Abstract
An enormous amount of long non-coding RNAs (lncRNAs) transcribed from eukaryotic genome are important regulators in different aspects of cellular events. Cytoplasm is the residence and the site of action for many lncRNAs. The cytoplasmic lncRNAs play indispensable roles with multiple molecular mechanisms in animal and human cells. In this review, we mainly talk about functions and the underlying mechanisms of lncRNAs in the cytoplasm. We highlight relatively well-studied examples of cytoplasmic lncRNAs for their roles in modulating mRNA stability, regulating mRNA translation, serving as competing endogenous RNAs, functioning as precursors of microRNAs, and mediating protein modifications. We also elaborate the perspectives of cytoplasmic lncRNA studies.
Keywords: lncRNA, mRNA stability, mRNA translation, ceRNA, MicroRNA
Introduction
Mammalian genome is pervasively transcribed into many different complex families of RNA. However, less than 2% of mammalian genome is transcribed into mRNA to encode proteins, whereas a major portion of the genome is transcribed into interweaved and overlapping transcripts that include thousands of non-coding RNA (ncRNA) transcripts [1], [2]. ncRNAs more than 200 nucleotides in length are called long ncRNAs (lncRNAs), which are often transcribed by RNA polymerase II [3], [4]. These lncRNAs are usually devoid of open reading frames (ORFs), with or without the 3′ polyadenylation [5], [6], [7], [8]. Interestingly, expression of lncRNA is more tissue-specific than that of mRNA [9].
In the last several years, a large number of nuclear lncRNAs have been discovered. These lncRNAs play diverse roles in the nucleus through various mechanisms [10]. For example, nuclear lncRNAs control the epigenetic state of particular genes [11], participate in transcriptional regulation [12], get involved in alternative splicing and constitute subnuclear compartments [13], [14].
Although for most if not all of the lncRNAs, nucleus is the place of biogenesis and processing, cytoplasm is the final residence and site of action for some lncRNAs. Biogenesis of lncRNAs is quite complicated and share many features of protein-coding RNAs. Within the nucleus, they occupy the chromatin fraction. 17% of lncRNAs vs. 15% of mRNAs are enriched in the nucleus, whereas 4% vs. 26%, respectively, are enriched in the cytoplasm [6]. Many lncRNA-mediated mechanisms of gene regulation have been identified in the cytoplasm [8], [15], [16]. In the last decade or so, thousands of cytoplasmic lncRNAs have been discovered, indicating their importance for multiple cellular activities. In this review we highlight the functions and underlying mechanisms of some important cytoplasmic lncRNAs that are responsible for posttranscriptional regulations such as on mRNA stability and translational control.
Modulation of mRNA stability
In the cytoplasm, several lncRNAs target mRNA transcripts and modulate mRNA stability. Some lncRNAs such as half-STAU1-binding site RNAs (1/2-sbsRNAs) and growth arrested DNA-damage inducible gene 7 (gadd7) decrease the stability of mRNA, while others such as antisense transcript for β-secretase 1 (BACE1-AS) and the terminal differentiation-induced ncRNA (TINCR) increase mRNA stability.
1/2-sbsRNAs
mRNAs can be degraded via staufen 1 (STAU1)-mediated mRNA decay (SMD), when their 3′ untranslated region (3′ UTR) binds to STAU1 [17]. STAU1 is a double-stranded RNA (dsRNA)-binding protein, which binds within 3′ UTR of translationally-active mRNA [14], [18]. STAU1 binds to a complex structure of 19-bp stem with 100 nt apex within the mRNA encoding ADP ribosylation factor 1 (ARF1) [18]. This stem region is conserved in 3′ UTRs of ARF1 mRNA of mouse and rat [6]. However, such stem structures were not identified in other STAU1 targets [18]. STAU1-binding sites can be formed by imperfect base-pairing between an Alu element in the 3′ UTR of an SMD target and another Alu element in a cytoplasmic, polyadenylated lncRNA [18]. These lncRNAs transactivate the binding of STAU1 to mRNA as only STAU1 could be immunoprecipitated with lncRNAs called 1/2-sbsRNAs, thus unveiling a pivotal strategy of recruiting proteins to mRNAs and mediating the mRNA decay (Figure1A). However, not all mRNAs containing Alu element in their 3′ UTR are targeted for SMD, despite the presence of complementary 1/2-sbsRNAs that target other mRNA for SMD [17]. One of the 378 identified 1/2-sbsRNAs in humans, 1/2-sbsRNA1 contains a single Alu element that base pairs with the Alu element in the 3′ UTR of plasminogen activator inhibitor type 1 (SERPINE1) and FLJ21870. 1/2-sbsRNA1 is present in the cytoplasm but absent in the nucleus of HeLa cells [18] and only STAU1 can be immunoprecipitated with 1/2-sbsRNA1. Two isoforms of 1/2-sbsRNA1, including 1/2- sbsRNA1(S) (short form) and 1/2-sbsRNA1(L) (long form), have been reported. Both isoforms contain the Alu element and 3’ UTR with poly (A) tail, although they differ at the 5′ end. Knocking down 1/2-sbsRNA1(S) increased the level of SERPINE1 and FLJ21870 mRNAs by 2–4.5-folds above normal. Other 1/2-sbsRNA members such as 1/2-sbsRNA2, 1/2-sbsRNA3, and 1/2-sbsRNA4 are largely cytoplasmic and polyadenylated as well, containing a single Alu element. Knocking down these 1/2-sbsRNAs led to upregulation of their mRNA targets [17]. Functional studies showed that 1/2-sbsRNA1 contributed to the reduced cell migration by targeting SERPINE1 and RAB11-family-interacing protein 1 (RAB11FIP1) mRNAs for SMD as confirmed by scarp injury repair assay [17].
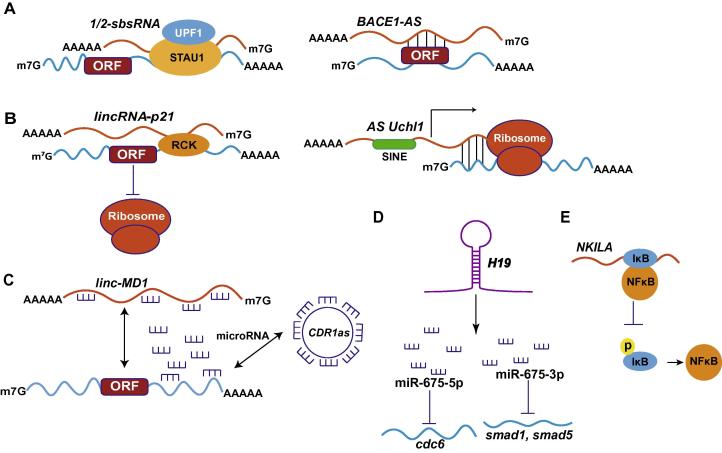
Figure 1.
Known working models of cytoplasmic lncRNA function
A. lncRNAs modify mRNA stability, with 1/2-sbsRNA as an example of decreasing the stability of mRNA and BACE1-AS as an example of increasing the stability of mRNA. B. lncRNAs regulate mRNA translation, with lincRNA-p21 as an example of inhibiting the translation and AS Uchl1 as an example of promoting translation. C. lncRNAs modulate gene expression by functioning as ceRNAs, with linc-MD1 as well as CDR1as shown as examples. D. lncRNAs can give rise to microRNAs, with H19 shown as an example. E. Some lncRNAs affect protein modification, with NKILA as one of such kind of cytoplasmic lncRNAs. ORF, open reading frame; 1/2-sbsRNA, half-STAU1-binding site RNA; BACE1-AS, antisense transcript for β-secretase 1; linc-MD1, long non-coding RNA muscle differentiation 1; CDR1as, cerebellar degeneration-related protein 1 antisense transcript; NKILA, NF-κB interacting lncRNA; AS Uchl1, antisense transcript for ubiquitin carboxy terminal hydrolase L1; SINE, short interspersed element; cdc6, cell division cycle 6.
gadd7
gadd7 is a 754-nt polyadenylated lncRNA isolated from Chinese hamster ovary (CHO) cells [15], [16]. Expression of gadd7 is induced by several types of DNA damage and growth arrest signals [19], [20], and gadd7 plays a pivotal role in regulating G1/S checkpoint post DNA damage. gadd7 also regulates lipid-induced oxidative and endoplasmic reticulum (ER) stress [21]. lncRNAs are known to bind to and regulate the functions of proteins. One such example is the binding of gadd7 with TAR DNA-binding protein (TDP-43), and this interaction is strengthened upon UV exposure [22], [23], [24]. TDP-43 is a member of the heterogeneous nuclear ribonucleoprotein (hnRNP) family. HnRNP family members are RNA/DNA binding proteins involved in transcription, splicing, mRNA transport, and mRNA stability [25], [26]. TDP-43 is known to repress the expression of cyclin-dependent kinase 6 (Cdk6) mRNA in Hela cells, which is important for G1-phase progression [27], [28]. Nonetheless, Cdk6 expression is found to be activated by TDP-43 in CHO cells [24]. UV-induced gadd7 directly interacts with TDP-43, thus leading to the decreased interaction between TDP-43 and Cdk6 mRNA. This results in Cdk6 mRNA degradation, and finally inhibition of cell cycle progression [24]. gadd7 is not highly conserved at the nucleotide level [29], [30]. Since the structure or the functional motif of lncRNAs may be more important, and thus would be more conserved than their nucleotide sequence [9], it remains possible to identify a functional gadd7 ortholog in humans. This may be important for unveiling the pathogenesis of diseases such as frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS), as dominant mutations in TDP-43 are causative of these two important neurodegenerative diseases [24], [31], [32].
BACE1-AS
Expression of the conserved non-coding BACE1-AS increases BACE1 mRNA stability when HEK-SW cells are exposed to cellular stressors like amyloid-β1–42 (Aβ1–42) [33]. BACE1-AS renders BACE1 mRNA stability by masking the binding site of miR-485-5p (Figure1A). BACE1-AS and miR-485-5p compete for binding in the sixth exon of BACE1 mRNA. The sense-antisense RNA duplex between BACE1 and BACE1-AS in the cytoplasm potentially perturb the interaction between miR-485-5p and BACE1 mRNA, which to some extent, explains the mRNA stabilization by BACE1-AS transcript [34].
TINCR
The TINCR gene resides on chromosome 19 in humans and encodes a predominantly cytoplasmic 3.7-kb lncRNA. TINCR regulates human epidermal differentiation by post transcriptional mechanism [35]. Previously found as an uncharacterized lncRNA, TINCR is now believed to be the most highly-induced lncRNA during epidermal differentiation [35], [36]. TINCR binds to mRNA through a 25-nt ‘TINCR box’ motif, which is robustly enriched in the interacted mRNAs. TINCR RNA has a strong affinity for STAU1 protein [17], [35], [37], [38]. TINCR–STAU1 complex mediates the stabilization of differentiation-related mRNAs, such as KRT80 encoding keratin 80 in an ultraviolet protection factor 1/2 (UPF1/2)-independent manner, however the exact mechanism remains obscure [39].
Modulation of translation
Gene expression control at translational level plays a crucial role in multiple biological systems and provides valuable means for the spatiotemporal management of complex protein dynamics in eukaryotic cells [40], [41], [42]. Some lncRNAs also get involved in such regulation at the translational level, which can either repress (as exemplified for lincRNA-p21 below) or promote (as exemplified for AS Uchl1 below) translation.
lincRNA-p21
The human lincRNA-p21 is also known as tumor protein p53 pathway corepressor 1 (Trp53cor1). lincRNA-p21 is ∼3.0 kb in length, and the encoding gene is located ∼15 kb upstream of p21/cdkn1a gene [23]. It is more abundant in cytoplasm compared to nucleus, known to co-distribute with ribosomes [43]. As a post-transcriptional modulator, lincRNA-p21 can negatively regulate the translation of CTNNB1 (β-catenin) and JUNB transcripts by imperfectly base pairing at different sites in the coding and untranslated regions (both 5′ and 3′ UTRs) of CTNNB1 (15 sites) and JUNB mRNAs (8 sites). When the level of Hu antigen R (HuR), a ubiquitous RNA binding protein, reduces, lincRNA-p21 becomes stable and interacts with its target transcripts including CTNNB1 and JUNB mRNAs. The resulting lincRNA-p21–mRNA complex can enhance the interaction between mRNAs and the translational repressors RCK as well as Fragile X mental retardation protein (FMRP). Consequently, translation of the target transcripts is repressed through reduced polysome sizes and ribosome drop-off (Figure 1B) [43], [44].
AS Uchl1
A recent study reported the discovery of a spliced nuclear-enriched antisense transcript (AS Uchl1) complementary to the mRNA that encodes mouse ubiquitin carboxy terminal hydrolase L1 (Uchl1) [45]. Uchl1 is an enzyme specifically expressed in dopaminergic neurons [24], [46], [47]. The activity of the AS Uchl1 depends on the presence of a 73-nt overlapping sequence complementary with 5′ end of Uchl1 mRNA and an embedded inverted SINEB2 repetitive element (Figure1B) [45]. Under normal physiological conditions, AS Uchl1 is enriched in the nucleus, and upon rapamycin treatment, inhibition of mTORC1 triggers the transport of AS Uchl1 to the cytoplasm, which then targets the overlapping Uchl1 mRNA to active polysomes for cap-independent translation. Exact molecular mechanism as to how AS Uchl1 promotes the translation of Uchl1 mRNA under stress conditions is still elusive.
Competing endogenous RNAs
Coding and non-coding RNAs can regulate each other through their ability to compete for miRNA binding. lncRNAs harboring multiple binding sites of identical miRNA are called competing endogenous RNAs (ceRNAs) [48]. ceRNA can sequester miRNAs and therefore protect their target mRNAs from repression [49], [50], [51], [52], [53]. This activity was first discovered in Arabidopsis thaliana and later in mammals [53], [54]. Multiple ceRNAs have been identified, and we present some as examples below.
HULC
Hepatocellular carcinoma (HCC) is one of the most fatal cancers [55]. Recent studies have indicated that a large number of lncRNAs are functionally deregulated in HCC [56], [57], [58], [59]. Among these, highly up-regulated in liver cancer (HULC) is a novel mRNA-like ncRNA. It is present in the cytoplasm, spliced, polyadenylated, and resembles the mammalian LTR transposon 1A [60]. As reflected by its name, HULC is highly upregulated in HCC, and it is also detected in gastric cancer and colorectal carcinomas that metastasize to the liver [60], [61], [62]. The HULC gene resides on chromosome 6p24.3 in humans and is conserved in primates. It is about 1.6 kb in length and contains two exons. Expression of HULC gene in Hep3B cells can be up-regulated by the transcription factor cAMP responsive element binding protein (CREB). HULC acts as endogenous sponge of miR-372 [63]. HULC binding to miR-372 reduces miRNA-mediated translational repression of protein kinase cAMP-activated catalytic subunit beta (PRKACB), one of the target genes of miR-372 [63]. PRKACB can induce phosphorylation of CREB, which in turn stimulates HULC expression, thus forming a feedforward loop [63].
linc-MD1
linc-MD1 is a muscle-specific lncRNA, which is indispensable for the timing of muscle differentiation and plays an important role in myogenesis [64]. linc-MD1 acts as a natural decoy for two muscle-specific miRNAs, miR-133 and miR-135 (Figure1C) [64]. Expression of mastermind-like-1 (MAML1) is controlled by miR-133, and myocyte-specific enhancer factor 2C (MEF2C) is the target of miR-135 [64]. MAML1 and MEF2C are important myogenic factors required for activation of muscle-specific genes. MEF2C binds to the promoter region of cardiac muscle genes and positively regulates the differentiation of muscle cells [65], [66], while MAML1 acts as a transcription coactivator in some signal transduction pathways (such as Notch signaling) related to muscle differentiation [67]. With the depletion of linc-MD1, expression of both MAML1 and MEF2C is repressed, whereas over expression of linc-MD1 resulted in high levels of MAML1 and MEF2C. These observations argue for a direct competition between linc-MD1 and mRNAs for miRNA binding [64].
linc-RoR
The lncRNA regulator of reprogramming (linc-RoR) functions as microRNA (miRNA) sponge against miR-145. Interaction between linc-RoR and miR-145 prevents mRNA of some important transcription factors (TFs) like Oct4, sox2, and Nanog in human embryonic stem cells (hESCs) from miRNA-mediated regulation [68], [69]. The expression of linc-RoR is positively correlated with the undifferentiated state of hESCs [69].
CDR1as and circSry
Recently, additional examples of ceRNA were found in circular RNAs (circRNAs), which represent a newly identified large class of lncRNAs [70], [71], [72]. circRNAs can be formed by back-splicing of the 5′ end of an upstream exon with the 3′ end of the same exon or a downstream exon. Although some circRNAs such as EIciRNAs are predominantly localized in the nucleus, circRNAs are generally cytoplasmic. circRNAs appear to be non-coding and lack the association with polysomes [71], [73]. Two cytoplasmic circRNAs have been reported to act as miRNA sponge. The first one is the cerebellar degeneration-related protein 1 antisense transcript (CDR1as, also called ciRS-7), which is a sponge for miR-7 (Figure1C). CDR1as contains 74 miR-7 seed matches, out of which 63 are conserved in mammals [72]. The other one is a testis-specific circRNA encoded by the gene sex-determining region Y (circSry), which contains 16 putative binding sites for miR-138 [71], [72], [74]. These two circRNAs may be special cases, and it may not be a general phenomenon for circRNAs to function as miRNA sponges [75].
Precursor of miRNAs
A genome-wide survey predicted that nearly 100 lncRNAs encode miRNAs [76]. These lncRNAs may not be predominantly cytoplasmic, but they may be processed in the nucleus and cytoplasm to give rise to functional miRNAs.
H19
H19 is one of the best known imprinting genes expressed from the maternal allele and required for proper muscle differentiation and muscle regeneration [77], [78], [79]. The H19 gene is present on chromosomes 11 and 7 in humans and mice, respectively [80], [81]. There is no conserved ORF sequence in H19 RNA between mice and human. Although the H19 gene is imprinted paternally, the H19 RNA itself does not take part in imprinting mechanism [82]. Studies based on structure prediction suggest that H19 is a ncRNA, 2.3-kb long, capped, spliced, and polyadenylated [82], [83]. It is reported that H19 lncRNA acts as a molecular sponge for let-7 family of miRNAs in a HEK293 cell line [84]. Depleting H19 causes accelerated muscle differentiation, which can be recapitulated by let-7 overexpression [84]. In the cytoplasm of undifferentiated multipotent mesenchymal C2C12 cells, H19 interacts with the K homology-type splicing regulatory protein (KSRP). Such binding favors KSRP-mediated destabilization of myogenin transcripts [85].
Besides the aforementioned roles of H19, exon 1 of H19 also gives rise to miR-675-3p and miR-675-5p (Figure1D). miR-675-3p targets the gene encoding the anti-differentiation TFs smad1 and smad5, which are crucial components of the bone morphogenetic protein (BMP) pathway [86], whereas miR-675-5p targets the gene encoding DNA replication initiation factor Cdc6 [86]. In this regard, H19 has a pro-differentiation function in primary myoblasts and regenerating skeletal muscles due to the resulting miR-675-3p and miR-675-5p [86], [87]. H19 is also found to regulate placenta growth. Insulin like growth factor 2 (Igf2), which is also targeted by miR-675-3p, is an important regulator of growth and is upregulated in H19-deficient placenta [88]. H19 is also found to modulate gastric cancer cell proliferation through miR-675, by targeting the gene encoding the tumor suppressor runt domain transcription factor1 (RUNX1). Thus H19/miR-675 regulates the expression of RUNX1 to modulate gastric cancer [89].
linc-MD1 (again)
We discussed linc-MD1 as a ceRNA before. However, linc-MD1 primary transcript also harbors the pri-miR-133b sequence. If cleaved by Drosha in the nucleus, linc-MD1 can give rise to a miRNA precursor. Recently, HuR protein is described as another component of linc-MD1 regulatory circuitry [90]. HuR is known to contribute to muscle differentiation [91]. HuR interacts with many coding and non-coding RNAs, indicating its pleiotropic RNA binding activity [92], [93]. HuR binds to and favors linc-MD1 accumulation at the expense of miR-133 biogenesis. HuR also recruits miR-133 onto linc-MD1 in the cytoplasm, thereby reinforcing this regulatory circuitry. There is an inverse correlation between levels of HuR and miR-133b. HuR binds the base of the pri-miR-133b stem loop, and physically interferes with microprocessor activity [90]. Further investigations have to be carried out to answer how the processing and function of linc-MD1 are regulated either as the pri-miR-133b in the nucleus or as sponge for miR-133b when exported to the cytoplasm as an unprocessed transcript.
Regulation of protein modification
In the recent years, several lncRNAs are identified to modulate modifications of cytoplasmic proteins such as ubiquitination/deubiquitination or phosphorylation/dephosphorylation.
lnc-DC
Expression of lnc-DC is almost exclusive to human conventional dendritic cells (DCs) [94]. lnc-DC could help activate STAT3 by binding to it in the cytoplasm, thus promoting the phosphorylation and preventing dephosphorylation of STAT3. Knockdown of lnc-DC inhibited the differentiation to the DC lineage as well as the functions of DCs [94].
NKILA
NF-κB interacting lncRNA (NKILA) binds directly to IκB and blocks IKK-induced IκB phosphorylation, thus inhibiting NF-κB activation (Figure1E) [95]. The expression of NKILA is also upregulated by NF-κB. NKILA interacts with the NF-κB/IκB complex, and seems to keep the NF-κB pathway from over-activation and to suppress cancer metastasis [95].
Another role of lincRNA-p21
lincRNA-p21 was reported to regulate the ubiquitination of HIF-1α, a transcription factor crucial to hypoxia-induced effects such as Warburg effect [96]. lincRNA-p21 is induced by HIF-1α under hypoxia condition, and binds to both HIF-1α and von Hippel–Lindau tumor suppressor (VHL) protein. Such binding blocks the interaction between VHL and HIF-1α, thus inhibiting VHL-mediated ubiquitination of HIF-1α. This positive feedback loop between HIF-1α and lincRNA-p21 promotes glycolysis under hypoxia [96].
Perspectives
lncRNAs are recognized as major regulators in life events such as gene expression, cell differentiation, and tumorigenesis. In this article, we summarized the roles of some lncRNAs in the cytoplasm. lncRNAs can function in the posttranscriptional gene expression such as mRNA stability and translation. Through RNA–protein or RNA–RNA interaction, cytoplasmic lncRNAs could also serve as ceRNAs, miRNA precursors, or modulators of protein phosphorylation.
Recent findings have shown that certain transcripts previously-annotated as lncRNAs in fact can be translated to produce small bioactive peptides [97], [98], [99], [100]. For instance, the conserved micropeptide myoregulin (MLN) was found to be encoded by a skeletal muscle-specific RNA, a previously putative lncRNA [100]. MLN shows structural and functional similarity with SERCA inhibitors, phospholamban and sarcolipin. Interacting directly with SERCA, MLN disrupts the Ca2+ uptake into the sarcoplasmic reticulum [100]. Similarly, the endogenous 34-amino acid micropeptide dwarf open reading frame (DWORF) is encoded by another putative muscle-specific lncRNA. DWORF enhances muscle performance by physically interacting with SERCA inhibitors such as phospholamban, sarcolipin, and MLN [100]. These examples demonstrated that some (although maybe limited in numbers) transcripts that are previously annotated as lncRNAs are actually coding, and thus should be considered as mRNAs. Given the vast amount of lncRNAs identified, and many of them are associated with noncoding functions, it is no doubt that more functions and functional working mechanisms are yet to be explored for the large number of cytoplasmic lncRNAs.
Regulation of lncRNA localization is important to coordinate their functions in the nucleus or in the cytoplasm. There should exist machinery either directly or indirectly to transport specific lncRNAs into the cytoplasm, and maybe further to special subcellular locations or complexes. The final localization, concentration, and functions of a specific lncRNA have to be fine tuned by the RNA biogenesis, transportation, degradation, and maybe even modifications. Substantial efforts are required to investigate these aspects.
A single lncRNA can have multiple roles. For example, both H19 and linc-MD1 can function as ceRNAs as well as precursors for miRNAs. How these different roles of the same lncRNA are coordinated remains to be addressed. On the other hand, there are undoubtedly more roles and functional mechanisms remain unknown for cytoplasmic lncRNAs. With the extensive investigations of the eukaryotic transcriptome by means of RNA sequencing, most of the lncRNAs including cytoplasmic ones may have already been described. Further studies on these lncRNAs may help to classify them into subclasses based on their biogenesis and functions.
Competing interests
The authors declare no competing interests.
Acknowledgments
This work is supported by the National Basic Research Program of China (973 Program; Grant No. 2015CB943000), the National Natural Science Foundation of China (Grant Nos. 91519333 and 31471225), and the Fundamental Research Funds for the Central Universities (Grant No. WK2070000034).
Handled by Er-Wei Song
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Mercer T.R., Dinger M.E., Mattick J.S. Long noncoding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 2.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu S., Wu J., Chen L., Shan G. Signals from noncoding RNAs: unconventional roles for conventional pol III transcripts. Int J Biochem Cell Biol. 2012;44:1847–1851. doi: 10.1016/j.biocel.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Ramskold D., Wang E.T., Burge C.B., Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran V.A., Perera R.J., Khalil A.M. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer T.R., Dinger M.E., Sunkin S.M., Mehler M.F., Mattick J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilusz J.E. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859:128–138. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung T., Wang Y., Lin M.F., Koegel A.K., Kotake Y. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T. The nuclear-retained non-coding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernard D., Prasanth K.V., Tripathi V., Colasse S., Nakamura T., Xuan Z. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingolia N.T., Lareau L.F., Weissman J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montes M., Nielsen M.M., Maglieri G., Jacobsen A., Hojfeldt J., Agrawal-Singh S. The lncRNA MIR31HG regulates p16INK4A expression to modulate senescence. Nat Commun. 2015;6:6967. doi: 10.1038/ncomms7967. [DOI] [PubMed] [Google Scholar]
- 17.Gong C., Maquat L.E. LncRNAs transactivate STAU1- mediated mRNA decay by duplexing with 39 UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y.K., Furic L., Parisien M., Major F., DesGroseillers L., Maquat L.E. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollander M.C., Alamo I., Fornace A.J., Jr. A novel DNA damage inducible transcript, gadd7, inhibits cell growth, but lacks a protein product. Nucleic Acids Res. 1996;24:1589–1593. doi: 10.1093/nar/24.9.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fornace A.J., Jr., Alamo I., Jr., Hollander M.C. DNA damage inducible transcripts in mammalian cells. Proc Natl Acad Sci U S A. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brookheart R.T., Michel C.I., Listenberger L.L., Ory D.S., Schaffer J.E. The non-coding RNA gadd7 is a regulator of lipid-induced oxidative and endoplasmic reticulum stress. J Biol Chem. 2009;284:7446–7454. doi: 10.1074/jbc.M806209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H., Wei L., Tao Q., Deng H., Ming M., Xu P. Decreased NURR1 and PITX3 gene expression in Chinese patients with Parkinson’s disease. Eur J Neurol. 2012;19:870–875. doi: 10.1111/j.1468-1331.2011.03644.x. [DOI] [PubMed] [Google Scholar]
- 25.Buratti E., Baralle F.E. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 26.Cohen T.J., Lee V.M., Trojanowski J.Q. TDP-43 functions and pathogenic mechanisms implicated in TDP-43 proteinopathies. Trends Mol Med. 2011;17:659–667. doi: 10.1016/j.molmed.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee M.H., Yang H.Y. Regulators of G1 cyclin-dependent kinases and cancers. Cancer Metastasis Rev. 2003;22:435–449. doi: 10.1023/a:1023785332315. [DOI] [PubMed] [Google Scholar]
- 28.Ayala Y.M., Misteli T., Baralle F.E. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin dependent kinase 6 expression. Proc Natl Acad Sci U S A. 2008;105:3785–3789. doi: 10.1073/pnas.0800546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang K.C., Frith M.C., Mattick J.S. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Hellwig S., Bass B.L. A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc Natl Acad Sci U S A. 2008;105:12897–12902. doi: 10.1073/pnas.0805118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabashi E., Valdmanis P.N., Dion P., Spiegelman D., McConkey B.J., Velde C.V. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 32.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faghihi M.A., Ming Z., Jia H., Farzaneh M., Van der Brug M.P., Michael A.N. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kretz M., Siprashvili Z., Chu C., Webster D.E., Zehnder A., Qu K. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan D., Gong Y., Qin W., Zhang P., Li J., Wei L. Large-scale cDNA transfection screening for genes related to cancer development and progression. Proc Natl Acad Sci U S A. 2004;101:15724–15729. doi: 10.1073/pnas.0404089101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiebler M.A., Hemraj I., Verkadi P., Kohrmann M., Fortes P., Marion R.M. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dugré-Brisson S., Elvira G., Boulay K., Chatel-Chaix L., Mouland A.J., DesGroseillers L. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kretz M. TINCR, staufen1, and cellular differentiation. RNA Biol. 2013;10:1597–1601. doi: 10.4161/rna.26249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gray N.K., Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 41.Sonenberg N., Hershey J.W.B., Mathews M.B. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2000. Translational control of gene expression; p. 1020. [Google Scholar]
- 42.Gebauer F., Hentze M.W. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon J.H., Abdelmohsen K., Srikantan S., Yang X., Martindale J.L., De S. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chu C.Y., Rana T.M. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y., Fallon L., Lashuel H.A., Liu Z., Lansbury P.T., Jr. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 47.Setsuie R., Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem Int. 2007;51:105–111. doi: 10.1016/j.neuint.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Salmena L., Poliseno L., Tay Y., Pandolfi PP. The ceRNA hypothesis: the Rosetta stone of a hidden RNA language. Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karreth F.A., Tay Y., Perna D., Ala U., Tan S.M., Rust A.G. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tay Y., Kats L., Salmena L., Weiss D., Tan S.M., Ala U. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng W., Si S., Zhang Q., Li C., Zhao F., Wang F. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79–89. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen M.T., Lin H.S., Shen C., Ma Y.N., Wang F., Zhao H.L. PU.1-regulated long noncoding RNA lnc-MC controls human monocyte/macrophage differentiation through interaction with microRNA 199a-5p. Mol Cell Biol. 2015;35:3212–3224. doi: 10.1128/MCB.00429-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 54.Poliseno L., Salmena L., Zhang J., Carver J., Haveman J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumor biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bosch F.X., Ribes J., Cléries R., Diaz M. Epidemiology of hepatocellular carcinoma. Clin Liver Dis. 2005;9:191–211. doi: 10.1016/j.cld.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 56.Matouk I.J., DeGroot N., Mezan S., Ayesh S., Abu-lail R., Hochberg A. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;9:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsang W.P., Kwok T.T. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene. 2007;26:4877–4881. doi: 10.1038/sj.onc.1210266. [DOI] [PubMed] [Google Scholar]
- 58.Ventura A., Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin R., Chen Z., Ding Y., Hao J., Hu J., Guo F. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma. 2013;60:486–492. doi: 10.4149/neo_2013_063. [DOI] [PubMed] [Google Scholar]
- 60.Panzitt K., Tschernatsch M.M., Guelly C., Moustafa T., Stradner M., Strohmaier H.M. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 61.Matouk I.J., Abbasi I., Hochberg A., Galun E., Dweik H., Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol. 2009;21:688–692. doi: 10.1097/meg.0b013e328306a3a2. [DOI] [PubMed] [Google Scholar]
- 62.Zhao Y., Guo Q., Chen J., Hu J., Wang S., Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31:358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 63.Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lilly B., Zhao B., Ranganayakulu G., Paterson B.M., Schulz R.A., Olson E.N. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 66.Lin Q., Schwarz J., Bucana C., Olson E.N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen H., McElhinny A.S., Cao Y., Gao P., Liu J., Bronson R. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20:675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loewer S., Cabili M.N., Guttman M., Loh Y.H., Thomas K., Park I.H. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y., Xu Z., Jiang J., Xu C., Kang J., Xiao L. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 70.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard K.C. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 72.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 73.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 74.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 75.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He S., Su H., Liu C., Skogerbo G., He H., He D. MicroRNA-encoding long non-coding RNAs. BMC Genomics. 2008;9:236. doi: 10.1186/1471-2164-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pachnis V., Belayew A., Tilghman S.M. Locus unlinked to a-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci U S A. 1984;81:5523–5527. doi: 10.1073/pnas.81.17.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poirier F., Chan C.T., Timmons P.M., Robertson E.J., Evans M.J., Rigby P.W. The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development. 1991;113:1105–1114. doi: 10.1242/dev.113.4.1105. [DOI] [PubMed] [Google Scholar]
- 79.Davis R.L., Weintraub H., Lassar A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 80.Bartolomei M.S., Zemel S., Tilghman S.M. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y., Tycko B. Monoallelic expression of the human H19 gene. Nat Genet. 1992;1:40–44. doi: 10.1038/ng0492-40. [DOI] [PubMed] [Google Scholar]
- 82.Brannan C.I., Dees E.C., Ingram R.S., Tilghman S.M. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Juan V., Crain C., Wilson C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000;28:1221–1227. doi: 10.1093/nar/28.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kallen A.N., Zhou X.B., Xu J., Qiao C., Ma J., Yan L. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giovarellia M., Bucci G., Ramos A., Bordo D., Wilusz C.J., Chen C.Y. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc Natl Acad Sci U S A. 2014;111:E5023–E5028. doi: 10.1073/pnas.1415098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dey B.K., Karl P., Anindya D. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cai X., Cullen B.R. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Keniry A., Oxley D., Monnier P., Kyba M., Dandolo L., Smits G. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhuang M., Gao W., Xu J., Wang P., Shu Y. The long non-coding RNA H19 derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315–322. doi: 10.1016/j.bbrc.2013.12.126. [DOI] [PubMed] [Google Scholar]
- 90.Legnini I., Morlando M., Mangiavacchi A., Fatica A., Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell. 2014;53:506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Von Roretz C., Beauchamp P., Di Marco S., Gallouzi I.E. HuR and myogenesis: being in the right place at the right time. Biochim Biophys Acta. 2011;1813:1663–1667. doi: 10.1016/j.bbamcr.2011.01.036. [DOI] [PubMed] [Google Scholar]
- 92.Lebedeva S., Jens M., Theil K., Schwanhäusser B., Selbach M., Landthaler M. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 93.Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 95.Liu B., Sun L., Liu Q., Gong C., Yao Y., Lv X. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 96.Yang F., Zhang H., Mei Y., Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 97.Kondo T., Plaza S., Zanet J., Benrabah E., Valenti P., Hashimoto Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 98.Andrews S.J., Rothnagel J.A. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 99.Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nelson B.R., Makarewich C.A., Anderson D.M., Winders B.R., Troupes C.D., Wu F. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]