Abstract
Thousands of sense-antisense mRNA-lncRNA gene pairs occur in the mammalian genome. While there is usually little doubt about the function of the coding transcript, the function of the lncRNA partner is mostly untested. Here we examine the function of the homeotic Evx1-Evx1as gene locus. Expression is tightly co-regulated in posterior mesoderm of mouse embryos and in embryoid bodies. Expression of both genes is enhanced by BMP4 and WNT3A, and reduced by Activin. We generated a suite of deletions in the locus by CRISPR-Cas9 editing. We show EVX1 is a critical downstream effector of BMP4 and WNT3A with respect to patterning of posterior mesoderm. The lncRNA, Evx1as arises from alternative promoters and is difficult to fully abrogate by gene editing or siRNA approaches. Nevertheless, we were able to generate a large 2.6 kb deletion encompassing the shared promoter with Evx1 and multiple additional exons of Evx1as. This led to an identical dorsal-ventral patterning defect to that generated by micro-deletion in the DNA-binding domain of EVX1. Thus, Evx1as has no function independent of EVX1, and is therefore unlikely to act in trans. We predict many antisense lncRNAs have no specific trans function, possibly only regulating the linked coding genes in cis.
Gastrulation is a critical early developmental process in which mesoderm and definitive endoderm are generated, and then specified. The three germ layers (ectoderm, mesoderm and definitive endoderm) eventually form all of the tissues of the adult organism. In the mouse, a proximal-distal (P-D) axis is established first from ~E5.0 in response to nodal signalling, then an anterior-posterior (A-P) axis is established during gastrulation in response to BMP, FGF and WNT morphogen gradients1,2. Specification of epiblast cells into mesendodermal cell fates occurs as they migrate through the primitive streak and activate transcriptional programs in response to these gradients3. The cells which migrate first are specified to a posterior fate in response to high local concentrations of BMPs, WNTs and FGFs. These cells form blood, the vasculature and heart. Cells which migrate last through the streak take on an anterior fate in response to high local concentrations of activin/nodal morphogens and inhibitors of BMPs and WNTs such as Chordin, Noggin, Dkk1, Cerberus-like 1 (Cer1) and Lefty 1 and 22. They form notochord, somites and definitive endoderm. Cells from the mid-streak region form intermediate mesoderm derivatives such as kidney.
Hox genes have an established role in patterning during bilaterian embryonic development4. Evx1 is located 50 kb downstream of the HoxA cluster5. Individual members of this cluster have graded anterior boundaries of expression and control rostral-caudal neural fates4. Evx1 is expressed in the posterior primitive streak from ~E6.56, which is earlier in development than any members of the HoxA cluster7. Therefore, it is considered not to be co-regulated with the HoxA cluster during gastrulation. In Xenopus and zebrafish, the homologs of EVX1 (xhox3, eve1) play roles in ‘posteriorization’ of nascent mesoderm8,9. In humans, EVX1 is thought to function in a regulatory network with GOOSECOID (GSC) and BRACHYURY (T) to control anterior-posterior cell fates10. Recent evidence also points to it being a direct target of the WNT signalling pathway, as β-catenin binds to the EVX1 promoter in differentiating hESCs11. Other related members of ‘non-clustered’ Hox gene families play master regulatory roles in A-P and dorsal-ventral (D-V) patterning in Xenopus laevis, zebrafish and mammals. For example, Mixl1 is essential for posterior fate specification in mice12, and many Mix/Bix family members are required for ventral specification in response to BMPs in frogs13,14,15,16,17. Mezzo18, dharma/bozozok19 and Mtx120 are required for formation or patterning of mesendoerm in zebrafish, and goosecoid or related anteriorly-expressed genes are required for anterior specification in most vertebrate species21.
Initially, EVX1 was thought to be critical for murine development, as EVX1−/− embryos were reported as failing to implant in the uterine wall22. However, other groups subsequently demonstrated that EVX1 is not required for viable embryonic development and suggested the initial EVX1−/− phenotype was due to disruption of another closely linked gene23 (Gail Martin, personal communication). In fact, EVX1−/− mice display no overt signs of gastrulation defects after birth, other than minor tail kinks in some offspring and genetic backgrounds (Gail Martin, personal communication). Therefore, the importance of EVX1 in the regulation of gastrulation remains controversial.
Evx1 is expressed from a complex locus, which also expresses a long non-coding RNA (lncRNA), known as Evx1as5. There are other well studied lncRNAs associated with the HoxA cluster, including Haunt/Halr1 which is located ~40 kb upstream of Hoxa124, and HOTTIP which is immediately downstream of Hoxa13, the most 3′ member of the cluster25 (Fig. 1). Both have complex functions in global regulation or fine tuning of expression of the HoxA cluster. LncRNAs are an emerging class of regulatory molecules which have been implicated in a number of developmental processes26,27,28,29. They have been proposed to function via regulation of transcription, splicing, and chromatin dynamics. Antisense/bidirectional transcription of protein coding genes and lncRNAs is a common feature in the mammalian genome, in particular at highly expressed and developmentally regulated genes30. We have previously reported Evx1 and Evx1as are dynamically and concomitantly expressed during embryoid body (EB) differentiation5, a commonly used model of early in vivo embryonic development and the function of lncRNAs24,31. Evx1as is abundant and has stability similar to Evx1 transcripts, suggesting function5.
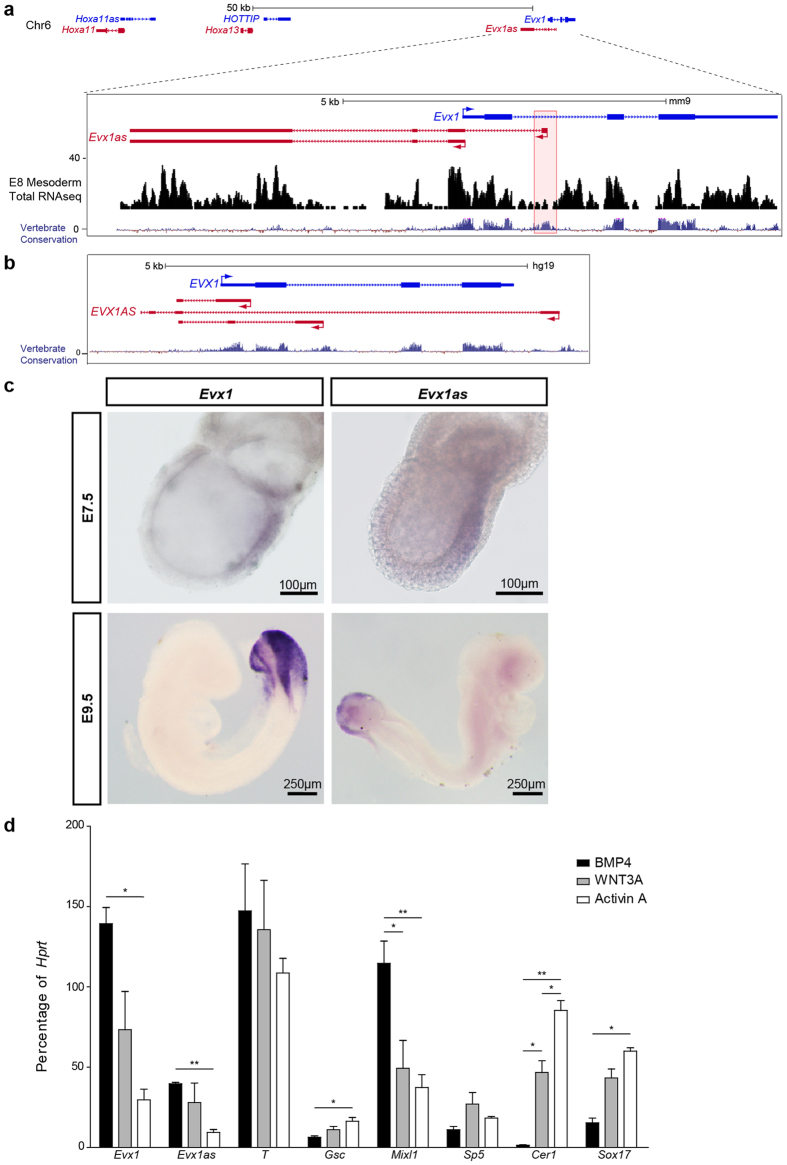
Figure 1. Evx1 and Evx1as are co-expressed in the primitive streak during gastrulation.
(a) Schematic of the Evx1/Evx1as locus, and its proximity to the HoxA cluster modified from the UCSC genome browser. Wiggle track of Total RNAseq from E8 mouse Pre-Somatic Mesoderm (PSM) and Vertebrate Conservation tracks are also shown. Conservation of the Evx1as P1 region is boxed in red. (b) UCSC browser shot of the human EVX1 locus and the syntenic EVX1AS transcripts. (c) WISH of E7.5 and E9.5 mouse embryos using probes against Evx1 and Evx1as. Purple indicates the presence of the transcript. (d) Expression profiling of mESCs differentiated to day 4 EBs under serum-free conditions with the addition of BMP4 (10 ng/ml), WNT3A (20 ng/ml) or Activin A (10 ng/ml). Three replicates were performed for each condition. *Indicates p-value < 0.05, **indicates p-value < 0.01. All error bars indicate SEM.
We sought to dissect the Evx1/Evx1as locus in order to elucidate the potential role for EVX1 and/or Evx1as during gastrulation, and to gain insights into the broader functional significance of antisense/bidirectional lncRNAs. To investigate the function of EVX1, we generated bi-allelic small frameshift deletions in the homeodomain-encoding region using CRISPR/Cas9. We generated stable murine ES cell clones and performed RNAseq at day 4 of embryoid body (EB) differentiation in direct comparison with parental non-edited ES cells. We found that disruption of EVX1 results in upregulation of anterior visceral endoderm (AVE) and definitive endoderm genes including Cer1 and Sox17, at the expense of posterior genes, such as Mixl1 and Kdr (Flk-1). We also show EVX1 is likely to function as a downstream effector of BMP4 and WNT signalling pathways, to regulate posterior tissue patterning.
To test whether Evx1as has a function independent of EVX1, we generated a suite of CRISPR-Cas9 mediated deletions using a similar approach to that recently reported for Haunt24. With all of these DNA manipulations, we were not able to disrupt expression of Evx1as without also disrupting expression of Evx1. We performed RNAseq to convincingly show loss of EVX1 and Evx1as produced identical aberrations in A-P gene expression patterns to those which we observed in the EVX1 loss-of-function cell lines. Together, our results strongly suggest there is no independent function for Evx1as beyond that of EVX1. However, we cannot rule out a function for Evx1as in the regulation of Evx1 in cis.
Results
Organization, conservation and expression of the Evx1/Evx1as locus
The Evx1 locus is located 50 kb downstream of the HoxA gene cluster on chromosome 6 (Fig. 1a). Evx1 and Evx1as are developmentally regulated, displaying peak and concordant expression during gastrulation5. They are both also highly expressed in vivo in the pre-somitic mesoderm (PSM)32. Whole mount in situ hybridization (WISH) of E7.5 and E9.5 embryos demonstrates the Evx1 and Evx1as are co-expressed in the primitive streak during gastrulation (Fig. 1c). At E7.5, both Evx1 and Evx1as are expressed at the posterior-proximal side of the embryo, which is the location of the primitive streak. At E9.5, both transcripts localize to the tail bud, which contains the embryological remnants of the primitive streak. Thus, Evx1 and Evx1as are co-expressed during gastrulation.
Like many lncRNA-coding gene pairs30, Evx1 and Evx1as are expressed from opposite DNA strands in a sense-antisense configuration (Fig. 1a). Interestingly, there are two other lncRNAs within the distal end of the HoxA cluster, Hox11as and HOTTIP25, which are expressed in an antisense direction with respect to Hox11 and Hox13 coding genes, respectively (Fig. 1a). Evx1as is expressed as at least two different isoforms according to EST and depostied cDNA data; the second exon overlaps with the start of Evx1 by ~70 bp and the first exon resides in the first intron of Evx1 (Fig. 1a). We confirmed only 1% of the Evx1as transcript contains the first exon in D4 EBs (Supplementary Figure 1A). EVX1 is highly conserved throughout the protein coding sequence, whereas Evx1as displays notable conservation at the promoter (Fig. 1a), like many lncRNAs33. The human EVX1-EVX1AS locus is syntenic with the murine locus. EVX1AS has two conserved promoters (intronic or bidirectional), but is also expressed from one additional promoter, located 3′ of EVX1 (Fig. 1b).
EVX1 regulates anterior-posterior patterning during gastrulation
In order to study the function of EVX1 and Evx1as, we used EB differentiation as a model for early in vivo mouse embryonic development. Day 4 EBs grown in serum or serum free media (SFM) with recombinant BMP4 display a very similar transcriptional program to embryos undergoing gastrulation7. Evx1 and Evx1as were most highly expressed when differentiated under BMP4 and WNT3A conditions (Fig. 1d), which suggests that they might be either downstream targets of the BMP4 and WNT3A signalling pathways or that BMP4 and WNT3A enhance the generation of posterior mesoderm cell types, wherein the Evx1 locus is expressed. Evx1 and Evx1as were expressed at a much lower level in EBs grown in SFM with Activin A, which is an anterior/dorsal mesoderm inducing factor34. Our results are consistent with expression of human EVX1 in these same growth factors10. Evx1 and Evx1as display a similar expression pattern to Mixl1 (Fig. 1d), a related homeodomain transcription factor which is also expressed in nascent posterior mesoderm12. Expression of pan-mesoderm genes such as Brachyury (T) and Sp5 was similar in BMP4, WNT3A and Activin A, while expression of anterior markers Cer1 and Sox17 was significantly higher in WNT3A and Activin A (Fig. 1d).
We generated EVX1 ‘knockout’ cell lines using CRISPR-Cas9 gene micro-editing of the homeodomain coding region in exon 2, thus leaving Evx1as unperturbed (Fig. 3a). Two independent compound heterozygous Evx1 gene edited clones were selected based on our previously described NGS screening strategy35. Both clones have small frameshift mutations in both alleles (Fig. 2a), which are predicted to result in a premature stop codons within 20 bps of the deletion site. Thus, no functional DNA-binding domain of EVX1 could be generated by either of these clones. We refer to these CRISPR-edited lines as ‘EVX1-Δfs’ from here on. EVX1-Δfs lines and W9.5 controls were differentiated in serum to D4 under suspension culture conditions to identify a potential regulatory role for EVX1 during gastrulation. EVX1-Δfs D4 EBs appeared phenotypically normal, and indistinguishable from the wildtype controls (Supplementary Figure 1B). However, qPCR analysis revealed that both EVX1-Δfs clones display differences in expression of key anterior and posterior patterning markers, including Mixl1 and Cer1 (Supplementary Figure 1C). There were no significant differences between the two clones, therefore only one was used for subsequent analyses (Clone 14) (Supplementary Figure 1C).
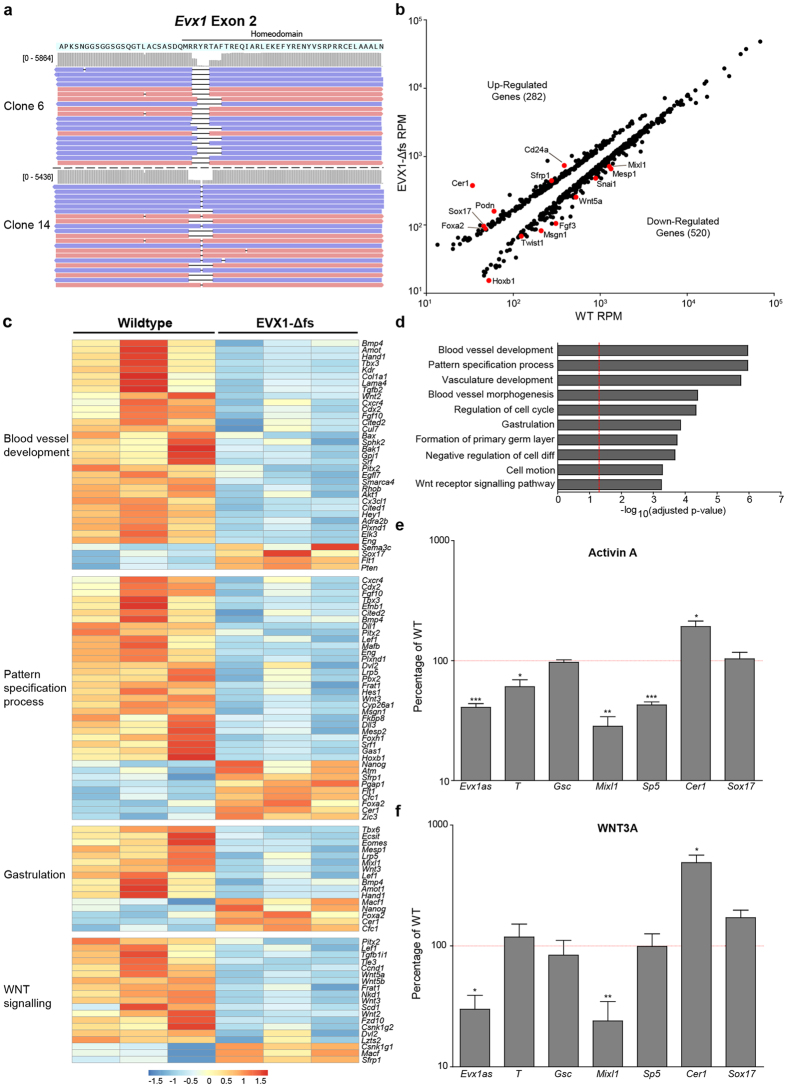
Figure 2. EVX1 is required to regulate anterior-posterior patterning during gastrulation.
(a) Visualization of CRISPR-induced mutations in two independent mESC clones (clone 6 and 14) using Integrated Genome Viewer (IGV). The peptide sequence of EVX1 is shown. Red and blue indicate different read strands. (b) Scatterplot of average counts of DEGs from mRNAseq comparing 3 replicates of WT and EVX1-Δfs Day 4 EBs. Known markers and regulators of tissue specification are shown in red. RPM = Reads per million reads.(c) Heatmap of counts for differentially expressed genes corresponding to particular Gene Ontology (GO) terms in (d). Each category is hierarchically clustered. Each value is normalized to its mean expression across all samples. Red indicates higher than average expression, blue indicates lower than average expression. (d) GO analysis (DAVID) of DEGs shows the biological processes disrupted in the EVX1-Δfs D4 EBs. Red line indicates an adjusted p-value of 0.05. (e,f) Expression profiling of EVX1-Δfs in SFM supplemented with (e) Activin A (10 ng/ul) or (f) WNT3A (20 ng/ul). Expression of each gene in each sample was first normalized to Hprt then normalized to WT expression. 4 biological replicates were performed. *Indicates a p-value < 0.05, **indicates a p-value < 0.01, ***indicates a p-value < 0.001 when compared to WT. All error bars indicate SEM.
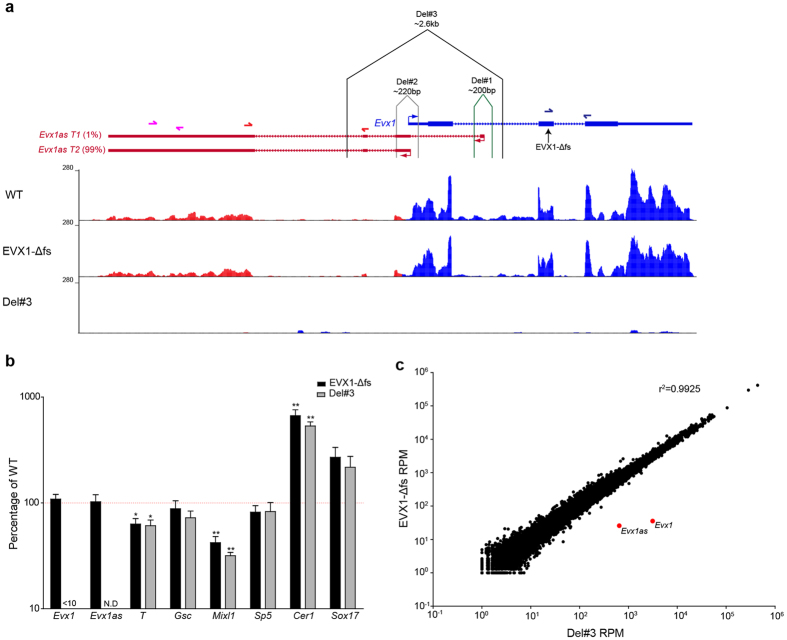
Figure 3. Evx1as does not have a function independent of EVX1.
(a) Schematic of the strategy for dissecting the Evx1/Evx1as locus modified from USCS Genome Browser. The three different deletions performed are shown on the diagram. Wiggle tracks of WT, EVX1-Δfs and Del#3 mRNAseq from D4 EBs are shown. Corresponding qPCR primers pairs are shown in the same colour. The location of the Evx1 deletion is indicated by the arrow. Del = deletion, T1 = transcript 1, T2 = transcript 2. (b) Expression profiling of EVX1-Δfs and Del#3. Expression of each gene in each sample was first normalized to Hprt then normalized to WT expression. 4 biological replicates were performed. Error bars show SEM. *Indicates a p-value < 0.05, **indicates a p-value < 0.01, when compared to WT. No significant differences were found when comparing EVX1-Δfs and Del#3. N.D = not detected. <10 = less than 10% of WT. (c) Scatter plot of average mRNAseq counts from three replicates comparing EVX1-Δfs and Del #3 D4 EBs. Evx1 and Evx1as are shown in red. A value of 1 was added to all RPM values to improve visualization. R2 was obtained from Pearson Correlation. RPM = Reads per million reads.
To determine whether EVX1 has a global role in anterior-posterior patterning, mRNA-seq was performed on three biological replicates of W9.5 and EVX1-Δfs EBs differentiated for 4 days in serum without LIF (see Methods). Using DESeq2, we identified 802 differentially expressed genes (DEGs). In the absence of functional EVX1, 282 genes were upregulated and 520 genes were downregulated (Fig. 2b). The upregulated genes included many which are preferentially expressed in anterior visceral endoderm and/or anterior mesendoderm (e.g. Sox17, Cer1, and Foxa2). Many of the down regulated genes are normally expressed in posterior mesoderm (e.g. Mixl1, Mesp1, Wnt5a and Fgf3) (Fig. 2b). This suggests there is an imbalance in cellular composition of EVX1-Δfs EBs with loss of posterior cell types and relative expansion of anterior cell types (both mesoderm and endoderm).
Using Gene Ontology (GO) we found the most highly significantly enriched cell biological process terms among the DEGs were: blood vessel development (35) and morphogenesis (28), pattern specification (38), regulation of the cell cycle (29), gastrulation (16) or formation of primary germ layer (12), cell differentiation (25), cell motion (37) and WNT signalling (20) (Fig. 2d). We undertook hierarchical clustering of the genes within each of the GO Ontology categories (Fig. 2c). Each category contains genes down-regulated (blue) or up-regulated (red) in EVX1-Δfs cells. Within the blood vessel development, patterning and gastrulation categories, there is down-regulation of many ligands, receptors, signalling molecules and transcription factors which are implicated in posterior patterning of mesoderm. These include Bmp4, Wnt2, Fgf10, Kdr (Flk1), Cxcr4, Cited1, Cited2, Cdx2, Hand1, Mixl1, Hes1, Tbx3, Tbx6, Mesp1, Eomes, Snai1 and others. There is also upregulation of a smaller set of key patterning genes which are normally expressed in the AVE, anterior mesendoderm or epiblast. These include Sox17, Nanog, Atm, Zic3, and Foxa2. We also found many genes in the WNT signalling pathway are downregulated in EVX1-Δfs EBs, including Wnt3, Wnt2, Wnt5a, Wnt5b, Lef1, Pitx2, Dvl2, Lrp5 and others. On the other hand, WNT pathway antagonists such as Cer1, Cfc1/cripto and Sfrp1 are upregulated. Interestingly, Evx1 is downregulated in WNT3A KO embryos36, demonstrating that there is a mutual dependence (direct or indirect) between WNT signalling and EVX1. There were no significant changes in expression of any members of the HoxA cluster under these differentiation conditions. Full lists of differentially expressed genes are provided in Supplementary Table 2.
Previously EVX1 has been shown to directly repress GSC in human ESCs under Activin A growth conditions10. Gsc was not upregulated in either EVX1-Δfs clone by qPCR or RNAseq under serum differentiation conditions (Supplemental Table 2, Supplementary Figure 1C). To ensure that the lack of Gsc upregulation was not due to differences in the differentiation conditions used, we differentiated EVX1-Δfs and W9.5 EBs in SFM with Activin A. We did not find significant upregulation of Gsc (Fig. 2e). Importantly, the A-P gene expression defect in EVX1-Δfs persisted in Activin A. Defective expression of Mixl1 and T remained, and additional significant reduction in expression of the pan mesoderm marker Sp5 was also present in EVX1-Δfs EBs (Fig. 2e). The upregulation of AVE and endoderm markers Cer1 and Sox17 was less dramatic in Activin A, presumably because selective expansion of AVE and anterior tissues dilutes the differences in anterior gene expression between wildtype and EVX1-Δfs EBs (Fig. 2f).
From this RNAseq data, it was unclear whether EVX1 exerts its A-P patterning function by directly regulating BMP4 and/or WNT signalling pathways or whether changes in Bmp4/Wnt expression are an indirect result of changes in cellular composition within the EVX1-Δfs EBs. BMP4 and WNT direct EB differentiation towards posterior cell types37, so the aberrant transcriptome in EVX1-Δfs EBs may be primarily due to downregulation of these factors. To address this possibility, we attempted to rescue EVX1-Δfs EBs by culture in SFM with exogenous recombinant WNT3A (20 ng/ml) or BMP4 (10 ng/ml). Under WNT3A conditions, we observed no rescue of Mixl1 gene expression; i.e. it remained significantly down regulated (Fig. 2f). Likewise there was no reversal of the up-regulation of Cer1, suggesting the A-P patterning defect was not rescued by WNT3A (Fig. 2f). Similar results were found for EBs grown in BMP4 (Supplementary Figure 1D). Thus, downregulation of Bmp4 and Wnt genes (Wnt3, Wnt5a and Wnt5b) is likely to be an indirect result of changes in cellular composition of EVX1-Δfs EBs and not due to direct regulation of these factors by EVX1. This data also shows EVX1 is required for appropriate transcriptional responses and differentiation downstream of BMP4 and WNT signalling pathways. This function appears to be conserved in humans, as β-catenin, the major effector of WNT signalling, has been shown to bind to regulatory regions in the EVX1 promoter in differentiated hESCs11. Together, these results confirm EVX1 plays a key role in A-P patterning of nascent mesoderm and endoderm and is essential for regulating the posteriorizing effects of WNTs and BMP4.
CRISPR/Cas9 mediated removal of the Evx1/Evx1as locus results in an identical D-V pattering defect as EVX1-Δfs
Bidirectional/antisense transcription from highly expressed, developmentally regulated genes is a common phenomenon in the mammalian genome30. However the functional significance and possible mechanisms of action of such antisense lncRNAs remain topics of debate38,39. The Evx1 locus contains a stable, highly expressed antisense lncRNA associated with Evx1, so it is an ideal locus to study. Visual inspection of RNAseq data from D4 EBs suggests that the majority of the Evx1as transcript is expressed as Evx1as T2 (Fig. 3a). We further validated this by qPCR using primers to distinguish between the different isoforms of Evx1as. Only 1% of the total transcript derived from the overlapping intronic promoter (Supplementary Figure 1A).
We initially used RNAi, delivered via shRNA containing lentiviral vectors40 to attempt stable knockdown of Evx1as without disturbing the genomic locus (Supplementary Figure 2A) (see Methods). Such an approach has been successful for some lncRNAs but challenging for others41,42. In some cases RNAi has obtained different and more severe phenotypes than constitutive deletions, which may be due to off target or toxicity effects in some cases or due to compensatory events in others43,44. We generated stable clones targeting Evx1as using three independent shRNAs (Supplementary Figure 2A). Unfortunately, none of the clones from any of the three siRNAs displayed knockdown of Evx1as to <50% of WT levels, so we could not make concrete conclusions about the potential function of Evx1as from these experiments. There were no significant changes in expression of key mesoderm genes such as Brachyury, Mixl1, or Evx1 itself (data not shown).
Genetic deletion or mutation remains the gold standard to demonstrate the requirement of a gene’s function43. We therefore performed 3 different CRISPR-Cas9 mediated manipulations of the Evx1/Evx1as locus (Fig. 3a). The first manipulation ‘Del#1’ removed the entire first intronic promoter of Evx1as, which is specific to isoform Evx1as T1 (1% of total transcript) and conserved (Fig. 1a). Removal of this promoter, which generates a small proportion of Evx1as RNA, did not affect expression of Evx1as or Evx1 (data not shown). ‘Del#2’ removed the shared promoter region between Evx1 and Evx1as. This deletion removed 220 bps centred around the beginning of Evx1 and Evx1as transcription, based on ESTs and RNAseq data. This deletion resulted in a dramatic loss of Evx1 reducing the transcript to ~16% of WT levels (Supplementary Figure 2B). However this deletion only decreased Evx1as to ~46% of WT levels. This result may be explained by a more malleable transcriptional start site and/or weaker constraints on transcriptional regulation at the lncRNA promoter. As expected, ‘Del#2’ displayed reduction of Mixl1 and significant upregulation of Cer1, supporting the results from the EVX1-Δfs RNAseq data (Supplemental Figure 2B).
Since Evx1as was still expressed at 46% of WT in ‘Del#2’, we could not draw any definitive conclusions about its potential function. Therefore, we generated a large ~2.6 kb deletion (‘Del#3’) that removed the promoters from both versions of the transcript (to prevent the possibility of isoform switching between the two alternative promoters), as well as the second/third exon (Fig. 3a). Successful deletion was validated by PCR and later by RNAseq (Fig. 3a, Supplementary Figure 2C). This deletion resulted in almost complete ablation of Evx1 by qRT-PCR (Fig. 3b). Since the Evx1as qPCR primers sit inside the deletion site (Fig. 3a), primers were designed for the last exon of the transcript to detect any residual expression. Approximately 15% of WT levels of expression of the last exon of Evx1as remained in the ‘Del#3’ D4 EBs (Supplementary Figure 2D).
To determine whether Evx1as has any function independent of EVX1, we performed a direct comparison between the transcriptome of EVX1-Δfs and the Evx1as/Evx1 double KO (‘Del#3’). We reasoned that if Evx1as has an independent function from EVX1, then differences should be observed between the two transcriptomes, regardless of whether it functions in transcriptional or post-transcriptional regulation. Transcriptional differences, if present, could be either due to direct transcriptional regulation by Evx1as or an indirect consequence of Evx1as regulating post-transcriptional pathways.
No significant differences were observed by qPCR between the two lines for a number of gastrulation and A-P patterning genes (Fig. 3b). Both the EVX1-Δfs and Evx1/Evx1as Δ2.6 kb (‘Del#3’) EBs displayed significantly different expression of Mixl1, T and Cer1 relative to WT EBs (Fig. 3b). To determine whether there was any trans function for Evx1as RNA independent of the function of EVX1, we searched for differences genome-wide by mRNAseq. Remarkably, differential gene expression analysis identified only 3 DEGs, two of which were Evx1 and Evx1as, themselves. In the Evx1/Evx1as double KO (‘Del#3’), Evx1 and Evx1as are dramatically reduced to 1% and 4% of EVX1-Δfs (in which Evx1 and Evx1as are expressed the same as WT under serum conditions). The other DEG (Grb10) has no reported role during gastrulation and is unlikely to be of any biological significance. The lack of differences is reflected in the very high correlation observed between the two samples (R2 = 0.9925, Pearson) (Fig. 3c). We therefore conclude that Evx1as does not have function independent of EVX1, and therefore is unlikely to function in trans. This does not rule out a potential function for Evx1as in the regulation of Evx1 in cis (see Discussion).
Discussion
Anterior-posterior patterning determines cell fates and tissue specification during gastrulation. Although, EVX1−/− mice are viable and display no dramatic gastrulation phenotype after birth, we show EVX1 is an important downstream effector of BMP4 and WNT signalling in EBs, and regulates A-P patterning. Genes which are normally highly expressed on the posterior side of the embryo are markedly reduced in EVX1-Δfs EBs. Thus, many genes associated with blood, blood vessel and cardiac outcomes (e.g. Kdr (Flk1), Hand1, Cited2, Tbx3)45,46,47,48,49 are markedly downregulated in the absence of EVX1. On the other hand, genes expressed throughout the mesoderm such as Brachyury (T) and Sp550 are only mildly affected. Genes expressed on the anterior-dorsal side of the embryo such as Cer1 and Sox1751 are increased. This is consistent with the functions of EVX1 in Xenopus laevis (xhox3), zebrafish (eve1) and humans (EVX1); i.e. they all play essential roles in posteriorization of nascent mesoderm8,9,10.
Members of the homeobox gene superfamily can act as either transcriptional activators and/or repressors depending on cofactors or context. EVX1 may function as a transcriptional activator of posterior mesoderm genes, such as Mixl1, and/or as a transcriptional repressor of anterior/dorsal genes such as Sox17 and Cer1. Thus, it could function to maintain the integrity of the posterior region of the embryo by preventing ectopic expression of anterior genes. Indeed, such a role has been proposed for human EVX110. Both scenarios are possible and not mutually exclusive. For example, EVX1 might co-operate with transcriptional activators or repressors, or recruit co-activators and/or co-repressors in different contexts. ChIPseq for EVX1 would be able to distinguish these possibilities; however, no specific mouse EVX1 antibodies are available and we found human antibodies did not work in the mouse.
As is the case for EVX1, disruption of some other key regulators of gastrulation does not always result in strong gastrulation phenotypes in the intact mouse. For example, the GSC, SP5 and CER1 knockout mice display no overt signs of gastrulation defects, despite each of these genes having well established roles in regulating patterning in ES cells or model organisms36,52,53,54,55. The lack of an overt phenotype may be explained by genetic redundancy and/or compensation by other genes and pathways. This is the case for Cer1, which has an overlapping expression profile and redundant function with Lefty156. Unlike either of the single knockout mice, the CER1−/−/LEFTY1−/− mouse has an expanded primitive streak or multiple primitive streaks56. Whether redundancy explains the lack of phenotype in the EVX1−/− mice is unclear. Evx1 has a paralog, Evx2, which is thought to have arisen from vertebrate specific duplication of Evx157. It is similar to Evx1 (82.5% at the nucleotide level), and only differs at one amino acid in the homeodomain58. However, Evx2 is not expressed during gastrulation in vivo57 or in EBs, so it is unlikely to act redundantly with Evx1.
Compensation within key signalling pathways has been shown to ‘buffer’ mis-regulation of some genes during development and dampen phenotypic consequences of gene knockouts59,60,61,62. Since BMP, WNT, TGFβ and FGF signalling pathways act on a large number of genes, it is possible that changes in expression or activity of components of these pathways could compensate for the loss of EVX1 in the context of an entire embryo1,59,61. This does not appear to occur in EVX1-Δfs EBs, since BMP4 and WNT3A cannot rescue the posterior mesoderm gene expression program. Perhaps only in the context of an intact embryo, can compensation be fully active. Alternatively, compensation may occur at the level of alternated expression of transcription factors (such as Mixl1) in vivo, but not in EBs. In short, it remains unclear why EVX1−/− mice display minimal defects, despite a clear function for EVX1 in regulating A-P patterning in EBs.
LncRNAs are an important new class of genetic material with a variety of biochemical activities. Some have proven key regulatory roles in development and differentiation. The abundance of sense-antisense coding-lncRNA gene pairs, such as Evx1/Evx1as, is widely appreciated but the functional significance of the associated lncRNA transcript remains mostly untested. This is partly because functional studies have focused primarily on intergenic lncRNAs to avoid disrupting overlapping coding transcripts63. Certainly, some lncRNAs have well established functions in trans28,63. However, despite being stable, spliced, abundant and developmentally regulated, Evx1as does not function to regulate gene expression independently of EVX1; it is therefore unlikely to function in trans. This does not rule out a possible cis function for Evx1as in the regulation of Evx1. Indeed, other examples of cis acting lncRNAs have been reported5,25,64,65. Further coding-lncRNA pairs need be carefully dissected using precise genetic tools to develop general principles which accurately describe their biological functions. We suggest that it is likely that some lncRNAs may function in trans to exert broad effects on gene expression, some function in cis to regulate their coding partner, and some may simply be non-functional bi-products of intense transcriptional activity at the partner coding gene’s promoter or enhancer.
Methods
ESC Culture and embryoid body differentiation
W9.5 mESCs were maintained on gelatanized plates in mESC media with 1000 U/ml LIF at 37 °C, 5% CO2, as described previously7. To generate EBs, mESCs were plated at a density of 2 × 104 cells/ml on STARTM low adherence plates and allowed to differentiate for 4 days in suspension. Serum free differentiation was conducted in ESGRO complete basal media (SF002, Millipore) as described previously66. Serum-free media was supplemented with recombinant BMP4 at 10 ng/ml, recombinant Activin A at 10 ng/ml and recombinant WNT3A at 20 ng/ml. BMP4, Activin A and WNT3A were purchased from R and D Biosystems. Differentiations performed for side by side comparisons between conditions were set up on the same day to minimize technical and biological variation. At least 3 differentiations (biological replicates) were performed for each comparison with each replicate set up on a different day.
RNA extraction and qRT-PCR
RNA was extracted from cells harvested at D4 using TrizolTM (Thermo-Fischer) according to manufacturer’s instructions. cDNA was synthesized using Superscript IIITM (Thermo-Fischer) according to manufacturer’s instructions using random hexamer primers. qRT-PCR was performed using SYBR-Green (Thermo-Fischer) on the AB7900 (Applied Biosystems). Delta Ct was calculated by comparing to Hprt as a house keeping gene. Delta Delta Ct was calculated by normalizing each condition to WT expression levels. Statistical significance was determined by t-tests. Each condition was tested for significantly different expression to WT. At least 3 biological replicates were performed for each condition in each experiment. For testing the relative abundance of each Evx1as isoform, standard curves were performed with known quantities of DNA to calculate primer efficiencies. These primers efficiencies were taken into account in the Delta Ct calculations. All graphs and statistical tests were performed in Prism 5 (GraphPad). Primer sequences are available in Supplementary Table 1.
CRISPR-Cas9 clone generation and screening
All sgRNAs were designed using the CRISPR design tool (crispr.mit.edu) and cloned into the pSpCas9(BB)-2A-GFP plasmid (#48138, Addgene) as described previously67. pSpCas9(BB)-2A-GFP (PX458) was a gift from Feng Zhang. For generating the EVX1-Δfs mESCs, a single guide was targeted to the second exon of Evx1 in the homeodomain region. mESCs were transfected with the pCas9-GFP plasmid also expressing the sgRNA targeting Evx1. Sorting and screening was conducted as described previously, with minor modifications35. Specifically, the top 5% of GFP + cells were sorted into a 10 cm2 dish and allowed to grow for two days before picking individual clones. This was done to prevent high numbers of mixed clones, which results from retention of the plasmid through cell division. Large deletions (‘Del#1′, ’Del#2′ and ‘Del#3′) were generated using the dual sgRNA strategy67. mESCs were transfected with two plasmids, each containing pCas9-GFP and the appropriate sgRNA. The top 5% of GFP + cells were sorted into a 10 cm2 dish and allowed to grow for two days before picking individual clones. Individual clones were transferred to a 96 well plate and allowed to grow until confluent. DNA was extracted and PCR was performed using primers outside and inside the deleted region to identify clones containing the intended deletions. Clones with the appropriate deletion were expanded and used for subsequent analyses. sgRNA and screening primer sequences are available in Supplementary Table 1.
Whole mount In Situ Hybridization (WISH)
WISH was conducted as described previously, with minor modifications to reduce Proteinase K digestion of E7.5 embryos5. Primers used to amplify and clone probes for Evx1 and Evx1as are available in Supplementary Table 1.
RNA Sequencing and data analysis
Total RNA was analysed on the Bioanalyser (Agilent) to confirm high RNA quality (Rinn value > 8). mRNA selection of total RNA was performed using Ambion Dynabeads© mRNA DIRECTTM Micro Purification Kit (#61021, Life Technologies). Barcoded RNAseq libraries were generated from the mRNA using the Ion Total RNAseq Kit v3 (#4475936) and Ion XpressTM RNAseq Barcode 1–16 Kit (#4475485, Life Technologies). RNAseq libraries were sequenced on the Ion Proton sequencing instrument using the Ion P1 Hi-Q Sequencing 200 Kit with the Ion P1 v2 chip. Each sample was sequenced to a depth of least 15 million reads. RNAseq data was mapped to the Mus Musculus reference genome (mm9) using TopHat2 and then TMAP68. Tophat2 is required to map spliced junctions, while TMAP is designed for IonTorrent sequencing data. Transcript counts were obtained by using HTSeq on the mapped data using the standard settings69. Differential gene expression analysis was performed on the HTSeq output using DESeq2 using the standard settings70. Hierarchically clustered heatmaps were generated in R using pHeatmap (Bioconductor). Scatterplots were generated in Prism 5 (GraphPad). Gene Ontology analysis was performed with all DEGs on DAVID71. Raw data is available via the GEO repository (http://www.ncbi.nlm.nih.gov/geo/) under the accession numbers: GSE75735, GSE75737.
Lentiviral Transduction of shRNAs
We employed the iRNAi v2.1 software to design siRNAs targeting the Evx1as gene. We tested for potential off targeting in the genome by performing BLAST searches to exclude candidates which bound to additional genomic sites. We selected three siRNAs for cloning (Supplementary Table 1). We purchased ssDNA oligos designed for annealing to leave 5′ and 3′ overhands for cloning into the Cla1 and Mlu1 restriction sites of pLVTHM40. We annealed 2 ul of sense and antisense oligos (1 ug/ul) in 1× DNA annealing solution (100 mM potassium acetate, 30 mM Hepes, 2 mM magnesium acetate) in a 50 final volume by heating to 94 °C for 5 mins and slowly cooling at 37 °C. dsDNA was phosphorylated with PNK and then ligated to pLVTHM cut with Mlu1 and Cla1 and dephosphorylated with CIAP. Ligations were performed by standard techniques. Plasmid preps were checked for inserts and concatamer exclusion by digestion with EcoR1 and Xba1 and Sanger sequencing. To generate lentiviruses, 293T cells were transduced Opti-MEM with Lipofectamine-2000 and shRNA-pLVTHM together with pCMV-dR8.2 and pMD2G-VSV-G, as described40. Viral stocks were harvested from media at 48–72 hours, filtered (0.45 μ) and snap frozen. For ES cell transductions, adherent ES cells were cultured for 48 hours with viral supernatant (20% in ES cell media) and polybrene (4 μg/ml). Transduced cells were sorted as eGFP + by FACS and seeded onto MEFs. GFP + clones were then selected for molecular and functional analyses.
Additional Information
How to cite this article: Bell, C. C. et al. The Evx1/Evx1as gene locus regulates anterior-posterior patterning during gastrulation. Sci. Rep. 6, 26657; doi: 10.1038/srep26657 (2016).
Supplementary Material
Acknowledgments
This work was funded by Australian NHMRC Project Grants, APP631644 and APP1045986.
Footnotes
Author Contributions C.C.B. generated the CRISPR/Cas9 vectors and ES cell lines, undertook ES cell differentiations, performed and analysed RNA-seq datasets and wrote the paper; P.P.A. designed experiments, generated plasmids, undertook ES cell culture and contributed to writing the manuscript; A.K. generated lentiviral stocks, ES cell lines and RT-PCR analyses; G.W.M. cultured ES cells and undertook bioinformatics analyses; K.R.G. co-supervised C.C.B. and supervised bioinformatics; P.T. performed RT-PCR on ES cell lines; L.d.L. generated stable shRNA ES cell lines and performed RT-PCR; S.W.C. performed ES cell differentiation experiments; F.G. undertook bioinformatics analyses of Evx1as conservation; J.F. generated plasmids; M.R.T. co-supervised P.T. and G.W.M.; K.-L.R. generated plasmids; J.C. maintained ES cell lines; J.S.M. co-designed and co-funded the project, co-supervised P.P.A. and contributed to writing the manuscript; M.E.D. discovered Evx1as, co-supervised P.P.A. and co-funded the project; A.C.P. managed and funded the project, contributed to ES cell experiments, co-supervised C.C.B. and P.P.A. and co-wrote the manuscript.
References
- Tam P. P. & Loebel D. A. Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet 8, 368–381 (2007). [DOI] [PubMed] [Google Scholar]
- Arnold S. J. & Robertson E. J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nature reviews. Molecular cell biology 10, 91–103, doi: 10.1038/nrm2618 (2009). [DOI] [PubMed] [Google Scholar]
- Kinder S. J. et al. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development 126, 4691–4701 (1999). [DOI] [PubMed] [Google Scholar]
- Pearson J. C., Lemons D. & McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6, 893–904, doi: 10.1038/nrg1726 (2005). [DOI] [PubMed] [Google Scholar]
- Dinger M. E. et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18, 1433–1445, doi: 10.1101/gr.078378.108 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dush M. K. & Martin G. R. Analysis of mouse Evx genes: Evx-1 displays graded expression in the primitive streak. Dev Biol 151, 273–287 (1992). [DOI] [PubMed] [Google Scholar]
- Bruce S. J. et al. Dynamic transcription programs during ES cell differentiation towards mesoderm in serum versus serum-freeBMP4 culture. BMC Genomics 8, 365, doi: 10.1186/1471-2164-8-365 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba A. & Melton D. A. Involvement of the Xenopus homeobox gene Xhox3 in pattern formation along the anterior-posterior axis. Cell 57, 317–326 (1989). [DOI] [PubMed] [Google Scholar]
- Seebald J. L. & Szeto D. P. Zebrafish eve1 regulates the lateral and ventral fates of mesodermal progenitor cells at the onset of gastrulation. Dev Biol 349, 78–89, doi: 10.1016/j.ydbio.2010.10.005 (2011). [DOI] [PubMed] [Google Scholar]
- Kalisz M., Winzi M., Bisgaard H. C. & Serup P. EVEN-SKIPPED HOMEOBOX 1 controls human ES cell differentiation by directly repressing GOOSECOID expression. Dev Biol 362, 94–103, doi: 10.1016/j.ydbio.2011.11.017 (2012). [DOI] [PubMed] [Google Scholar]
- Funa N. S. et al. beta-Catenin Regulates Primitive Streak Induction through Collaborative Interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell 16, 639–652, doi: 10.1016/j.stem.2015.03.008 (2015). [DOI] [PubMed] [Google Scholar]
- Hart A. H. et al. Mixl1 is required for axial mesendoderm morphogenesis and patterning in the murine embryo. Development 129, 3597–3608 (2002). [DOI] [PubMed] [Google Scholar]
- Mead P. E., Brivanlou I. H., Kelley C. M. & Zon L. I. BMP-4-responsive regulation of dorsal-ventral patterning by the homeobox protein Mix.1. Nature 382, 357–360 (1996). [DOI] [PubMed] [Google Scholar]
- Rosa F. M. Mix.1, a homeobox mRNA inducible by mesoderm inducers, is expressed mostly in the presumptive endodermal cells of Xenopus embryos. Cell 57, 965–974 (1989). [DOI] [PubMed] [Google Scholar]
- Tada M., Casey E. S., Fairclough L. & Smith J. C. Bix1, a direct target of Xenopus T-box genes, causes formation of ventral mesoderm and endoderm. Development 125, 3997–4006 (1998). [DOI] [PubMed] [Google Scholar]
- Lemaire P., Darras S., Caillol D. & Kodjabachian L. A role for the vegetally expressed Xenopus gene Mix.1 in endoderm formation and in the restriction of mesoderm to the marginal zone. Development 125, 2371–2380 (1998). [DOI] [PubMed] [Google Scholar]
- Henry G. L. & Melton D. A. Mixer, a homeobox gene required for endoderm development. Science 281, 91–96 (1998). [DOI] [PubMed] [Google Scholar]
- Poulain M. & Lepage T. Mezzo, a paired-like homeobox protein is an immediate target of Nodal signalling and regulates endoderm specification in zebrafish. Development 129, 4901–4914 (2002). [DOI] [PubMed] [Google Scholar]
- Fekany K. et al. The zebrafish bozozok locus encodes Dharma, a homeodomain protein essential for induction of gastrula organizer and dorsoanterior embryonic structures. Development 126, 1427–1438 (1999). [DOI] [PubMed] [Google Scholar]
- Kikuchi Y. et al. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev 14, 1279–1289 (2000). [PMC free article] [PubMed] [Google Scholar]
- Cho K. W., Blumberg B., Steinbeisser H. & De Robertis E. M. Molecular nature of Spemann’s organizer: the role of the Xenopus homeobox gene goosecoid. Cell 67, 1111–1120 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulos D. D. & Capecchi M. R. Targeted disruption of the even-skipped gene, evx1, causes early postimplantation lethality of the mouse conceptus. Genes Dev 8, 1949–1961 (1994). [DOI] [PubMed] [Google Scholar]
- Moran-Rivard L. et al. Evx1 is a postmitotic determinant of v0 interneuron identity in the spinal cord. Neuron 29, 385–399 (2001). [DOI] [PubMed] [Google Scholar]
- Yin Y. et al. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell 16, 504–516, doi: 10.1016/j.stem.2015.03.007 (2015). [DOI] [PubMed] [Google Scholar]
- Wang K. C. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124, doi: 10.1038/nature09819 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E. & Mattick J. S. Long non-coding RNAs: insights into functions. Nat Rev Genet 10, 155–159, doi: 10.1038/nrg2521 (2009). [DOI] [PubMed] [Google Scholar]
- Mattick J. S., Amaral P. P., Dinger M. E., Mercer T. R. & Mehler M. F. RNA regulation of epigenetic processes. Bioessays 31, 51–59, doi: 10.1002/bies.080099 (2009). [DOI] [PubMed] [Google Scholar]
- Rinn J. L. & Chang H. Y. Genome regulation by long noncoding RNAs. Annual review of biochemistry 81, 145–166, doi: 10.1146/annurev-biochem-051410-092902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A. & Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15, 7–21, doi: 10.1038/nrg3606 (2014). [DOI] [PubMed] [Google Scholar]
- Sigova A. A. et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci USA 110, 2876–2881, doi: 10.1073/pnas.1221904110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani S., Sauer S., Bolotin E. & Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell 43, 1040–1046, doi: 10.1016/j.molcel.2011.08.019 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Werber M., Wittler L., Timmermann B., Grote P. & Herrmann B. G. The tissue-specific transcriptomic landscape of the mid-gestational mouse embryo. Development 141, 2325–2330, doi: 10.1242/dev.105858 (2014). [DOI] [PubMed] [Google Scholar]
- Derrien T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789, doi: 10.1101/gr.132159.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. B. & Smith J. C. Graded changes in dose of a Xenopus activin A homologue elicit stepwise transitions in embryonic cell fate. Nature 347, 391–394, doi: 10.1038/347391a0 (1990). [DOI] [PubMed] [Google Scholar]
- Bell C. C., Magor G. W., Gillinder K. R. & Perkins A. C. A high-throughput screening strategy for detecting CRISPR-Cas9 induced mutations using next-generation sequencing. BMC Genomics 15, 1002, doi: 10.1186/1471-2164-15-1002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunty W. C. Jr., Kennedy M. W., Chalamalasetty R. B., Campbell K. & Yamaguchi T. P. Transcriptional profiling of Wnt3a mutants identifies Sp transcription factors as essential effectors of the Wnt/beta-catenin pathway in neuromesodermal stem cells. PLoS One 9, e87018, doi: 10.1371/journal.pone.0087018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nostro M. C., Cheng X., Keller G. M. & Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell 2, 60–71 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J., Ponting C. P. & Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Pelechano V., Jarvelin A. I. & Steinmetz L. M. Functional consequences of bidirectional promoters. Trends Genet 27, 267–276, doi: 10.1016/j.tig.2011.04.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M. & Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol 77, 8957–8961 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paralkar V. R. et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood 123, 1927–1937, doi: 10.1182/blood-2013-12-544494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N. et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 53, 1005–1019, doi: 10.1016/j.molcel.2014.01.021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko A. E., Guenzl P. M., Barlow D. P. & Pauler F. M. Gene regulation by the act of long non-coding RNA transcription. BMC biology 11, 59, doi: 10.1186/1741-7007-11-59 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A. et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233, doi: 10.1038/nature14580 (2015). [DOI] [PubMed] [Google Scholar]
- Risebro C. A. et al. Hand1 regulates cardiomyocyte proliferation versus differentiation in the developing heart. Development 133, 4595–4606, doi: 10.1242/dev.02625 (2006). [DOI] [PubMed] [Google Scholar]
- Li Q., Pan H., Guan L., Su D. & Ma X. CITED2 mutation links congenital heart defects to dysregulation of the cardiac gene VEGF and PITX2C expression. Biochem Biophys Res Commun 423, 895–899, doi: 10.1016/j.bbrc.2012.06.099 (2012). [DOI] [PubMed] [Google Scholar]
- Hoogaars W. M. et al. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovascular research 62, 489–499, doi: 10.1016/j.cardiores.2004.01.030 (2004). [DOI] [PubMed] [Google Scholar]
- Choi K., Kennedy M., Kazarov A., Papadimitriou J. C. & Keller G. A common precursor for hematopoietic and endothelial cells. Development 125, 725–732 (1998). [DOI] [PubMed] [Google Scholar]
- Huber T. L., Kouskoff V., Fehling H. J., Palis J. & Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 432, 625–630 (2004). [DOI] [PubMed] [Google Scholar]
- Treichel D., Becker M. B. & Gruss P. The novel transcription factor gene Sp5 exhibits a dynamic and highly restricted expression pattern during mouse embryogenesis. Mech Dev 101, 175–179 (2001). [DOI] [PubMed] [Google Scholar]
- Arkell R. M. & Tam P. P. Initiating head development in mouse embryos: integrating signalling and transcriptional activity. Open biology 2, 120030, doi: 10.1098/rsob.120030 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada G. et al. Targeted mutation of the murine goosecoid gene results in craniofacial defects and neonatal death. Development 121, 2917–2922 (1995). [DOI] [PubMed] [Google Scholar]
- Meno C. et al. lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell 94, 287–297 (1998). [DOI] [PubMed] [Google Scholar]
- Rivera-Perez J. A., Mallo M., Gendron-Maguire M., Gridley T. & Behringer R. R. Goosecoid is not an essential component of the mouse gastrula organizer but is required for craniofacial and rib development. Development 121, 3005–3012 (1995). [DOI] [PubMed] [Google Scholar]
- Simpson E. H. et al. The mouse Cer1 (Cerberus related or homologue) gene is not required for anterior pattern formation. Dev Biol 213, 202–206, doi: 10.1006/dbio.1999.9372 (1999). [DOI] [PubMed] [Google Scholar]
- Perea-Gomez A. et al. Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell 3, 745–756 (2002). [DOI] [PubMed] [Google Scholar]
- Dolle P., Fraulob V. & Duboule D. Developmental expression of the mouse Evx-2 gene: relationship with the evolution of the HOM/Hox complex. Dev Suppl, 143–153 (1994). [PubMed] [Google Scholar]
- Bastian H. & Gruss P. A murine even-skipped homologue, Evx 1, is expressed during early embryogenesis and neurogenesis in a biphasic manner. Embo J 9, 1839–1852 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A. et al. Robustness of the BMP morphogen gradient in Drosophila embryonic patterning. Nature 419, 304–308, doi: 10.1038/nature01061 (2002). [DOI] [PubMed] [Google Scholar]
- Zhu L. et al. Cerberus regulates left-right asymmetry of the embryonic head and heart. Curr Biol 9, 931–938 (1999). [DOI] [PubMed] [Google Scholar]
- Goentoro L. & Kirschner M. W. Evidence that fold-change, and not absolute level, of beta-catenin dictates Wnt signaling. Mol Cell 36, 872–884, doi: 10.1016/j.molcel.2009.11.017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead A. M. & Wright C. V. Disrupting Foxh1-Groucho interaction reveals robustness of nodal-based embryonic patterning. Mech Dev 136, 155–165, doi: 10.1016/j.mod.2014.12.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I. & Bartel D. P. lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46, doi: 10.1016/j.cell.2013.06.020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Liu Y., Liu R., Zhang K. & Zhang Y. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell reports 11, 137–148, doi: 10.1016/j.celrep.2015.03.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Weissman S. M. & Newburger P. E. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA biology 11, 777–787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D. et al. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell 3, 508–518, doi: 10.1016/j.stem.2008.09.013 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nature protocols 8, 2281–2308, doi: 10.1038/nprot.2013.143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36, doi: 10.1186/gb-2013-14-4-r36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T. & Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, doi: 10.1093/bioinformatics/btu638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W. et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8, R183, doi: 10.1186/gb-2007-8-9-r183 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.