal cell carcinoma: a meta-analysis. Sci Rep 5, 17921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y. et al. Polymorphism in protein tyrosine phosphatase receptor delta is associated with the risk of clear cell renal cell carcinoma. Gene 512, 64–69 (2013). [DOI] [PubMed] [Google Scholar]
- Tan X. et al. Genetic variation in the GSTM3 promoter confer risk and prognosis of renal cell carcinoma by reducing gene expression. Br J Cancer 109, 3105–3115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z. et al. IGFBP3 mRNA expression in benign and malignant breast tumors. Breast Cancer Res 9, R2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.