Abstract
Background
Hepatectomy generally offers the best chance of long-term survival for patients with hepatocellular carcinoma (HCC). Many studies have shown that hepatectomy accelerates tumor metastasis, but the mechanism remains unclear.
Methods
An orthotopic nude mice model with palliative HCC hepatectomy was performed in this study. Metastasis-related genes in tumor following resection were screened; HCC invasion, metastasis, and some molecular alterations were examined in vivo and in vitro. Clinical significance of key gene mRNA expression was also analyzed.
Results
Metastasis suppressor 1 (MTSS1) located in the central position of gene function net of residual HCC. MTSS1 was up-regulated in residual tumor after palliative resection. In hepatitis B-related HCC patients undergone palliative hepatectomy, those with higher MTSS1 mRNA expression accompanied by activation of matrix metalloproteinase 2 (MMP2) in residual HCC, had earlier residual HCC detection after hepatectomy and poorer survival when compared to those with lower MTSS1. In different cell lines, the levels of MTSS1 mRNA increased in parallel with metastatic potential. MTSS1 down regulation via siRNA decreased MMP2 activity, reduced invasive potentials of HCC by 28.9 % in vitro, and averted the deteriorated lung metastatic extent in vivo.
Conclusions
The poor prognosis of hepatitis B-related HCC patients following palliative hepatectomy associates with elevated MTSS1 mRNA expression; therefore, MTSS1 may provide a new research field for HCC diagnosis and treatment.
Electronic supplementary material
The online version of this article (doi:10.1186/s13046-016-0361-8) contains supplementary material, which is available to authorized users.
Keywords: Metastasis suppressor 1, Hepatocellular carcinoma, Invasion, Metastasis
Background
Liver cancer in men is the fifth most frequently diagnosed cancer worldwide but the second most frequent cause of cancer-related deaths; in women, it is the sixth most commonly diagnosed cancer and the sixth leading cause of cancer-related deaths [1]. Among primary liver cancers, hepatocellular carcinoma (HCC) represents the major histological subtype, accounting for 70 to 85 % of the total liver cancer burden worldwide [2]. Hepatic resection is a standard treatment for patients with HCC. However, even in patients with HCC undergoing curative resection, it is only potentially curative due to the existence of tumor cells or clinically undetectable residual intrahepatic lesions [3]. Our clinical data showed that the proportion of cases with palliative hepatectomy was 34.0 % from 1958 to 2008 (2754/8107, unpublished data). Our follow-up research suggests that HCC patients treated with palliative hepatectomy suffered from dramatically worsened metastases, suggesting that palliative hepatectomy enhances the metastatic potential of residual HCC. Although surgery is associated with tumor suppression and prolonged survival in a few cases [4], increasing number of reports indicate that hepatic resection exacerbates tumor growth and triggers tumor metastasis [5–7]. We found that palliative resection enhances the metastatic potential of residual HCC [5]. The present study sought to explore the underlying molecular mechanism of this metastasis-enhancing effect.
The incidence, development, invasion, and metastasis of HCC are regulated by the activation, inactivation, or dysregulation of several genes. This study employed an orthotopic human HCC model with high metastatic potential in nude mice. This model was developed at Liver Cancer Institute, Fudan University [5, 8]. Using Human Tumor Metastasis Microarray, we screened the metastasis-related genes in tumor tissues following palliative resection, and found that up-regulated metastasis suppressor 1 (MTSS1) was located in the central position of gene function net of residual HCC in liver.
MTSS1 is also known as “missing in metastasis” (MIM) gene. Among the multiple MIM gene products including MIM-A, MIM-B and MIM-C, MIM-B is the longest and most abundant protein in the cell, which is representative of MIM protein [9]. MTSS1 has been proposed as a potential metastasis suppressor gene in some studies of HCC [10, 11]. However, other studies have shown that MTSS1 is highly expressed in various tumors [12, 13], including HCC [14]. MTSS1 may have an important role in tumor metastasis [12, 15, 16]. MTSS1/MIM over-expression is associated with enhanced cell migration, resulting in tumorigenesis, invasion and metastasis [17–19], and predicts poor prognosis in colorectal cancer [20], cervical carcinoma [21], and lung cancer [22]. Recently, Mertz et al. reported that MTSS1 promotes the metastasis of melanocytes, and high MTSS1 expression defines a subgroup of primary melanomas with unfavorable prognosis [23]. It remains unclear whether or not MTSS1 plays a role in metastasis of residual HCC following palliative resection. HCC metastasis involves basement membrane invasion following matrix metalloproteinase (MMP) activation [24]. Previously, we found that palliative resection activates MMP2 in nude mouse models with HCC [5]. In this study, we first screened the metastasis-related genes in residual HCC tissues, and found that MTSS1 was located in the central position of the tumor gene network. We investigated the MTSS1 mRNA expression in residual tumor and analyzed its association with prognosis in patients with hepatitis B-related HCC after palliative resection. Subsequently, using in vitro and in vivo studies, we found that MTSS1 enhanced the invasive and metastatic potential of HCC cells via MMP2 activation. To our knowledge, the current study provides the first evidence that elevated MTSS1 mRNA expression exacerbates lung metastasis after palliative resection in an HCC model, with poor prognosis of hepatitis B-related patients with HCC treated with palliative hepatectomy.
Methods
Patients, specimens and follow-up
The inclusion criteria for patients in this study were (a) patients with hepatitis B from 2000 to 2010; (b) pathologically proven HCC based on WHO criteria; (c) no anticancer treatment prior to first hepatectomy; (d) surgical resection twice in 6 months; (e) availability of frozen biopsy and/or resected tissues expressing HCC and residual HCC; and (f) availability of follow-up data. This study was approved by the Research Ethics Committee of the Shanghai Jiaotong University Affiliated Sixth People’s Hospital, and informed consent was obtained from each patient. HCC patients with hepatectomy were followed up every 2 months during the first postoperative year and at least every 3 to 4 months subsequently, until December 2014 by monitoring serum α-fetoprotein (AFP) levels, abdominal ultrasonography, chest X-ray or computed tomography every 1 to 6 months depending on the patient’s condition. Residual HCC was suspected 2 months after first hepatectomy (HCC-P I group, n = 37) in all patients and was confirmed by fine needle autopsy and/or secondary surgery (HCC-P II group, n = 37) in 6 months after the first resection. Forty patients with radical hepatectomy (complete resection of tumor with the cut surface free of cancer histologically) and without recurrence at least 12 months after surgery (HCC-R group), and 20 liver tissues from patients with hepatic hemangioma combined with hepatitis B who underwent liver resection were also collected (Liver-T group). All specimens were collected in the operating room immediately (≤15 min) after tissue removal and were snap frozen in liquid nitrogen and stored at −80 °C. General data, metastatic characteristics, pathologic characteristics and survival were compared among the groups.
Animals, tumor models and cell lines
Five-week-old male athymic BALB/c nu/nu mice weighing 18–20 g each were obtained from the Shanghai Institute of Materia Medica, Chinese Academy of Science. All mice used in these investigations were handled in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council, 1996), and were approved by the Institutional Animal Care and Use committee of the Shanghai Jiaotong University Affiliated Sixth People’s Hospital. A stepwise metastatic human HCC model system was established in Liver Cancer Institute of Fudan University, which included a metastatic HCC model in nude mice LCI-D20 [8], a HCC cell line MHCC97 with high metastatic potential that originated from LCI-D20 tumor, a high metastatic subclone (MHCC97H with a pulmonary metastatic rate up to 100 % using orthotopic inoculation) and a lower metastatic subclone (MHCC97L with a lung metastatic rate up to 40 % using orthotopic inoculation) established through in vivo selection of MHCC97 [25]. The noninvasive human liver cell line of L02 (normal), human HCC cell lines of Hep3B with very low invasiveness [26], and highly invasive MHCC97H [25], SMCC7721, MHCC97L, HCCLM3 and HCCLM6 were prepared in this study.
Mice grouping and treatment
Human HCC tumor models produced by MHCC97H were established in nude mice by orthotopic inoculation [8]. Briefly, under anesthesia, the left lobe of the liver was exposed, and part of the liver surface was mechanically injured with scissors. A piece of MHCC97H tumor tissue (size 2 × 2 × 2 mm) was fixed within the liver tissue. Thirty-six nude mice were used in this study.
In the trial experiment, we treated MHCC97H cells with mouse plasma extracted at different time intervals after palliative hepatectomy in nude mice bearing HCC. We found that these cells showed two trans-membrane peaks: one occurring at the 12th hour post-hepatectomy and the other occurring on day 14 post-hepatectomy. In our mouse model, by the day 14 post-hepatectomy, lung metastasis occurred in a majority of the cases. Therefore, we selected the day 14 as a cut-off point to study residual HCC and related lung metastasis, subsequently. Eighteen nude mice bearing HCC xenografts were randomized into three groups 14 daysays after orthotopic implantation: palliative resection group (mice undergoing partial HCC resection with preservation of 2 mm of tumor pedicles), sham operation group (mice with exposed liver but without resection), and blank control group (mice without further surgical intervention). All mice were sacrificed by cervical dislocation 14 days after palliative resection based on pre-experimental results (Additional file 1: Figure. S1A and B). The HCC tissues excised first were designated as tumor tissues T1; tissues excised next from sham operation group were named T2; tissues from blank control group were named tumor tissues T3; and those derived from palliative resection group were denoted T4 that represented residual HCC. Five randomly selected tumor specimens from each group were used for the screening of genes related to metastasis by microarray techniques. Another 18 nude mice treated with palliative resection were randomly injected at multiple points with 100 μL borate-buffered saline (BBS, BBS group), 2 × 107 U Lenti-GFP (Lenti-GFP group) or Lenti-MTSS1 (Lenti-MTSS1 group), respectively, under ultrasound guidance, and the procedure was repeated 3 h later. All mice were sacrificed by cervical dislocation 14 d post-resection for the evaluation of early metastasis.
Parameters, sample preparations and grading of lung metastasis
Mice were sacrificed for gene profiling of pulmonary metastatic nodules. Lung tissues, tumor tissues, mRNA and protein extracts were harvested for further studies. Serial sections of lung tissue measuring 5 μm in thickness were obtained, and all metastases were verified histologically. The degree of lung metastasis was graded by the maximum number of tumor cells counted in the solitary pulmonary metastatic nodules (grades): grade I, < 20; grade II, 20–50; grade III, 50–100; and grade IV, > 100 [27].
Tumor metastasis-related gene profiling
In recent years, more and more researches employed Human Tumor Metastasis Microarray to dissect the gene function and form gene regulatory network in various of cancers [28–30]. Thus, the Human Tumor Metastasis Microarray is useful in searching for new therapeutic targeted genes and helpful in dissect mechanism of the gene in several of cancers [31, 32]. The OHS-028 OligoGEArray Human Tumor Metastasis Microarray (Super Array, Bethesda, MD) was used to characterize the metastasis-related gene expression profiles of HCC tissues in this study. GEArray Expression Analysis Suitesoft ware was used for gene analysis. The methods of Support Vector Machine (SVM), gene significance analysis and gene correlation degree analysis were adopted to detect the potential markers involved in HCC metastasis. To further explore the relationship between gene function and gene function, we built the GO-network (gene function net). In the GO-network, the degree represented the relationship between GO terms. The key GO terms in the network were signal transduction, multicellular organismal development, ion transport, and signal transduction in the GO terms. Among them, multicellular organismal development, GO terms had the highest degree in the network. Then, a gene function net was constructed based on specific gene clustering analysis and multi-dimensional scale [33, 34].
This study describes the advantages of using Support Vector Machines algorithm for the classification of gene expression data. Using the RVM model, we found the different genes including the d genes from microarray data. Our conclusions are as follows. The characteristic of samples, which amount to n was allocated the different gene set, and we obtain a matrix (X)n × d from the samples. The liver cancers samples, which are high or low, constituted the dataset of {xi, yi}(i = 1, 2, ⋯, n), where the xi represents the d dimension vector, and yi ∈ {high = 1, low = − 1} is the class label. We obtained an optimal hyperplane derived from the SVM algorithm. The optimal hyperplane that is high and low is as follows: Minimizing and Subject to yi × 〈xi, ω〉 + b ≥ 1. From the quadratic form, we selected the Support Vectors which carried weights, and decision functions as follows: . We transformed the nonlinear Microarray data into linear format using the gauss and ploy kernel functions. Gauss kernel function was as follows: . The ploy kernel function was as follows: k(xi, xj) = (〈xTi, xj〉 + 1)d = 〈Φ(xi), Φ(xj)〉, where 〈Φ(xi), Φ(xj)〉 is a scalar product. Because the {x1, x2, ⋯, xn} represents nonlinear space, we transformed the vector xi into high-dimensional feature space using the kernel function. The prediction function is as follows: . We demonstrated that the gauss kernel function was effective with a predictive accuracy close to 90 %.
The expression datasets presented in this publication have been deposited in NCBIs Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE75965.
Hematoxylin and eosin (H&E) stains
Paraffin blocks of 10 % buffered formalin-fixed samples of tumor and lung tissues were prepared, and 5–μm-thick serial sections were obtained. Pulmonary metastatic nodules were verified via Hematoxylin and Eosin (H&E) stains.
Real-time fluorescent quantitative polymerase chain reaction (Real-time PCR)
The primers used for real-time polymerase chain reaction (PCR) were designed using Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA): human MTSS1 forward, 5′-tagctggaaggactgggcta-3′, and reverse, 5′-agtcatgctccgtggtctct-3′. GAPDH forward and reverse primers were 5′-ggtgaaggtcggagtcaacg-3′ and 5′-accatgtagttgaggtcaatgaagg-3′, respectively. PCR was performed in the Rotor-Gene 3000 PCR system (Corbett Research, Sydney, Australia). Conditions for PCR were 37 °C for 2 min, 94 °C for 3 min, 40 cycles for GAPDH of 94 °C for 5 s, 60 °C for 40 s, followed by 37 °C for 5 s, and 95 °C for 30 s, 95 °C for 30 s, 40 cycles for MTSS1 of 95 °C for 15 s, 60 °C for 15 s, 72 °C for 30 s. Finally, baseline and threshold values of these genes were set using the Rotor-Gene 6.0 (Corbett Research) for analysis.
Western blot
Proteins were separated by 10 % sodium dodecyl sulfate -polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, MA). The membrane was blocked with 5 % non-fat dried milk in TBST (20 mM Tris–HCl, 150 mM NaCl, and 0.1 % Tween 20, pH 7.5) for 2 h and incubated overnight with antibodies against MTSS1 (Abnova, Caltag-Medsystems Ltd., Buckingham, UK) at 4 °C. After washing with TBST buffer, membranes were incubated with horseradish peroxidase-conjugated anti-mouse IgG secondary antibodies for 1 h at room temperature and detected by enhanced chemiluminescence detection system (Amersham-Pharmacia Biotech, Braunschweig, Germany). GAPDH was used as an internal control (Santa-Cruz Biotechnologies, California, USA).
siRNA design and lentivirus construction
The small interfering RNA (siRNA) sequences targeting MTSS1 were designed according to a modified Tuschel standard, and the detailed sequences were as follows:
Target 1:
(+) 5′-CGGCCAGTGATTGAAGAAGAA-3′,
(−) 5′-TTCTTCTTCAATCACTGGCCG-3′;
Target 2:
(+) 5′-GCTGGATAAAGACCACGCAAA-3′,
(−) 5′-TTTGCGTGGTCTTTATCCAGC-3′;
Target 3:
(+) 5′-CCTTCCAGACTACGCTCATTA-3′,
(−) 5′-TAATGAGCGTAGTCTGGAAGG-3′;
Target 4:
(+) 5′-CCCATGACTCAGGATTCATAT-3′,
(−) 5′-ATATGAATCCTGAGTCATGGG-3′
and Target 5:
(+) 5′-GACCATCTCGGAAGATCTAAA-3′,
(−) 5′-TTTAGATCTTCCGAGATGGTC-3′.
The target 2 sequences were selected for further study. The siDNA sequences matching the designed siRNA were then amplified with PCR and inserted into a mouse U6 promoter-driven lentivirus plasmid (purchased from Genechem Co., Shanghai, China), in which a luciferase expressing construct was then incorporated, forming the pGCL-MTSS1-GFP plasmid. In brief, the infectious titer was determined by fluorescence-activated cell sorting analysis of GFP-positive in 293 T cells. The virus titers were in the range of 107 transducing units/mL medium.
Stable lentiviral transfection of HCC cells
HCC cells were plated in 24-well plates. Subconfluent cells were infected with 3 mL/well three times (about 3 h per infection). The cells were divided into three groups: the knockdown cells transfected with MTSS1 siRNA lentivirus (KD group), the negative control cells transfected with empty lentivirus (NC group) and the blank control cells not transfected (CON group).
MTSS1 overexpression
The full sequence of MTSS1 was amplified by the standard PCR procedure. The primer sequences for cloning include sense primer: ATGGAGGCTGTGATTGAG; and antisense primer: CTAAGAAAAGCGAGGGG. We transfected the pCMV6-empty and pCMV6-MTSS1 into Hep3B cancer cells and RNA and protein were extracted. MTSS1 overexpression was confirmed, and invasive potential assessed.
In vitro invasion assay
The MHCC97H cells (5 × 104 cells/well) treated with MTSS1 siRNA lentivirus, empty lentivirus or with control (no treatment) were added to the upper chamber (100 μL DMEM), and 600 μL conditioned medium was added to the lower chamber. After 24 h of incubation, the invaded cells were fixed with methanol and stained with crystal violet solution. The results were expressed as the number of penetrated cells under a microscope at 200× magnification in five random fields and presented as the means ± SD of three assays.
Gelatin zymography assay
Zymography for MMP2 activity was conducted via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) [35], with modification. The MHCC97H cells (5 × 104 cells/well) treated with or without MTSS1 siRNA lentivirus were serum-starved for 24 h before cell supernatant collection. The molecular weights of these bands indicating MMP2 activity were determined by molecular weight standards (Bio-Rad Laboratories, Hercules, CA). The assays were conducted in triplicate.
MMP2 activity assay
Human MMP2 activity was quantified with an enzyme-linked immunosorbent assay (ELISA) System (Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer’s instructions. The plate was read at 450 nm in a SPECTRAmax 250 Microplate Spectrophotometer (Molecular Devices, Sunnyvale, CA). The assays were conducted in triplicate.
Statistical analysis
All continuous variables were expressed as means ± SD or means ± SE. ANOVAs, Student t test and Cochran-Mantel-Haenszel (CMH) test (Row Mean Scores Differ) were used for statistical comparison among groups. MTSS1 expression and clinicopathological parameters were analyzed by Pearson chi-square test or Fisher’s exact test. The survival analysis was conducted using the Kaplan-Meier method. The statistical SAS software package (Version 8.2, SAS Institute, Inc., Cary, NC, USA) was adopted for data analysis, and the statistical significance was defined as P < 0.05.
Results
MTSS1 located in the central position of the gene function net of residual HCC
Palliative HCC resection promoted tumor metastasis. Therefore, we first screened the metastasis-related genes in HCC tissues to detect the altered genes in residual tumor. Gene cluster and sample cluster analysis were based on tumor-related genes found in the HCC tissues (Fig. 1a). Gene function nets were constructed successfully (Fig. 1b, c, d and e). MTSS1 was located in the central position of the gene function net of residual HCC (Fig. 1e). Analysis of differential gene expression showed that the density of the net (Fig. 1f), which reflects the relevance of different genes, was higher in the palliative group (T4 group, 0.0670) than in controls (T1 group, 0.0145; T2 group, 0.0210; T3 group, 0.0146), and the condensation degree of the net (Fig. 1g) in residual HCC (T4 group, 0.1940) was substantially higher than in the corresponding control group (T1 group, 0.0098). MTSS1 mRNA expression (Fig. 1h) was up-regulated in residual HCC (T4 group) when compared with controls (T1, T2, and T3 groups, P = 0.001, 0.034, and 0.002, respectively).
Fig. 1.
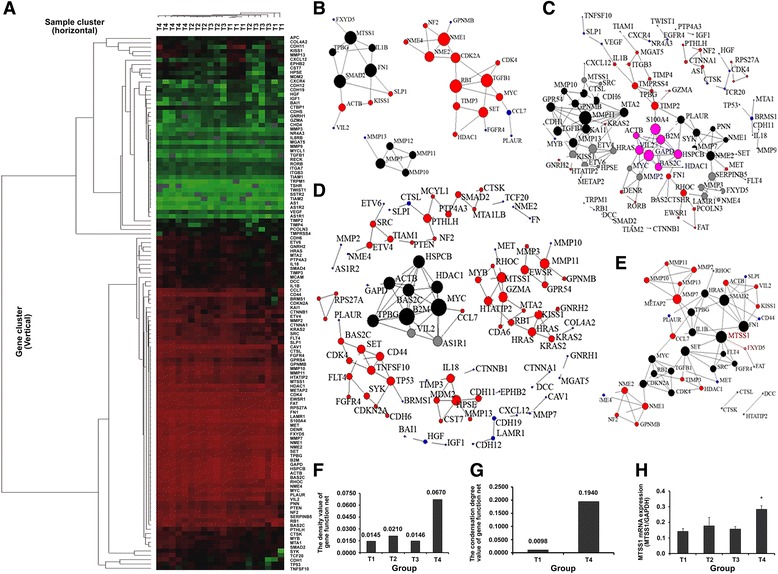
MTSS1 located in the central position of the gene function net of residual HCC. a the cluster heat map of gene cluster (vertical) and sample cluster (horizontal) were based on tumor-related genes in HCC. The color red and green represents high and low gene expression, respectively; samples with expression above mean level (red) are clustered. Gene function nets of HCC were constructed successfully. b gene function nets of HCC from palliative resection group (HCC tissues T1). c gene function nets of HCC from sham operation group (HCC tissues T2). d gene function nets of HCC from blank control group (HCC tissues T3). e gene function nets shown that MTSS1 was located in the central position of the gene function net of residual HCC (HCC tissues T4). f the density value of the gene function net. g the condensation degree of the net. h qRT-PCR analysis shown that palliative resection up-regulated MTSS1 expression, *P = 0.001, 0.034, and 0.002, respectively, when compared with controls (T1, T2, and T3 groups). The bars indicate the means ± s.d
Elevated MTSS1 mRNA expression is associated with poor prognosis
We found that MTSS1 was situated in the central position of gene function net of residual HCC in the nude mice model. We tested MTSS1 expression in tumor tissues from HCC patients. HCC tissues expressed higher MTSS1 mRNA level than the control liver tissues (all P = 0.000, Fig. 2a). MTSS1 mRNA level was significantly increased in residual HCC tissues (HCC-P II group) compared with controls (all P = 0.000, Fig. 2a). In the meantime, MMP2 was also significantly activated (P = 0.000, Fig. 2b). There was no difference in MTSS1 mRNA level or MMP2 activity between tissues derived from HCC-P I and HCC-R groups.
Fig. 2.
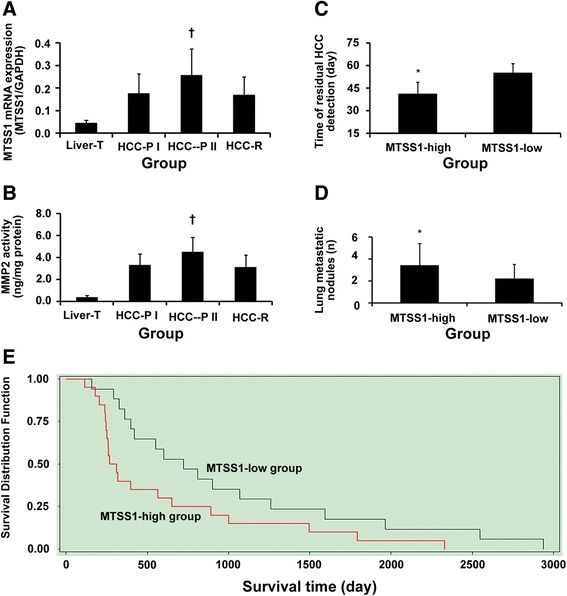
Elevated MTSS1 mRNA expression is associated with poor prognosis. a the qRT-PCR analysis of MTSS1 expression in different groups, P = 0.000, when compared with controls. b MMP2 activity in residual HCC (HCC-P II group); †P = 0.000, when compared with controls; no difference in MTSS1 mRNA level or MMP2 activity between HCC-P I and HCC-R groups. c earlier detection of residual HCC in MTSS1-high group than in MTSS1-low group, *P = 0.000. d higher number of lung metastatic nodules in MTSS1-high group than in MTSS1-low group, *P = 0.039, when compared with controls. e longer survival in MTSS1-low group compared with MTSS1-high group, P = 0.023
The mean MTSS1/GADPH mRNA level of 0.18 was calculated out in the HCC-P I group. Then, we split the patients following palliative resection by MTSS1 expression in residual HCC: > 0.18 (MTSS1-high group, n = 20); and ≤ 0.18 (MTSS1-low group, n = 17). Correlation of MTSS1 mRNA expression with their clinicopathological parameters is displayed in Table 1. A higher level of MTSS1 mRNA was significantly correlated with tumor number (P = 0.035), presence of HCC satellite (P = 0.019), incomplete or no encapsulation (P = 0.008), presence of vascular invasion (P = 0.042) and advanced TNM stage (P = 0.033). Further, most of the AFP-negative patients (9 of 11, 81.8 %) showed higher MTSS1 mRNA levels (P = 0.036). No significant association between MTSS1 mRNA and age, sex, liver cirrhosis, tumor size, or γ-glutamyl transferase was found.
Table 1.
Correlation of tumor MTSS1 mRNA expression level with clinicopathological parameters of HCC patients
| Clinicopathological parameters | Number | MTSS1/GAPDH mRNA (n) | P | |
|---|---|---|---|---|
| MTSS1-low group | MTSS1-high group | |||
| (≤0.18, n = 17) | (>0.18, n = 20) | |||
| Age | 37 | 0.900 | ||
| ≤ 52 y | 20 | 9 | 11 | |
| > 52 y | 17 | 8 | 9 | |
| Sex | 37 | 0.588 | ||
| Female | 8 | 3 | 5 | |
| Male | 29 | 14 | 15 | |
| Cirrhosis | 37 | 0.855 | ||
| Yes | 30 | 14 | 16 | |
| No | 7 | 3 | 4 | |
| Tumor number | 37 | 0.035* | ||
| Multiple | 20 | 6 | 14 | |
| Single | 17 | 11 | 6 | |
| Satellite | 36a | 0.019* | ||
| Yes | 15 | 3 | 12 | |
| No | 21 | 13 | 8 | |
| Tumor size, cm | 37 | 0.212 | ||
| ≤ 5 | 7 | 5 | 2 | |
| > 5 | 30 | 12 | 18 | |
| Encapsulation | 37 | 0.008** | ||
| Complete | 15 | 11 | 4 | |
| Incomplete or none | 22 | 6 | 16 | |
| Vascular invasion | 36a | 0.042* | ||
| Yes | 21 | 6 | 15 | |
| No | 15 | 11 | 4 | |
| TNM stage | 37 | 0.033* | ||
| Early stage (II) | 7 | 6 | 1 | |
| Advanced stage (III-IVA) | 30 | 11 | 19 | |
| AFP level, ng/mL | 37 | 0.036* | ||
| ≤ 20 | 11 | 2 | 9 | |
| > 20 | 26 | 15 | 11 | |
| γ-Glutamyl transferase, units/L | 37 | 0.134 | ||
| ≤ 54 | 19 | 11 | 8 | |
| > 54 | 18 | 6 | 12 | |
| Adjuvant TACE | 37 | 0.815 | ||
| Yes | 16 | 7 | 9 | |
| No | 21 | 10 | 11 | |
NOTE: HCC hepatocellular carcinoma, TACE transcatheter arterial chemoembolization
Significant difference: * < 0.05, ** < 0.01
aNumber less than 37 due to missing data
Patients with higher MTSS1 mRNA expression revealed earlier residual HCC (P = 0.000, Fig. 2c), a higher number of lung metastatic nodules (P = 0.039, Fig. 2d), and lower 1-year survival rate after the first resection [40.0 % (8/20) vs. 76.5 % (13/17), P = 0.026]. The survival of the MTSS1-high group was significantly shorter than that of the MTSS1-low group (P = 0.023, Fig. 2e). In the MTSS1-high group, when sub-grouping patients according to the mean value of MTSS1/GADPH mRNA expression (0.33, n = 20), only 1 out of 9 in those with MTSS1/GADPH mRNA higher than 0.33 survived for more than 1 year, whereas 7 out of 11 with MTSS1/GADPH mRNA lower than 0.33 survived for more than 1 year (P = 0.028). There was no significant difference in the 3-year or 5-year survival rates.
The cumulative survival rates were 56.8 % (21/37) for 1 year and 21.6 % (8/37) for 3 years. The 5-year survival in these patients was 10.8 % (4/37), which was lower than that of all patients with palliative resection performed in our hospital between the years 1958 and 2008 (30.0 %, 826/2754, P = 0.010, unpublished). None of the 37 patients in the current study survived longer than 10 years. Transcatheter arterial chemoembolization (TACE) did not significantly affect MTSS1 mRNA expression and survival between groups or subgroups.
Altered MTSS1 expression is correlated with metastatic potential
To identify the relationship between MTSS1 expression and metastatic potential of HCC, we examined the MTSS1 expression in different cell lines. Significant differences in MTSS1 mRNA levels were found between the L02 cell line without metastatic potential, the Hep3B cell line with very low metastatic potential, and the MHCC97H cell line with higher metastatic potential (P = 0.000, 0.001, respectively, Fig. 3a). The metastatic potential of cell lines increased in parallel with increased levels of MIM-B protein, encoded by MTSS1 (Fig. 3b).
Fig. 3.
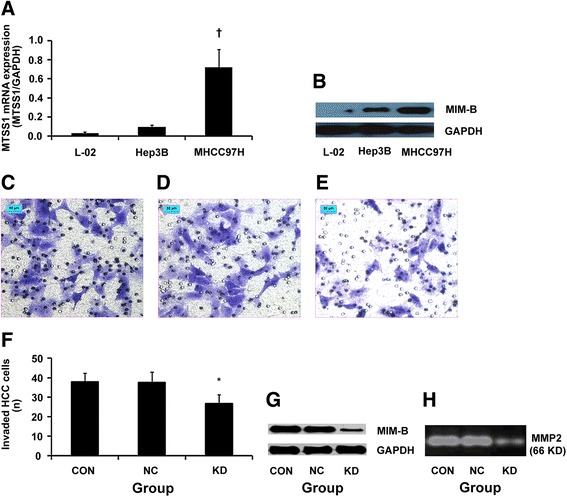
MTSS1/MIM-B expression in different cell lines. a, qRT-PCR analysis of MTSS1 mRNA expression; †P = 0.000, 0.001, respectively, when compared with cell lines of L02 and Hep3B. b, western blot analysis of MIM-B protein level; the bars indicate the means ± s.d. Stable lentiviral transfection of MHCC97H cells with siRNA targeting MTSS1, ×200, bar, 50 μm. Invasion assay analysis shown that the number of transfected HCC cells (original magnification, × 200) after treatment with MTSS1 siRNA lentivirus (e) was lower than that of controls (c, d), bar, 50 μm. f, the difference in invasiveness, *P = 0.000, when compared with controls. The assays were conducted in triplicate. The bars indicate the means ± s.d. g, western blot analysis demonstrated that MIM-B was down-regulated by siRNA. h, results of gelatin zymography assay shown that MMP2 activity was inhibited (0.6-fold) by siRNA, P = 0.000, compared with controls
The siRNA targeting MTSS1 was constructed successfully with high transfection efficiency, which markedly inhibited the invasive ability of MHCC97H cells by 28.9 % (Fig. 3c, d and e). The number of invaded HCC cells in MTSS1 knockdown group (KD group) was less than in the empty lentivirus group (NC group) and the blank control group (CON group, all P = 0.000, Fig. 3f). The MIM-B protein level in KD group was down-regulated when compared with controls (Fig. 3g). Gelatin zymography assay showed that MMP2 activity was decreased significantly (0.6-fold, P = 0.000, Fig. 3h). Further, in vitro studies showed that MTSS1 overexpression in Hep3B cells promoted cell invasion by 18.2 % (P = 0.021, Additional file 1: Figure. S2A and B). The effects of MTSS1 downregulation and overexpression in HCC cells were validated by several other HCC cell lines. MTSS1 downregulation inhibited the invasive ability of HCCLM3 and HCCLM6 cells by 25.7 % (P = 0.002) and 23.1 % (P = 0.001), respectively, while MTSS1 upregulation increased the invasive ability of SMCC7721 and MHCC97L cells by 14.8 % (P = 0.017) and 12.5 % (P = 0.011), respectively.
SiRNA targeting MTSS1 attenuates pulmonary metastasis following palliative resection in nude mice bearing human HCC xenograft
Subsequently, we used in vivo studies to validate the in vitro effects of siRNA targeting MTSS1. Two weeks following palliative HCC resection, the lung metastasis (Fig. 4a) was upgraded when compared with controls (P = 0.039, Table 2). In Lenti-MTSS1 group, the pulmonary metastasis was downgraded after MTSS1 knock down (P = 0.041, Table 2). MMP2 activity of the residual HCC was also significantly inhibited along with MTSS1 inhibition in the Lenti-MTSS1 group when compared with controls (P = 0.003, 0.039, respectively, Fig. 4b and c). Further, no animal experienced therapy-related side effects following siRNA administration.
Fig. 4.
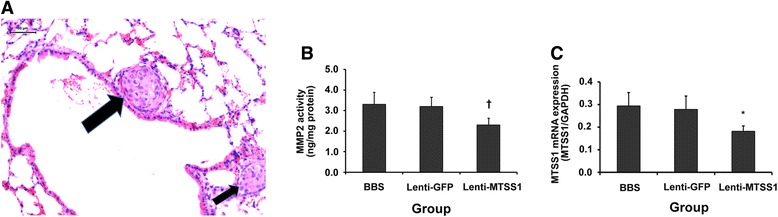
SiRNA targeting MTSS1 attenuates pulmonary metastasis following palliative resection in nude mice bearing human HCC xenograft. a, two pulmonary metastatic nodules (arrows, original magnification, × 200), bar, 50 μm. b, MMP2 activity was inhibited by siRNA targeting MTSS1 in vivo, †P = 0.003 compared with controls. c, qRT-PCR analysis demonstrated that MTSS1 expression was inhibited by siRNA in vivo, *P = 0.039 compared with controls. The bars indicate the means ± s.d
Table 2.
Effects of palliative resection and Lenti-MTSS1 on pulmonary metastasis
| Group | The total number of solitary pulmonary nodule | Lung metastatic extent (Grade)* | ||
|---|---|---|---|---|
| (n) | I | II | III-IV | |
| Palliative Resection | 35 | 12 | 11 | 12 |
| Sham Operation | 33 | 19 | 8 | 6 |
| Black Control | 32 | 20 | 7 | 5 |
| BBS | 32 | 9 | 10 | 13 |
| Lenti-GFP | 31 | 8 | 11 | 12 |
| Lenti-MTSS1 | 30 | 15 | 10 | 5 |
NOTE: All pulmonary metastases were verified via histological examination when mice were sacrificed 2 weeks following palliative resection, or receiving BBS, Lenti-GFP, or Lenti-MTSS1. Lung metastatic extent was graded by number of tumor cells counted in the maximum section of the solitary pulmonary metastatic nodule. *P = 0.039 or 0.041 when Palliative Resection group or Lenti-MTSS1 group was compared among Control groups, respectively
Discussion
HCC is one of the deadliest types of cancers, with a mortality of almost 100 % [36]. The mortality of HCC remains high because the disease is typically diagnosed when it is already at an advanced stage, when most potential curative therapies such as resection and transplantation are of limited efficacy. Recent studies reported that different therapies including surgery [5–7], hepatic artery ligation [37], insufficient radiofrequency ablation [38–40], and sublethal heat treatment [41] promoted residual tumor progression. It is especially important to investigate the underlying mechanism. Our research team has developed a safe and reliable method of palliative HCC resection in an orthotopic nude mouse model, and reported that palliative resection enhances metastatic potential of residual HCC in liver [5]. In the present study, using this method, we investigated the precise mechanism mediating this metastasis.
Data from our institution (1958–2008, unpublished) revealed that in HCC patients who underwent hepatectomy the proportion of cases with palliative HCC resection was 34.0 % (2754/8107). The 5-year survival of patients who underwent palliative HCC resection (30.0 %, n = 2754) was much lower than in those who underwent curative resection (52.6 %, n = 5353). It suggested that HCC palliative resection may promote HCC aggressive metastasis in residual tumor. Using gene microarray techniques and bioinformatics tools, we first found that MTSS1 was upregulated and situated in the central position of gene function net of residual HCC in nude mouse models. We tested the MTSS1 expression in HCC tissues from HCC patients who underwent palliative hepatectomy. We found that patients with high MTSS1 expression manifested residual liver tumors early, higher number of metastatic lung nodules and lower 1-year survival rates, which suggests that HCC patients with high MTSS1 expression have a poor prognosis and a higher mortality following palliative hepatectomy. No significant changes in the survival rates were detected between high- and low-MTSS1 groups after 3 or 5 years, which might be an artifact of the small sample size as few patients survived beyond the first year, especially in high-MTSS1 high patients. Notably, the MTSS1 levels were higher in cancers at advanced stage than in early stage, in our study. In contrast, Ma et al. reported that patients with early pTNM stage (I-II) manifested high MIM-B mRNA expression [14]. Consistent with our study, other studies reported that the positive expression rates of MTSS1 were significantly higher in advanced stages of colorectal cancer [20] and cervical carcinoma [21]. Interestingly, most AFP negative patients (9 of 11, 81.8 %) showed higher MTSS1 mRNA levels in this study, suggesting that MTSS1 may be another early predictor of residual HCC, which merits investigation with a large group of patients.
The cumulative 5-years-survival rate in this group of 37 cases (10.8 %, 4/37) was lower than in all patients undergoing palliative resection at our hospital during 1958 – 2008 (30.0 %, 826/2754). The reasons may be related partially to reoperation 37 cases. In another group of 2754 cases, patients underwent reoperation, TACE or other therapeutic methods, which suggested that palliative reoperation decreases 5-years-survival rate. Therefore, we may increase the 5-years-survival rate with reasonable therapeutic care of patients undergoing palliative resection. However, this hypothesis requires further investigation in a larger number of cases.
Our in vitro studies found that the MTSS1/MIM-B expression was increased in parallel with the metastatic potential of cell lines. Downregulation of MTSS1 reduced the invasive potential of HCC cells and prevented the exacerbated of lung metastasis following palliative resection in nude mice bearing HCC xenografts. MTSS1/MIM-B is also found upregulated in the early stages of several cancers, including melanomas [42], head and neck squamous cell carcinoma [43], and lung cancer [22]. MIM-B mRNA and protein is also proved overexpressed in HCC [14]. Therefore, we believe that advanced lung metastasis after palliative surgery may correlate with upregulated MTSS1 expression.
In our study, patients had chronic hepatitis B infection which progressed to HCC, and we found that the poor prognosis of hepatitis B-related HCC patients following palliative hepatectomy associates with elevated MTSS1 mRNA expression. Thus, to dissect the regulation of MTSS1 expression in HCC may provide a new research field for HCC diagnosis and treatment. Previous study demonstrated that p63 could control the cell migration and invasion by regulating the MTSS1 expression in breast tumor cells [44]. Recent reports revealed that chronic hepatitis B infection could induce gene expression in several of cancers [45, 46]. Therefore, there is a possibility that the virus infection plays some role in the expression of MTSS1, which needs further study to identify the role of chronic hepatitis B infection in MTSS1 regulation in HCC. Furthermore, p63 may affect cell migration and invasion by regulating the MTSS1 expression in HCC that remains to be fully elucidated.
The mechanism of MTSS1 upregulation in HCC after palliative surgery is unclear. The altered microenvironment may be a key player. Predina et al. reported the absence of any changes in the tumor cells per se but rather in the surrounding tumor microenvironment after surgery [47]. Surgery generates a sequence of events to induce wound healing and is characterized by the release of VEGF, PDGF, prostaglandins, TGF-beta, clotting factors, and complement [47–49], which may contribute to the high levels of MTSS1 by as unknown mechanism.
Cancer metastasis involves basement membrane invasion by MMP activation [50]. A higher level of MMP2 has been related to a poorer prognosis in HCC patients [35, 51–53]. Our results revealed that MTSS1 expression, MMP2 activity and lung metastasis were increased after palliative hepatectomy both in nude mice models and in HCC patients. After down-regulating MTSS1 expression, the MMP2 activity and lung metastasis were also decreased. The synchronous alteration of MMP2 with MTSS1 suggests that MTSS1 promotes tumor metastasis by targeting MMP2. Based on our in vitro and in vivo studies, we believe that the metastasis-enhancing effect of palliative resection may partially be due to the dysregulation of the MIM-B/MMP2 pathway. We found MMP2 was activated after palliative HCC resection [5]. However, the mechanism underlying MTSS1-mediated MMP2 activation is still unclear. Further, MTSS1 encodes an intracellular MIM that is implicated in actin cytoskeletal reorganization, and MTSS1 represents a novel signaling pathway from PDGF receptor to the actin cytoskeleton via Src-related kinases [54]. Therefore, MTSS1 depletion affects the cytoskeletal reorganization. Giacobbe et al. reported that MTSS1 enhances breast tumor cell migration resulting in poor prognosis [44]. We concluded that MTSS1 depletion affects the migratory capabilities of HCC cells, which needs further investigation.
Currently, there are several limitations in - and resistance to the therapy of HCC, thus, there is a necessary need for development of new and more effective therapeutic alternatives [55, 56]. In this study, patients with increased MTSS1 expression in residual HCC showed the worst prognosis. Downregulation of MTSS1 successfully averted pulmonary metastasis in our mouse model. The findings suggest that individualized treatment targeting high MTSS1 expression may be an effective strategy to treat HCC.
There are some limitations in this study. The nature of the nude mice lowers the strength of evidence. The size of our patient population is relatively small, which may lead to selection bias. Finally, we cannot exclude the possibility that other downstream pathways of MTSS1 might also mediate HCC metastasis after palliative hepatectomy. Therefore, the precise mechanism triggering metastasis remains to be fully elucidated. Despite these limitations, we believe that the current study provides preliminary and powerful data underscoring the value of MTSS1/MIM-B expression in HCC diagnosis and treatment.
Conclusions
In summary, palliative HCC resection upregulates MTSS1 mRNA expression, activates MMP2 activation and enhances residual HCC metastasis to lung. Patients with hepatitis B-related HCC exhibiting high MTSS1 mRNA levels in the residual tumor show poor prognosis after hepatectomy. However, additional investigations into the mechanisms underlying the role of MTSS1/MIM-B in metastasis are needed, to facilitate the search for novel therapeutic strategies for HCC, and improve prognosis of patients with HCC undergoing hepatectomy.
Abbreviations
AFP, α-fetoprotein; BBS, borate-buffered saline; ELISA, enzyme-linked immunosorbent assay; GEO, Gene expression omnibus; H&E, Hematoxylin and eosin; HCC, hepatocellular carcinoma; MIM, missing in metastasis; MMP2, matrix metalloproteinase 2; MTSS1, metastasis suppressor 1; PCR, polymerase chain reaction; PDGF, platelet derived growth factor; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; siRNA, small interfering RNA; SVM, Support vector machine; TACE, Transcatheter arterial chemoembolization; TGF-beta, transforming growth factor beta; VEGF, vascular endothelial growth factor.
Acknowledgments
The authors acknowledge the contribution of all investigators at all participating study sites. We thank Dr. Ju-Hong Yang, Metabolic Disease Hospital & Tianjin Institute of Endocrinology, Tianjin Medical University (Tianjin, People’s Republic of China), for expert technical assistance with statistical analysis.
Authors’ Contributions
Conceived and designed the experiments: X-YH X-YH Z-LH BX ZC T-JR; performed the experiments: X-YH Z-LH BX; analyzed the data: X-YH Z-LH BX ZC T-JR; contributed reagents/materials/analysis tools: X-YH Z-LH QZ Z-YT; and wrote the paper: X-YH Z-LH X-YH BX ZC T-JR. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Ethics approval
This study was approved by the Research Ethics Committee of the Shanghai Jiaotong University Affiliated Sixth People’s Hospital, and informed consent was obtained from each patient.
Additional file
Figure S1. The invasive ability (% of control) of MHCC97H cells. A, 0 h-24 h. B, 1 day (24 h)-35 day. Preliminary experiment was carried in 204 nude mice bearing HCC xenografts. 14 days after orthotopic implantation these mice were randomized into two groups (102 mice/group): palliative resection group and sham operation group (control). At 0 h, 1 h, 3 h, 5 h, 7 h, 9 h, 12 h, 18 h, 24 h (1 d), 3 d, 5 d, 7 d, 9 d, 14 d, 21 d, 28 d and 35 d after palliative resection, 6 mice from each group were humanely killed by cervical dislocation to harvest serum for cell culture studies. MHCC97H cells were treated with serum taken from above mentioned mice or cell culture medium supplemented with 10% human AB serum as control for 72 h, then these cells were added to the upper chamber (100 μL DMEM, 5 × 104 cells/well) and 600 μL conditioned medium was added to the lower chamber. The invaded cells were fixed with methanol and stained with crystal violet solution after 24-hour incubation. The results were expressed as the number of penetrated cells under microscope at ×200 magnification on five random fields and were presented as means ± SD of three assays. MHCC97H cells treated with serum from the palliative resection group had the most invasive potential through Matrigel at 12 h and 14 d, as compared to cells in controls (P =0.027, 0.019, respectively). The bars indicate the means ± s.d. Figure S2. MTSS1 promoted metastatic potential of Hep3B via transwell invasion assay. The number of penetrated Hep3B cells in the Vector-MTSS1 Group (A) was more than that in the Control Vector Group (B, P = 0.021). The assays were conducted in triplicate. (DOCX 3113 kb)
Footnotes
Xiu-Yan Huang, Zi-Li Huang, and Bin Xu are co-first authors.
Contributor Information
Xiu-Yan Huang, Phone: +86-21-64701361, FAX: +86-21-64701361, Email: xyhuang1119@163.com.
Xin-Yu Huang, Phone: +86-21-64701361, FAX: +86-21-64701361, Email: xinyuhuang9@163.com.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45(4):529–38. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Zhou XD, Tang ZY, Ma ZC, Fan J, Wu ZQ, Qin LX, et al. Twenty-year survivors after resection for hepatocellular carcinoma-analysis of 53 cases. J Cancer Res Clin Oncol. 2009;135(8):1067–72. doi: 10.1007/s00432-009-0546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokoyama H, Goto S, Chen CL, Pan TL, Kawano K, Kitano S. Major hepatic resection may suppress the growth of tumors remaining in the residual liver. Br J Cancer. 2000;83(8):1096–101. doi: 10.1054/bjoc.2000.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang XY, Huang ZL, Wang L, Xu YH, Huang XY, Ai KX, et al. Herbal compound “Songyou Yin” reinforced the ability of interferon-alfa to inhibit the enhanced metastatic potential induced by palliative resection of hepatocellular carcinoma in nude mice. BMC Cancer. 2010;10:580. doi: 10.1186/1471-2407-10-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picardo A, Karpoff HM, Ng B, Lee J, Brennan MF, Fong Y. Partial hepatectomy accelerates local tumor growth: potential roles of local cytokine activation. Surgery. 1998;124(1):57–64. doi: 10.1016/S0039-6060(98)70075-3. [DOI] [PubMed] [Google Scholar]
- 7.García-Alonso I, Palomares T, Alonso A, Portugal V, Castro B, Caramés J, et al. Effect of hepatic resection on development of liver metastasis. Rev Esp Enferm Dig. 2003;95(11):771. [PubMed] [Google Scholar]
- 8.Sun FX, Tang ZY, Liu KD, Ye SL, Xue Q, Gao DM, et al. Establishment of a metastatic model of human hepatocellular carcinoma in nude mice via orthotopic implantation of histologically intact tissues. Int J Cancer. 1996;66(2):239–43. doi: 10.1002/(SICI)1097-0215(19960410)66:2<239::AID-IJC17>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Liu J, Smith E, Zhou K, Liao J, Yang G, et al. Downregulation of missing in metastasis gene (MIM) is associated with the progression of bladder transitional carcinomas. Cancer Invest. 2007;25(2):79–86. doi: 10.1080/07357900701205457. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Li J, Shen J, Wang C, Yang L, Zhang X. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer. 2012;12:227. doi: 10.1186/1471-2407-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan H, Chen L, Zhang F, Quan Y, Su X, Qiu X, et al. MTSS1, a novel target of DNA methyltransferase 3B, functions as a tumor suppressor in hepatocellular carcinoma. Oncogene. 2012;31(18):2298–308. doi: 10.1038/onc.2011.411. [DOI] [PubMed] [Google Scholar]
- 12.Bompard G, Sharp SJ, Freiss G, Machesky LM. Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J Cell Sci. 2005;118(Pt22):5393–403. doi: 10.1242/jcs.02640. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Quevedo R, Shoffer M, Horng L, Oro AE. Receptor tyrosine phosphatase-dependent cytoskeletal remodeling by the hedgehog-responsive gene MIM/BEG4. J Cell Biol. 2005;168(3):453–63. doi: 10.1083/jcb.200409078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma S, Guan XY, Lee TK, Chan KW. Clinicopathological significance of missing in metastasis B expression in hepatocellular carcinoma. Hum Pathol. 2007;38(8):1201–6. doi: 10.1016/j.humpath.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Machesky LM, Johnston SA. MIM: a multifunctional scaffold protein. J Mol Med (Berl) 2007;85(6):569–76. doi: 10.1007/s00109-007-0207-0. [DOI] [PubMed] [Google Scholar]
- 16.Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, et al. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 2005;24(2):240–50. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamagishi A, Masuda M, Ohki T, Onishi H, Mochizuki N. A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J Biol Chem. 2004;279(15):14929–36. doi: 10.1074/jbc.M309408200. [DOI] [PubMed] [Google Scholar]
- 18.Callahan CA, Ofstad T, Horng L, Wang JK, Zhen HH, Coulombe PA, et al. MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev. 2004;18(22):2724–9. doi: 10.1101/gad.1221804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasper M, Regl G, Frischauf AM, Aberger F. GLI transcription factors: mediators of oncogenic Hedgehog signaling. Eur J Cancer. 2006;42(4):437–45. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 20.Wang D, Xu MR, Wang T, Li T, Zhu JW. MTSS1 overexpression correlates with poor prognosis in colorectal cancer. J Gastrointest Surg. 2011;15(7):1205–12. doi: 10.1007/s11605-011-1546-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Tong Y, Ren L, Li CD. Expression of metastasis suppressor 1 in cervical carcinoma and the clinical significance. Oncol Lett. 2014;8(5):2145–9. doi: 10.3892/ol.2014.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kayser G, Csanadi A, Kakanou S, Prasse A, Kassem A, Stickeler E, et al. Downregulation of MTSS1 expression is an independent prognosticator in squamous cell carcinoma of the lung. Br J Cancer. 2015;112(5):866–73. doi: 10.1038/bjc.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mertz KD, Pathria G, Wagner C, Saarikangas J, Sboner A, Romanov J, et al. MTSS1 is a metastasis driver in a subset of human melanomas. Nat Commun. 2014;5:3465. doi: 10.1038/ncomms4465. [DOI] [PubMed] [Google Scholar]
- 24.Chen CL, Wu JC, Chen GY, Yuan PH, Tseng YW, Li KC, et al. Baculovirus-mediated miRNA regulation to suppress hepatocellular carcinoma tumorigenicity and metastasis. Mol Ther. 2015;23(1):79–88. doi: 10.1038/mt.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Tang Y, Ye L, Liu B, Liu K, Chen J, et al. Establishment of a hepatocellular carcinoma cell line with unique metastatic characteristics through in vivo selection and screening for metastasis–related genes through cDNA microarray. J Cancer Res Clin Oncol. 2003;129(1):43–51. doi: 10.1007/s00432-002-0396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giannelli G, Fransvea E, Marinosci F, Bergamini C, Colucci S, Schiraldi O, et al. Transforming growth factor-beta1 triggers hepatocellular carcinoma invasiveness via alpha3beta1 integrin. Am J Pathol. 2002;161(1):183–93. doi: 10.1016/S0002-9440(10)64170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang XY, Wang L, Huang ZL, Zheng Q, Li QS, Tang ZY. Herbal extract “Songyou Yin” inhibits tumor growth and prolongs survival in nude mice bearing human hepatocellular carcinoma xenograft with high metastatic potential. J Cancer Res Clin Oncol. 2009;135(9):1245–55. doi: 10.1007/s00432-009-0566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng G, Fan X, Hao M, Wang J, Zhou X, Sun X. Higher levels of TIMP-1 expression are associated with a poor prognosis in triple-negative breast cancer. Mol Cancer. 2016;15(1):30. doi: 10.1186/s12943-016-0515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambele MA, Dessels C, Durandt C, Pepper MS. Genome-wide analysis of gene expression during adipogenesis in human adipose-derived stromal cells reveals novel patterns of gene expression during adipocyte differentiation. Stem Cell Res. 2016;16(3):725–34. doi: 10.1016/j.scr.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Englund E, Bartoschek M, Reitsma B, Jacobsson L, Escudero-Esparza A, Orimo A, et al. Cartilage oligomeric matrix protein contributes to the development and metastasis of breast cancer. Oncogene. 2016 doi: 10.1038/onc.2016.98. [DOI] [PubMed] [Google Scholar]
- 31.Billard MJ, Fitzhugh DJ, Parker JS, Brozowski JM, Mcginnis MW, Timoshchenko RG, et al. G protein coupled receptor kinase 3 regulates breast cancer migration, invasion, and metastasis. PLoS One. 2016;11(4):e0152856. doi: 10.1371/journal.pone.0152856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Jing X, Cheng X, Ma D, Jin Z, Yang W, et al. FGFR3 promotes angiogenesis-dependent metastasis of hepatocellular carcinoma via facilitating MCP-1-mediated vascular formation. Med Oncol. 2016;33(5):46. doi: 10.1007/s12032-016-0761-9. [DOI] [PubMed] [Google Scholar]
- 33.Prieto C, Risueño A, Fontanillo C, Delas Rivas J. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. PLoS One. 2008;3(12):e3911. doi: 10.1371/journal.pone.0003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nayak RR, Kearns M, Spielman RS, Cheung VG. Coexpression network based on natural variation in human gene expression reveals gene interactions and functions. Genome Res. 2009;19(11):1953–62. doi: 10.1101/gr.097600.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sze KM, Wong KL, Chu GK, Lee JM, Yau TO, Ng IO. Loss of phosphatase and tensin homolog enhances cell invasion and migration through AKT/Sp-1 transcription factor/matrix metalloproteinase 2 activation in hepatocellular carcinoma and has clinicopathologic significance. Hepatology. 2011;53(5):1558–69. doi: 10.1002/hep.24232. [DOI] [PubMed] [Google Scholar]
- 36.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 37.Liu L, Ren ZG, Shen Y, Zhu XD, Zhang W, Xiong W, et al. Influence of hepatic artery occlusion on tumor growth and metastatic potential in a human orthotopic hepatoma nude mouse model: relevance of epithelial-mesenchymal transition. Cancer Sci. 2010;101(1):120–8. doi: 10.1111/j.1349-7006.2009.01363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong J, Kong J, Pan B, Ke S, Dong S, Li X, et al. Insufficient radiofrequency ablation promotes angiogenesis of residual hepatocellular carcinoma via HIF-1α/VEGFA. PloS One. 2012;7(5):e37266. doi: 10.1371/journal.pone.0037266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong J, Kong L, Kong J, Ke S, Gao J, Ding X, et al. After insufficient radiofrequency ablation, tumor associated endothelial cells exhibit enhanced angiogenesis and promote invasiveness of residual hepatocellular carcinoma. J Transl Med. 2012;10:230. doi: 10.1186/1479-5876-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang N, Wang L, Chai ZT, Zhu ZM, Zhu XD, Ma DN, et al. Incomplete radiofrequency ablation enhances invasiveness and metastasis of residual cancer of hepatocellular carcinoma cell HCCLM3 via activating β-catenin signaling. PLoS One. 2014;9(12):e115949. doi: 10.1371/journal.pone.0115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida S, Kornek M, Ikenaga N, Schmelzle M, Masuzaki R, Csizmadia E, et al. Sublethal heat treatment promotes epithelial-mesenchymal transition and enhances the malignant potential of hepatocellular carcinoma. Hepatology. 2013;58(5):1667–80. doi: 10.1002/hep.26526. [DOI] [PubMed] [Google Scholar]
- 42.Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell. 2011;20(1):92–103. doi: 10.1016/j.ccr.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawson JC, Timpson P, Kalna G, Machesky LM. Mtss1 regulates epidermal growth factor signaling in head and neck squamous carcinoma cells. Oncogene. 2012;31(14):1781–93. doi: 10.1038/onc.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giacobbe A, Compagnone M, Bongiorno-Borbone L, Antonov A, Markert EK, Zhou JH, et al. P63 controls cell migration and invasion by transcriptional regulation of MTSS1. Oncogene. 2016;35(12):1602–8. doi: 10.1038/onc.2015.230. [DOI] [PubMed] [Google Scholar]
- 45.Sarkar N, Chakravarty R. Hepatitis B virus infection, microRNAs and liver disease. Int J Mol Sci. 2015;16(8):17746–62. doi: 10.3390/ijms160817746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu M, Guo J, Li W, Xia H, Lu Y, Dong X, et al. HBx induced AFP receptor expressed to activate PI3K/AKT signal to promote expression of Src in liver cells and hepatoma cells. BMC Cancer. 2015;15:362. doi: 10.1186/s12885-015-1384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Predina J, Eruslanov E, Judy B, Kapoor V, Cheng G, Wang LC, et al. Changes in the local tumor microenvironment in recurrent cancers may explain the failure of vaccines after surgery. Proc Natl Acad Sci U S A. 2013;110(5):E415–24. doi: 10.1073/pnas.1211850110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shakhar G, Ben-Eliyahu S. Potential prophylactic measures against postoperative immunosuppression: could they reduce recurrence rates in oncological patients? Ann Surg Oncol. 2003;10(8):972–92. doi: 10.1245/ASO.2003.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Govaert KM, Emmink BL, Nijkamp MW, Cheung ZJ, Steller EJ, Fatrai S, et al. Hypoxia after liver surgery imposes an aggressive cancer stem cell phenotype on residual tumor cells. Ann Surg. 2014;259(4):750–9. doi: 10.1097/SLA.0b013e318295c160. [DOI] [PubMed] [Google Scholar]
- 50.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Théret N, Musso O, Turlin B, Lotrian D, Bioulac-Sage P, Campion JP, et al. Increased extracellular matrix remodeling is associated with tumor progression in human hepatocellular carcinomas. Hepatology. 2001;34(1):82–8. doi: 10.1053/jhep.2001.25758. [DOI] [PubMed] [Google Scholar]
- 52.Chen R, Cui J, Xu C, Guo D, Liu Y, Ye S, et al. The significance of MMP-9 over MMP-2 in HCC invasiveness and recurrence of hepatocellular carcinoma after curative resection. Ann Surg Oncol. 2012;19(Suppl 3):S375–84. doi: 10.1245/s10434-011-1836-7. [DOI] [PubMed] [Google Scholar]
- 53.Fang JH, Zhou HC, Zeng C, Yang J, Liu Y, Huang X, et al. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54(5):1729–40. doi: 10.1002/hep.24577. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Zhou K, Zeng X, Lin J, Zhan X. Tyrosine phosphorylation of missing in metastasis protein is implicated in platelet-derived growth factor-mediated cell shape changes. J Biol Chem. 2007;282(10):7624–31. doi: 10.1074/jbc.M608448200. [DOI] [PubMed] [Google Scholar]
- 55.El-Shemi A, Ashshi A, Na Y, Li Y, Basalamah M, Al-Allaf F, et al. Combined therapy with oncolytic adenoviruses encoding TRAIL and IL-12 genes markedly suppressed human hepatocellular carcinoma both in vitro and in an orthotopic transplanted mouse model. J Exp Clin Cancer Res. 2016;35:74. doi: 10.1186/s13046-016-0353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lippolis C, Refolo MG, D’Alessandro R, Carella N, Messa C, Cavallini A, et al. Resistance to multikinase inhibitor actions mediated by insulin like growth factor-1. J Exp Clin Cancer Res. 2015;34:90. doi: 10.1186/s13046-015-0210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


