Abstract
Catalase (CAT), one antioxidant enzyme, may provide resistance against many diseases. Many previous studies reported predictive and prognostic values of CAT C262T polymorphism in cancers, with divergent results. This study aimed to summarize the overall relationships between CAT C262T polymorphism and cancer risk or survival. A total of 27 eligible publications were included in susceptibility analysis, while 8 publications contained survival outcomes. The results revealed significant relationship between CAT C262T polymorphism and cancer risk(TT + CT vs CC: OR = 1.05, 95%CI = 1.00–1.10, P = 0.036), subgroup analyses indicated the CAT C262T polymorphism was significantly correlated with an increased risk for prostate cancer (TT vs CC + CT: OR = 1.43, 95%CI = 1.20–1.70, P < 0.001) and increased risk among Caucasians (TT vs CC + CT: OR = 1.19, 95%CI = 1.09–1.31, P < 0.001), while no associations between the polymorphism and Asian or mixed population were established. In the survival analysis, no interactions were identified between this polymorphism and cancer survival (TT + CT vs CC: HR = 1.37, 95%CI = 0.70–2.70, P = 0.36). In conclusion, the CAT C262T polymorphismmay be a candidate markerfor cancer risk with type-specific and population-specific effects but not a fine prognostic factor for cancer survival.
The molecular mechanisms of carcinogenesis have not been wellunderstood, but growing studies have reported that oxidative stress played a significant role in the progression of many diseases, including cancers1. Oxidative stress could contribute to imbalance between the reactive oxygen species (ROS) and antioxidant defense system2. When present at high and/or sustained level, ROS may induce severe DNA damage and chromosomal aberrations3,4,5, which may be followed by abnormal expression of proto-oncogenes and tumor suppressor genes. However, antioxidant defense system could prevent or combat the negative effects caused by ROS, including myeloperoxidase (MPO), glutathione peroxidase (GPX), catalase (CAT), and mitochondrial manganese superoxide dismutase (MnSOD)6,7,8.
Catalase is an important endogenous antioxidant enzyme thatcatalyzes hydrogen peroxide into oxygen and water, thus neutralizing the deleterious effects of ROS9. The CAT gene, which is located on chromosome11p13, consists of 12 introns and 13 exons10. There are several single nucleotide polymorphisms (SNPs) identified in the CAT gene, of which the rs1001179 polymorphism (C262T) was the most extensively studied11,12. The CAT C262T polymorphism is encoded on the promoter region, influencing transcriptional and splicing regulation13. In comparison with the variant C allele, the variant T allele of the CAT C262T polymorphism has been reported to indicate lower enzyme activity, thus raising the levels of ROS and might lead to cancer development or progression14. Recently, a series of studies has demonstrated the associations between the CAT C262T polymorphism and risk for multiple cancers, such as breast cancer15, prostate cancer16, hepatocellular carcinoma11, chronic myeloid leukemia17, etc. So far, some studies have indicated the CAT C262T polymorphismcould increase prostate cancer risk6,16,18. However, the final results were not consistent or conclusive. In terms of survival, no studies confirmed whether the CAT C262T polymorphism could be a prognostic factor of cancer patients. Here, we conducted this updated meta-analysis to comprehensively estimate the relationships between the CAT C262T polymorphism and susceptibility or survival of cancers.
Results
Eligible studies
The initial search yielded 1676 articlesthrough the databases of Pubmed, Embase and China National Knowledge Infrastructure (CNKI). After screening the titles and abstracts, 82 potentially relevant articles were retrieved for the full-text. 49 articles were excluded: 3 were reviews; 9 were conference abstracts; 4 were related to other SNPs of the CAT gene; 11 did not report extractable data; 22 were irrelevant papers. Finally, a total of 33 articles6,7,8,11,12,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42 published from 2005 to 2015met the inclusion criteria and were included in our meta-analysis. There were 27 publications6,7,8,11,12,15,16,17,18,19,21,23,24,25,26,27,28,29,30,31,33,34,35,37,38,39,40 regarding susceptibility analysis, which involved 35 case-control or cohort studies with 15531 cancer patients and 41816 controls, while 8 publications6,20,22,29,32,36,41,42 contained the survival data. The search process was presented in Fig. 1 and the clinical characteristics of the studies or other relevant information were listed in Table 1.
Figure 1. Flow chart of study inclusion and exclusion in this meta-analysis.
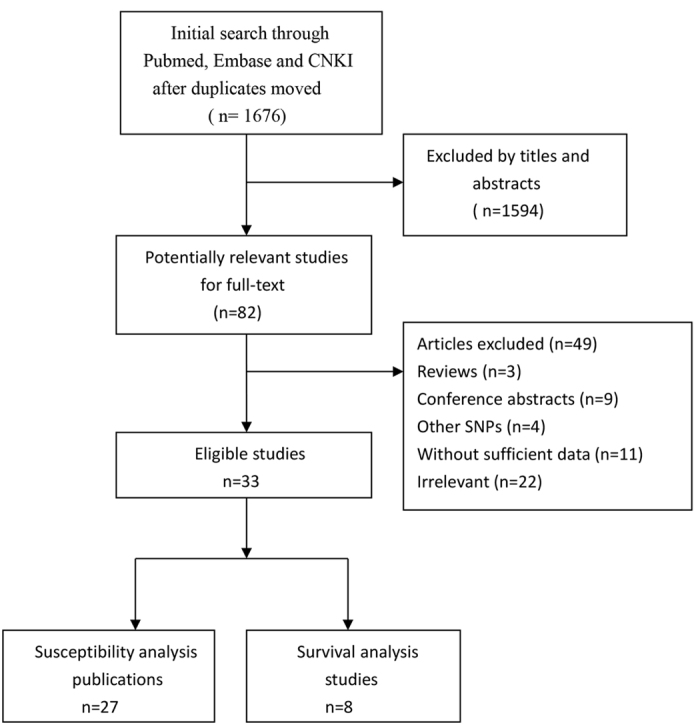
Table 1. Baseline characteristics of eligible studies (N = 33).
| First Author | #* | Year | Country | Ethnicity | Source of Controls | Quality Control | Cancer Type | Case/Control | Genotyping Method | HWE |
|---|---|---|---|---|---|---|---|---|---|---|
| Ahn19 | 2005 | USA | Caucasian | PB | Yes | Breast cancer | 1008/1056 | MALDI-TOF | Yes | |
| Ambrosone20 | 2005 | USA | Mixed | PB | NA | Breast cancer | 279/NA | MALDI-TOF | NA | |
| Aynali21 | 2013 | Turkey | Caucasian | HB | NA | Laryngeal cancer | 25/23 | PCR | Yes | |
| Banescu17 | 2014 | Romania | Caucasian | HB | NA | CML | 168/321 | PCR-RFLP | Yes | |
| Belotte22 | 2015 | USA | Mixed | NA | NA | Ovarian cancer | NA | TaqMan | NA | |
| Bhatti23 | 1 | 2009 | USA | Caucasian | HB | Yes | Glioma | 362/494 | TaqMan | NA |
| Bhatti23 | 2 | 2009 | USA | Caucasian | HB | Yes | Glioblastoma multiforme | 176/494 | TaqMan | NA |
| Bhatti23 | 3 | 2009 | USA | Caucasian | HB | Yes | Meningioma | 134/494 | TaqMan | NA |
| Castaldo12 | 2015 | Portugal | Caucasian | HB | NA | Cervical cancer | 120/107 | PCR | No | |
| Cebrian24 | 2006 | UK | Caucasian | PB | Yes | Breast cancer | 2171/2262 | TaqMan | Yes | |
| Cheng25 | 2011 | USA | mixed | PB | NA | Prostate cancer | 150/761 | PCR | NA | |
| Choi7 | 2007 | USA | Mixed | PB | Yes | Prostate cancer | 508/1403 | MALDI-TOF | Yes | |
| Ding26 | 2012 | China | Asian | PB | NA | Prostate cancer | 1417/1008 | HapMap | Yes | |
| Ezzikouri27 | 2010 | France | Caucasian | HB | Yes | Hepatocellular carcinoma | 96/222 | PCR-RFLP | Yes | |
| Farawela28 | 2012 | Egypt | Caucasian | HB | Yes | NHL | 100/100 | PCR-RFLP | Yes | |
| Funke29 | 2009 | Germany | Caucasian | PB | Yes | Colorectal Cancer | 632/605 | Pyrosequencing Technology | Yes | |
| Geybels6 | 2014 | Netherland | Caucasian | PB | Yes | Prostate cancer | 1527/25184 | PCR | No | |
| He30 | 1 | 2010 | USA | Caucasian | PB | NA | BCC | 270/796 | TaqMan | Yes |
| He30 | 2 | 2010 | USA | Caucasian | PB | NA | Melanoma | 211/796 | TaqMan | Yes |
| He30 | 3 | 2010 | USA | Caucasian | PB | NA | SCC | 266/796 | TaqMan | Yes |
| Ho31 | 2006 | China | Asian | HB | NA | Lung cancer | 230/240 | PCR-RFLP | Yes | |
| Kakkoura15 | 2015 | Cyprus | Caucasian | PB | Yes | Breast cancer | 1057/1141 | TaqMan | Yes | |
| Karunasinghe16 | 2012 | New Zealand | Caucasian | HB | NA | Prostate cancer | 258/434 | TaqMan | Yes | |
| Koistinen32 | 2006 | Finland | Caucasian | NA | Yes | AML | 89/NA | PCR | NA | |
| Li33 | 2009 | USA | Caucasian | PB | Yes | Breast cancer | 497/493 | TaqMan | Yes | |
| Lightfoot34 | 2006 | USA/UK | Caucasian | PB | NA | NHL | 928/1446 | TaqMan | Yes | |
| Liu35 | 2015 | China | Asian | PB | Yes | Hepatocellular carcinoma | 266/248 | PCR-RFLP | Yes | |
| Nahon36 | 2009 | France | Caucasian | NA | NA | Hepatocellular carcinoma | 190/NA | PCR | NA | |
| Quick37 | 1 | 2008 | USA | Mixed | PB | Yes | Breast cancer | 57/108 | MALDI-TOF | Yes |
| Quick37 | 2 | 2008 | USA | Caucasian | PB | Yes | Breast cancer | 569/974 | MALDI-TOF | Yes |
| Rajaraman8 | 1 | 2008 | USA | Mixed | HB | Yes | Acoustic neuroma | 69/494 | TaqMan | Yes |
| Rajaraman8 | 2 | 2008 | USA | Mixed | HB | Yes | Glioma | 362/494 | TaqMan | Yes |
| Rajaraman8 | 3 | 2008 | USA | Mixed | HB | Yes | Meningioma | 134/494 | TaqMan | Yes |
| Saadat38 | 2015 | Iran | Caucasian | PB | NA | Breast cancer | 407/395 | PCR | Yes | |
| Su11 | 2015 | China | Asian | HB | Yes | Hepatocellular carcinoma | 400/480 | PCR-RFLP | Yes | |
| Tang39 | 2010 | USA | Mixed | HB | NA | Pancreatic cancer | 551/602 | TaqMan | Yes | |
| Tefik18 | 2013 | Turkey | Caucasian | HB | NA | Prostate cancer | 155/195 | PCR | Yes | |
| Tsai40 | 2012 | China | Asian | HB | Yes | Breast cancer | 260/224 | PCR | Yes | |
| Ulder41 | 2007 | England | Caucasian | PB | Yes | Breast cancer | NA | TaqMan | NA | |
| Van Blarigan42 | 2014 | USA | Caucasian | PB | NA | Prostate cancer | NA | MALDI-TOF | NA |
*Number of data separately reported by articles.
HWE: Hardy-Weinberg equilibrium; MALDI-TOF: Matrix-Assisted Laser Desorption/ Ionization Time of Flight Mass Spectrometry; PCR: polymerase chain reaction; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; PB: population-based; HB: hospital-based; NA: not available. CML: Chronic myeloid leukemia; NHL: non-Hodgkin lymphoma; BCC: Basal cell carcinoma; SCC: Squamous cell carcinoma; AML: Acute myeloid leukemia.
C262T polymorphism and susceptibility to cancer
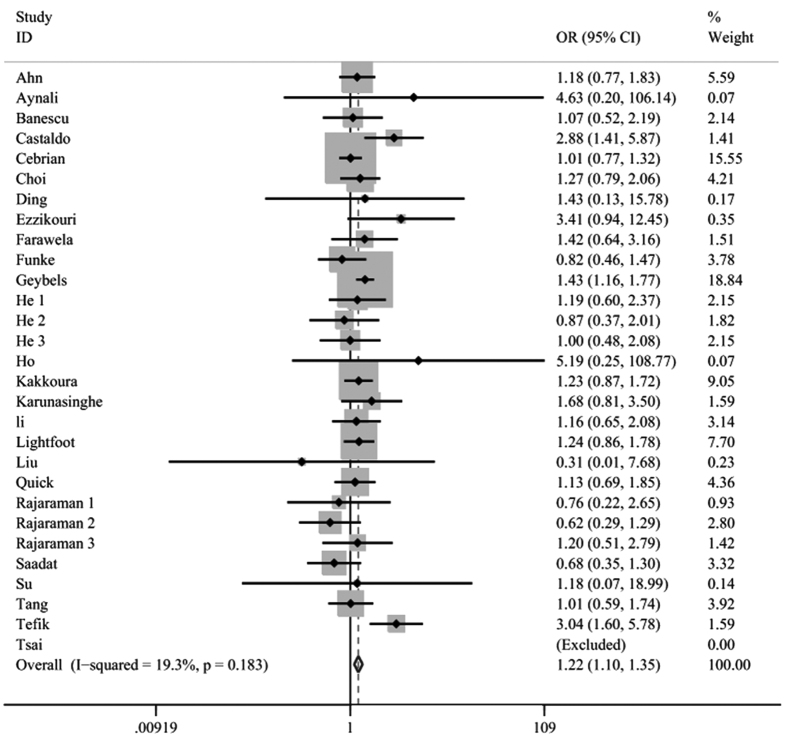
The meta-analysis of the 27 articles6,7,8,11,12,15,16,17,18,19,21,23,24,25,26,27,28,29,30,31,33,34,35,37,38,39,40 with 35 case-control or cohort studies suggested there was a positive correlation between the CAT C262T polymorphism and cancer risk (TT + CT vs CC: OR = 1.05, 95%CI = 1.00–1.10, P = 0.036; TT vs CT + CC: OR = 1.18, 95%CI = 1.08–1.29, P < 0.001; TT vs CC: OR = 1.22, 95%CI = 1.10–1.35, P < 0.001; T vs C: OR = 1.07, 95%CI = 1.03–1.11, P = 0.001 Fig. 2). In the studies which were not derived from the Hardy-Weinberg equilibrium (HWE), the pooled ORs also showed the significance of CAT C262T polymorphism in susceptibility to cancers (TT vs CT + CC: OR = 1.15, 95%CI = 1.02–1.28, P = 0.019; TT vs CC: OR = 1.14, 95%CI = 1.02–1.28, P = 0.026). Furthermore, a subgroup analysis was also performed stratified by cancer types and ethnicity. There was a significant association between CAT C262T polymorphism and the development of prostate cancer6,7,16,18,25,26 (TT vs CT + CC: OR = 1.43, 95%CI = 1.20–1.70, P < 0.001; TT vs CC: OR = 1.52, 95%CI = 1.27–1.81, P < 0.001; CT vs CC: OR = 1.15, 95%CI = 1.05–1.26, P = 0.002; T vs C: OR = 1.21, 95%CI = 1.05–1.40, P = 0.01). The association between the polymorphism of the CAT C262T gene and increased skin cancer risk was also confirmed30 (CT + TTvs CC: OR = 1.19, 95%CI = 1.00–1.41, P = 0.04; CT vs CC: OR = 1.21,95%CI = 1.02–1.44, P = 0.03). Meanwhile, the CAT C262T polymorphism retained its high position for predicting the susceptibility to cervical cancer12 (TT vs CT + CC: OR = 2.85, 95%CI = 1.44–5.65, P = 0.003; TT vs CC: OR = 2.88, 95%CI = 1.41–5.87, P= 0.004; T vs C: OR = 1.96, 95%CI = 1.31–2.93, P = 0.001). However, no evidence of statistical significance could be detected in other cancer types. In terms of subgroup analysis by ethnicity (Caucasian, Asian and Mixed), the assessment of the results revealed that the CAT C262T polymorphism was associated with cancer risk in Caucasians (TT vs CT + CC: OR = 1.19, 95%CI = 1.09–1.31 P < 0.001; TT vs CC: OR = 1.24, 95%CI = 1.12–1.38, P < 0.001; T vs C: OR = 1.08, 95%CI = 1.01–1.16, P = 0.02). No relationship could be found in Asian or mixed population. The pooled results were shown in Table 2.
Figure 2. Forest plot for the association between the CAT C262T polymorphism and cancer risk (TT vs CC).
Significant association was observed between the CAT C262T polymorphism and cancer susceptibility.
Table 2. The results of evidence synthesis of susceptibility analysis.
| Variables | Dominant model (TT + CT vs CC) |
Recessive model (TT vs CT + CC) |
Homozygote model (TT vs CC) |
Heterozygote model (CT vs CC) |
Allel contrast model (T vs C) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | P | I2 (%) | OR(95%CI) | P | I2 (%) | OR(95%CI) | P | I2 (%) | OR(95%CI) | P | I2 (%) | OR(95%CI) | P | I2 (%) | |
| All | 1.05(1.00–1.10) | 0.036 | 39.80 | 1.18(1.08–1.29) | <0.001 | 2.20 | 1.22(1.10–1.35) | <0.001 | 19.30 | 1.03(0.98–1.08) | 0.23 | 28.90 | 1.07(1.03–1.11) | 0.001 | 47.60 |
| By cancer type | |||||||||||||||
| Breast cancer | 1.02(0.95–1.10) | 0.58 | 30.40 | 1.08(0.92–1.27) | 0.36 | 0.00 | 1.08(0.92–1.27) | 0.37 | 0.00 | 1.01(0.94–1.09) | 0.75 | 25.40 | 1.03(0.97–1.09) | 0.42 | 26.70 |
| Hematological malignancies | 0.92(0.79–1.07) | 0.30 | 46.20 | 1.30(0.98–1.74) | 0.07 | 0.00 | 1.23(0.91–1.66) | 0.18 | 0.00 | 0.82(0.60–1.13) | 0.23 | 51.90 | 0.99(0.88–1.12) | 0.92 | 26.50 |
| Brain cancer | 0.86(0.69–1.06) | 0.16 | 0.00 | 1.02(0.85–1.23) | 0.80 | 0.00 | 0.80(0.48–1.34) | 0.40 | 0.00 | 0.86(0.69–1.08) | 0.2 | 2.30 | 0.88(0.73–1.05) | 0.17 | 0.00 |
| Prostate cancer | 1.15(0.98–1.36) | 0.09 | 58.10 | 1.43(1.20–1.70) | <0.001 | 0.00 | 1.52(1.27–1.81) | <0.001 | 26.20 | 1.15(1.05–1.26) | 0.002 | 22.30 | 1.21(1.05–1.40) | 0.01 | 61.90 |
| Digestive system cancer | 0.92(0.79–1.06) | 0.24 | 0.00 | 1.05(0.73–1.50) | 0.81 | 15.10 | 1.01(0.70–1.46) | 0.95 | 9.40 | 0.91(0.80–1.05) | 0.19 | 0.00 | 0.94(0.83–1.07) | 0.36 | 0.00 |
| Skin cancer | 1.19(1.00–1.41) | 0.04 | 0.00 | 0.96(0.63–1.47) | 0.86 | 0.00 | 1.03(0.67–1.58) | 0.90 | 0.00 | 1.21(1.02–1.44) | 0.03 | 0.00 | 1.13(0.98–1.30) | 0.10 | 0.00 |
| By ethnicity | |||||||||||||||
| Caucasian | 1.06(0.98–1.15) | 0.13 | 50.20 | 1.19(1.09–1.31) | <0.001 | 14.10 | 1.24(1.12–1.38) | <0.001 | 31.00 | 1.04(0.98–1.09) | 0.18 | 39.80 | 1.08(1.01–1.16) | 0.02 | 58.00 |
| Asian | 1.04(0.85–1.28) | 0.72 | NA | 1.41(0.40–5.00) | 0.60 | 0.00 | 1.40(0.39–4.98) | 0.60 | 0.00 | 1.03(0.84–1.27) | 0.78 | 0.00 | 1.05(0.86–1.28) | 0.66 | 0.00 |
| Mixed | 0.91(0.72–1.16) | 0.45 | 52.40 | 0.94(0.65–1.35) | 0.73 | 0.00 | 0.89(0.62–1.29) | 0.55 | 0.00 | 0.96(0.70–1.31) | 0.78 | 64.90 | 0.93(0.81–1.06) | 0.27 | 49.10 |
| By HWE | |||||||||||||||
| Yes | 1.01(0.96–1.07) | 0.58 | 0.13 | 1.15(1.02–1.28) | 0.02 | 0.70 | 1.14(1.02–1.28) | 0.03 | 0.50 | 1.00(0.95–1.05) | 0.93 | 0.20 | 1.03(0.99–1.07) | 0.19 | 0.12 |
| No | 1.23(1.11–1.37) | <0.001 | 0.26 | 1.82(0.88–3.75) | 0.10 | 0.04 | 1.86(0.96–3.63) | 0.07 | 0.06 | 1.18(1.06–1.32) | 0.003 | 0.70 | 1.47(0.91–2.38) | 0.11 | 0.02 |
P: P-value of Z-test to evaluate the significance of the ORs; NA: not available.
C262T polymorphism and cancer survival
The meta-analysis included 8 studies investigating CAT C262T polymorphism and cancer survival6,20,22,29,32,36,41,42. No overall survival (OS) difference was detected between patients with CT/TT genotypes and those with CC genotype (HR = 1.37, 95%CI = 0.70–2.70, P = 0.36), or between patients with TT genotype and allele C carrier (HR = 0.90, 95%CI = 0.44–1.83, P = 0.77). Furthermore, when compared to CC genotype, CT or TT genotype didn’t suggest poorer OS (HR = 1.07, 95%CI = 0.95–1.20, P = 0.29; HR = 1.04, 95%CI = 0.81–1.34, P = 0.74, respectively). In addition, cancer patients with T allele showed similar survival compared to those with C allele (HR = 1.07, 95%CI = 0.97–1.18, P = 0.21). The main results were summarized in Table 3.
Table 3. The results of evidence synthesis of overall survival analysis.
| Model | Variables | N* | HR(95%CI) | P | I2(%) |
|---|---|---|---|---|---|
| Dominant model | CC | 3 | Reference | 0.358 | 66.7% |
| CT/TT | 1.37(0.70–2.70) | ||||
| Recessive model | CC/CT | 2 | Reference | 0.77 | 0% |
| TT | 0.90(0.44–1.83) | ||||
| Homozygote model | CC | 6 | Reference | 0.744 | 17.1% |
| TT | 1.04(0.81–1.34) | ||||
| Heterozygote model | CC | 6 | Reference | 0.29 | 0% |
| CT | 1.07(0.95–1.20) | ||||
| Allelic model | C | 4 | Reference | 0.21 | 9.6% |
| T | 1.07(0.97–1.18) |
*Number of studies in analysis.
Publication bias and sensitivity analysis
We didn’t detect any significant publication bias by Begg’ test (Pr > |z| = 0.775 Fig. 3a) or Egger’ test (P > |t| = 0.548 Fig. 3b), which indicated the reliability of our meta-analysis. Furthermore, no significant change was detected when we sequentially dropped out each included study and thus the results of our study were stable.
Figure 3. Begg’s funnel plot and Egger’s on publication bias for included studies on the association of the CAT C262T polymorphism and cancer risk (TT vs CC).
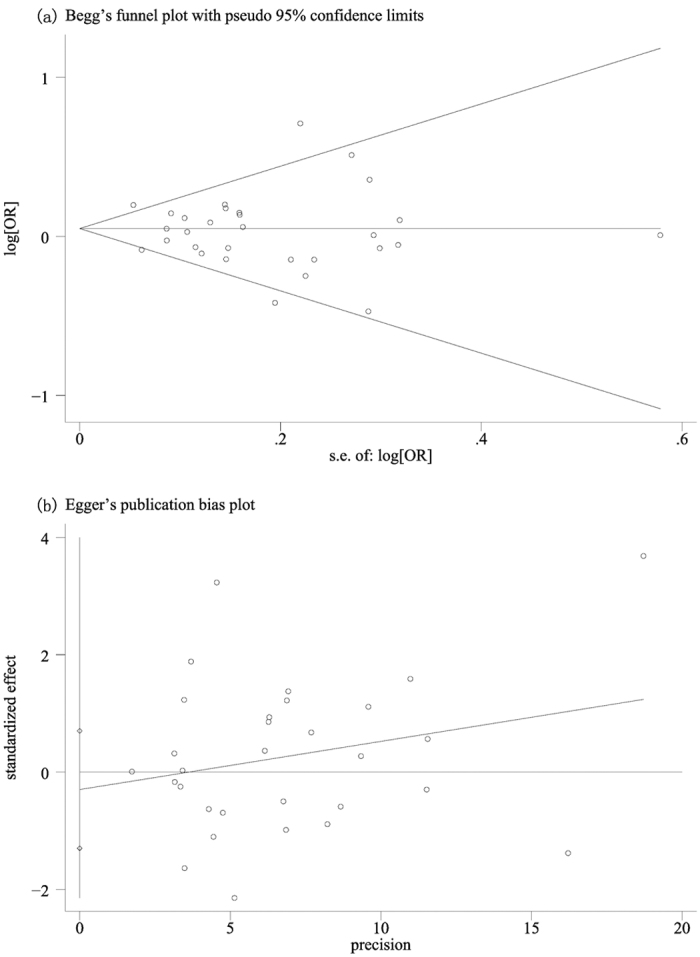
The funnel plot seemed symmetrical, indicating absence of publication bias.
Discussion
ROS are naturally generated fromaerobic metabolism3. The human body develops a sophisticated set of antioxidant molecules to prevent the toxic accumulation of these species43. CAT belongs to the antioxidant molecules and is present in all aerobic cells while the highest levels of the enzyme are found in the liver, kidneyand erythrocytes44. CAT is a heme enzyme that plays a very important role in avoiding hydrogen peroxide concentration by converting H2O2 into H2O and O2, and protects cells from detrimental effects of oxidative stress45. Allelic variants of CAT gene may contribute to lower CAT enzymatic activity and higher sensitivity to ROS, and alter ROS detoxification and increase oxidative stress, thereby implicating oxidative DNA damage and modulating disease risk46. 245 CAT SNPs have been identified, with most studies investigating the relationships between multiple diseases and rs1001179, a C > T substitution at position −262 from the transcription start site44. Previous studies indicated thatCAT C262T gene polymorphism had an influence on transcription factors binding thus altering the basal transcription and consequent expression of this enzyme and hence influenced the oxidative status of cells and its microenvironment25,26. Consequently, this polymorphism was believed to play a key role in the pathogenesis of cancer25,26. The growing studies investigated the relation of CAT C262T gene polymorphism to breast cancer, lung cancer, diabetic neuropathy, non-Hodgkin lymphoma, liver cancer and colorectal cancer43, however, these results did not reach an agreement. A meta-analysis is a useful strategy because it potentially investigates a large number of individuals and could evaluate the effect of a genetic factor oncancer risk. We performed the current meta-analysis to combine the eligible studies and data to precisely estimate the role of CAT C262T polymorphism in the susceptibility and survival of cancers.
The present meta-analysis, including 15531 cancer patients and 41816 controls from 35 case–control or cohort studies, investigated the association between the CAT C262T polymorphism and cancer risk. Based on current accessible evidences, the individuals who carry the TT homozygote have 17% increased risk of cancer compared with the C allele carriers, revealing that the CAT C262T gene polymorphism may be a risk factor for cancer47. For tumor origin could influence the results from meta analysis, we performed subgroup analyses by cancer type. However, we did not find any positive relationship in the studies of breast cancer, head and neck cancer, hematological malignancies, digestive system cancer or brain cancer. Interestingly, the significant association between the CAT C262T gene and prostate cancer6,7,16,18,25,26 was the opposite in most genetic models. The relationships between the CAT C262T gene and skin cancer31 or cervical cancer12 were opposite in part genetic models. Meanwhile, in the stratified analysis by ethnicity, significantly elevated cancer risks were indicated in Caucasian group but not in Asian population. The underlying genetic backgrounds and/or environmental and social factors may account for the ethnic discrepancy.
It is worth mentioning that the current study was the first meta-analysis to investigate the survival outcomes. While the TT genotype was associated with increased cancer risk especially in prostate cancer and Caucasian population, however, neither of TT or CT genotype contributed to poorer survival of cancer patients. These results indicated that CAT C262T polymorphism might only influence susceptibility to cancer instead of cancer prognosis. In addition, the association between C262T polymorphism and treatment efficiency such as chemotherapy and radiotherapy remained unclear and those data were insufficient to reach a pooled result. Further studies could focus on the role of CAT C262T polymorphism on treatment strategy. The exact mechanisms of the C262T polymorphism on cancer development and progression were warranted to investigate in future.
In interpreting the current results, several limitations of the meta-analysis should be addressed. Only if literatures that were indexed by the selected databases were included for the current study, and some relevant published studies or unpublished studies with null results were missed or ongoing studies were not sought, which might have influenced our results. Secondly, the numbers of published studies were not large to identify possible associations, especially in survival analysis. Thirdly, part studies investigated several cases with the same control, which might reduce the statistical power to identify possible associations. Fourthly, lacking the original data of the reviewed studies limited our further evaluation of the potential interaction. However, our current study also had some merits. On one hand, over 30 case-control or cohort studies from different publications significantly increased statistical power of the analyses. On the other hand, on the basis of our studies, we find a novel mechanism to predict cancer risk. In addition, the current study is the first to investigate the survival outcomes.
To sum up, the results from the current study suggest that the CAT C262T polymorphism may contribute to genetic susceptibility to cancer, supporting the hypothesis that the polymorphism serves as a potential susceptibility tumor marker. However the CAT C262T polymorphismmay not be a fine prognostic factor for cancer survival. Further well-designed, multicenter epidemiological studies including a wider spectrum of subjects should be performed to investigate the role of this functional polymorphism in other populations and biological mechanism of CAT C262T polymorphism, which should lead to better, comprehensive interpretation of the association between the CAT C262T polymorphism and cancer risk.
Methods
Identification and Eligibility of Relevant Studies
Two investigators performed a comprehensive and systematic search through the databases of Pubmed, Embase and CNKI for relevant studies with the following terms: “catalase” or “CAT”, “polymorphism” or “variant” or “mutation”, and “cancer” or “carcinoma” or “malignancy” (Last search update December 2015). The publication language and publication date were not restricted in our search. Some potential publications were obtained from a manual search of the references of retrieved articles.
The inclusion criteria were: (1) case-control studies or cohort studies; (2) evaluating the associations between the CAT C262T polymorphism and cancer risk or survival outcomes; (3) detailed data on genotype frequency of the CAT C262T for calculating the odds ratios (ORs), available hazards ratios (HRs) and 95% confidence intervals (95%CIs). The exclusion criteria were: (1) reviews, conference abstracts, case reports, animal studies or editorials; (2)without available genotype frequency of the CAT C262T; (3) when the same or overlapped population and duplicated studies were met, only the most recent studies with sufficient information were included.
Data extraction
Two investigators extracted data independently and consensus on all the items was reached after discussion. The main information included the first author, publication year, country, ethnicity, source of controls, sample, quality control, quality health, cancer type, number of cases and controls, genotype distributions of cases and controls, genotyping method, HWE of the control groups, and HR with 95%CI of this polymorphism in survival analysis.
Statistical Analysis
All statistical analyses were conducted with STATA 12.0 (Stata Corp, College Station, TX, USA). The statistical heterogeneity among the studies was calculated by the I2 statistics. If I2 > 50%, the random-effects model was applied to analysis; otherwise, the fixed-effects model was adopted48,49. ORs with 95% CIs were used to stabilize risk estimates, while HRs with 95% CIs were required to predict whether the CAT C262T gene polymorphism had influence on OS of cancer patients. The following genetic models were used to evaluate the susceptibility: dominant model (TT + CT vs CC), recessive model (TT vs CT + CC), homozygote model (TT vs CC), heterozygote model (CT vs CC), and allelic contrast model (T vs C). We also performed the subgroup analyses based on cancer type and ethnicity. The significance of the pooled OR was assessed by Z test and the statistically significant outcome was defined as P < 0.05. HWE was evaluated by the chi-square test in control groups for each study, where P < 0.05 was considered significant50. Both Egger’s and Begg’s tests were used to evaluate the publication bias51. Sensitivity analysis, which aimed to identify whether the heterogeneity across these studies was from one individual study, was also performed to ensure the reliability of the results.
Additional Information
How to cite this article: Wang, C.-D. et al. The Role of Catalase C-262T Gene Polymorphism in the Susceptibility and Survival of Cancers. Sci. Rep. 6, 26973; doi: 10.1038/srep26973 (2016).
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No. 81171320).
Footnotes
Author Contributions C.D. and Y.L. proposed the project. Y.S. and C.D. searched the databases and obtained the data. N.C. and C.D. performed the statistical analysis and assessed the results. Y.S. And N.C. wrote the manuscript draft. L.H. and J.W. did the data analysis. C.D., Y.L., L.H., J.W., M.Z. and T.W. commented on the manuscript. All authors revised and approved of the final manuscript.
References
- Dalle-Donne I., Giustarini D., Colombo R., Rossi R. & Milzani A. Protein carbonylation in human diseases. Trends Mol Med 9, 169–176 (2003). [DOI] [PubMed] [Google Scholar]
- Klaunig J. E. & Kamendulis L. M. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol 44, 239–267 (2004). [DOI] [PubMed] [Google Scholar]
- Ziech D. et al. The role of reactiveoxygen species and oxidative stress in environmental carcinogenesisand biomarker development. Chem Biol Interact 188, 334–339 (2010). [DOI] [PubMed] [Google Scholar]
- Kang D. H. Oxidative stress, DNA damage, and breast cancer. AACN Clin Issues 13, 540–549 (2002). [DOI] [PubMed] [Google Scholar]
- Bensaad K. & Vousden K. H. Savoir and Slayer: the two faces of p53. Nat. Med. 11, 1278–1279 (2005). [DOI] [PubMed] [Google Scholar]
- Geybels M. S., Van Den Brandt P. A., Van Schooten F. J. & Verhage B. A. J. Oxidative stress-related genetic variants, pro-and antioxidant intake and status, and advanced prostate cancer risk. Cancer Epidemiology Biomarkers and Prevention 24, 178–186 (2014). [DOI] [PubMed] [Google Scholar]
- Choi J. Y. et al. Polymorphisms in oxidative stress-related genes are not associated with prostate cancer risk in heavy smokers. Cancer Epidemiol Biomarkers Prev 16, 1115–1120 (2007). [DOI] [PubMed] [Google Scholar]
- Rajaraman P. et al. Oxidative response gene polymorphisms and risk of adult brain tumors. Neuro Oncol 10, 709–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent A. et al. Controlling tumor growth by modulating endogenous production of reactive oxygen species. Cancer Res 65, 948–956 (2005). [PubMed] [Google Scholar]
- Jiang Z. et al. A polymorphism in the promoter region of catalase is associated with blood pressure levels. Hum Genet 109, 95–98 (2001). [DOI] [PubMed] [Google Scholar]
- Su S. et al. Genetic polymorphisms in antioxidant enzyme genes and susceptibility to hepatocellular carcinoma in Chinese population: a case-control study. Tumour Biol 36, 4627–4632 (2015). [DOI] [PubMed] [Google Scholar]
- Castaldo S. A. et al. The role of CYBA (p22phox) and catalase genetic polymorphisms and their possible epistatic interaction in cervical cancer. Tumour Biol 36, 909–914 (2015). [DOI] [PubMed] [Google Scholar]
- Forsberg L., Lyrenas L., de Faire U. & Morgenstern R. A common functional C-T substitution polymorphism in the promoter region of the human catalase gene influences transcription factor binding, reporter gene transcription and is correlated to blood catalase levels. Free Radic Biol Med 30, 500–505 (2001). [DOI] [PubMed] [Google Scholar]
- Ahn J. et al. Associations between catalase phenotype and genotype: modification by epidemiologic factors. Cancer Epidemiol Biomarkers Prev 15, 1217–1222 (2006). [DOI] [PubMed] [Google Scholar]
- Kakkoura M. G. et al. MnSOD and CAT polymorphisms modulate the effect of the Mediterranean diet on breast cancer risk among Greek-Cypriot women. Eur J Nutr, doi: 10.1007/s00394-015-0971-5(2015). [DOI] [PubMed] [Google Scholar]
- Karunasinghe N. et al. Prostate disease risk factors among a New Zealand cohort. J Nutrigenet Nutrigenomics 5, 339–351 (2012). [DOI] [PubMed] [Google Scholar]
- Banescu C. et al. CAT, GPX1, MnSOD, GSTM1, GSTT1, and GSTP1 genetic polymorphisms in chronic myeloid leukemia: a case-control study. Oxidative Medicine and Cellular Longevity 2014, 875861 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefik T. et al. Manganese superoxide dismutase Ile58Thr, catalase C-262T and myeloperoxidase G-463A gene polymorphisms in patients with prostate cancer: relation to advanced and metastatic disease. BJU Int. 112, E406–414 (2013). [DOI] [PubMed] [Google Scholar]
- Ahn J. et al. Associations between breast cancer risk and the catalase genotype, fruit and vegetable consumption, and supplement use. Am J Epidemiol 162, 943–952 (2005). [DOI] [PubMed] [Google Scholar]
- Ambrosone C. B. et al. Polymorphisms in genes related to oxidative stress (MPO, MnSOD, CAT) and survival after treatment for breast cancer. Cancer Res 65, 1105–1111 (2005). [PubMed] [Google Scholar]
- Aynali G. et al. Polymorphic variants of MnSOD Val16Ala, CAT-262 C < T and GPx1 Pro198Leu genotypes and the risk of laryngeal cancer in a smoking population. J Laryngol Otol 127, 997–1000 (2013). [DOI] [PubMed] [Google Scholar]
- Belotte J. et al. A Single Nucleotide Polymorphism in Catalase Is Strongly Associated with Ovarian Cancer Survival. PloS one 10, e0135739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatti P. et al. Lead exposure, polymorphisms in genes related to oxidative stress, and risk of adult brain tumors. Cancer Epidemiol Biomarkers Prev 18, 1841–1848 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian A. et al. Tagging single-nucleotide polymorphisms in antioxidant defense enzymes and susceptibility to breast cancer. Cancer Res 66, 1225–1233 (2006). [DOI] [PubMed] [Google Scholar]
- Cheng T. Y. et al. Genetic variation in myeloperoxidase modifies the association of serum alpha-tocopherol with aggressive prostate cancer among current smokers. J Nutr 141, 1731–1737 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G. et al. The association between polymorphisms in prooxidant or antioxidant enzymes (myeloperoxidase, SOD2, and CAT) and genes and prostate cancer risk in the Chinese population of Han nationality. Clin Genitourin Cancer 10, 251–255 (2012). [DOI] [PubMed] [Google Scholar]
- Ezzikouri S. et al. Polymorphisms in antioxidant defence genes and susceptibility to hepatocellular carcinoma in a Moroccan population. Free radical research 44, 208–216 (2010). [DOI] [PubMed] [Google Scholar]
- Farawela H. et al. The association between hepatitis C virus infection, genetic polymorphisms of oxidative stress genes and B-cell non-Hodgkin’s lymphoma risk in Egypt. Infect Genet Evol 12, 1189–1194 (2012). [DOI] [PubMed] [Google Scholar]
- Funke S., Hoffmeister M., Brenner H. & Chang-Claude J. Effect modification by smoking on the association between genetic polymorphisms in oxidative stress genes and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev 18, 2336–2338 (2009). [DOI] [PubMed] [Google Scholar]
- He C., Qureshi A. A. & Han J. Polymorphisms in genes involved in oxidative stress and their interactions with lifestyle factors on skin cancer risk. J Dermatol Sci 60, 54–56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J. C. et al. Manganese superoxide dismutase and catalase genetic polymorphisms, activity levels, and lung cancer risk in Chinese in Hong Kong. J Thorac Oncol 1, 648–653 (2006). [PubMed] [Google Scholar]
- Koistinen P. et al. An association between manganese superoxide dismutase polymorphism and outcome of chemotherapy in acute myeloid leukemia. Haematologica 91, 829–832 (2006). [PubMed] [Google Scholar]
- Li Y. et al. Oxidative stress-related genotypes, fruit and vegetable consumption and breast cancer risk. Carcinogenesis 30, 777–784 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot T. J. et al. Polymorphisms in the oxidative stress genes, superoxide dismutase, glutathione peroxidase and catalase and risk of non-Hodgkin’s lymphoma. Haematologica 91, 1222–1227 (2006). [PubMed] [Google Scholar]
- Liu Y. et al. Association between catalase gene polymorphisms and risk of chronic hepatitis B, hepatitis B virus-related liver cirrhosis and hepatocellular carcinoma in Guangxi population. Medicine 94, e702 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahon P. et al. Myeloperoxidase and superoxide dismutase 2 polymorphisms comodulate the risk of hepatocellular carcinoma and death in alcoholic cirrhosis. Hepatology 50, 1484–1493 (2009). [DOI] [PubMed] [Google Scholar]
- Quick S. K. et al. Effect modification by catalase genotype suggests a role for oxidative stress in the association of hormone replacement therapy with postmenopausal breast cancer risk. Cancer Epidemiol Biomarkers Prev 17, 1082–1087 (2008). [DOI] [PubMed] [Google Scholar]
- Saadat M. & Saadat S. Genetic Polymorphism of CAT C-262 T and Susceptibility to Breast Cancer, a Case-Control Study and Meta-Analysis of the Literatures. Pathol Oncol Res 21, 433–437 (2015). [DOI] [PubMed] [Google Scholar]
- Tang H., Dong X., Day R. S., Hassan M. M. & Li D. Antioxidant genes, diabetes and dietary antioxidants in association with risk of pancreatic cancer. Carcinogenesis 31, 607–613 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. M. et al. Oxidative stress-related enzyme gene polymorphisms and susceptibility to breast cancer in non-smoking, non-alcohol-consuming Taiwanese women: a case-control study. Ann Clin Biochem 49, 152–158 (2012). [DOI] [PubMed] [Google Scholar]
- Udler M. et al. Common germline genetic variation in antioxidant defense genes and survival after diagnosis of breast cancer. J Clin Oncol 25, 3015–3023 (2007). [DOI] [PubMed] [Google Scholar]
- Van Blarigan E. L. et al. Plasma antioxidants, genetic variation in SOD2, CAT, GPX1, GPX4, and prostate cancer survival. Cancer Epidemiology Biomarkers and Prevention 23, 1037–1046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. Oxidant signals and oxidative stress. Cur Opin Cell Biol 15, 247–254 (2003). [DOI] [PubMed] [Google Scholar]
- Crawford A. et al. Relationships between single nucleotide polymorphisms of antioxidant enzymes and disease. Gene 501, 89–103 (2012). [DOI] [PubMed] [Google Scholar]
- Bauer G. Tumor cell-protective catalase as a novel target for rational therapeutic approaches based on specific intercellular ROS signaling. Anticancer Res 32, 2599–2624 (2012). [PubMed] [Google Scholar]
- Forsberg L., de Faire U. & Morgenstern R. Oxidative stress, humangenetic variation, and disease. Arch Biochem Biophys 389, 84–93 (2001). [DOI] [PubMed] [Google Scholar]
- Shen Y. et al. The catalase C-262T gene polymorphism and cancer risk: a systematic review and meta-analysis. Medicine 94, e679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R. & Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials 28, 105–114 (2007). [DOI] [PubMed] [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–748 (1959). [PubMed] [Google Scholar]
- Thakkinstian A., McElduff P., D’Este C., Duffy D. & Attia J. A method for meta-analysis of molecular association studies. Stat Med 24, 1291–1306 (2005). [DOI] [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.