Abstract
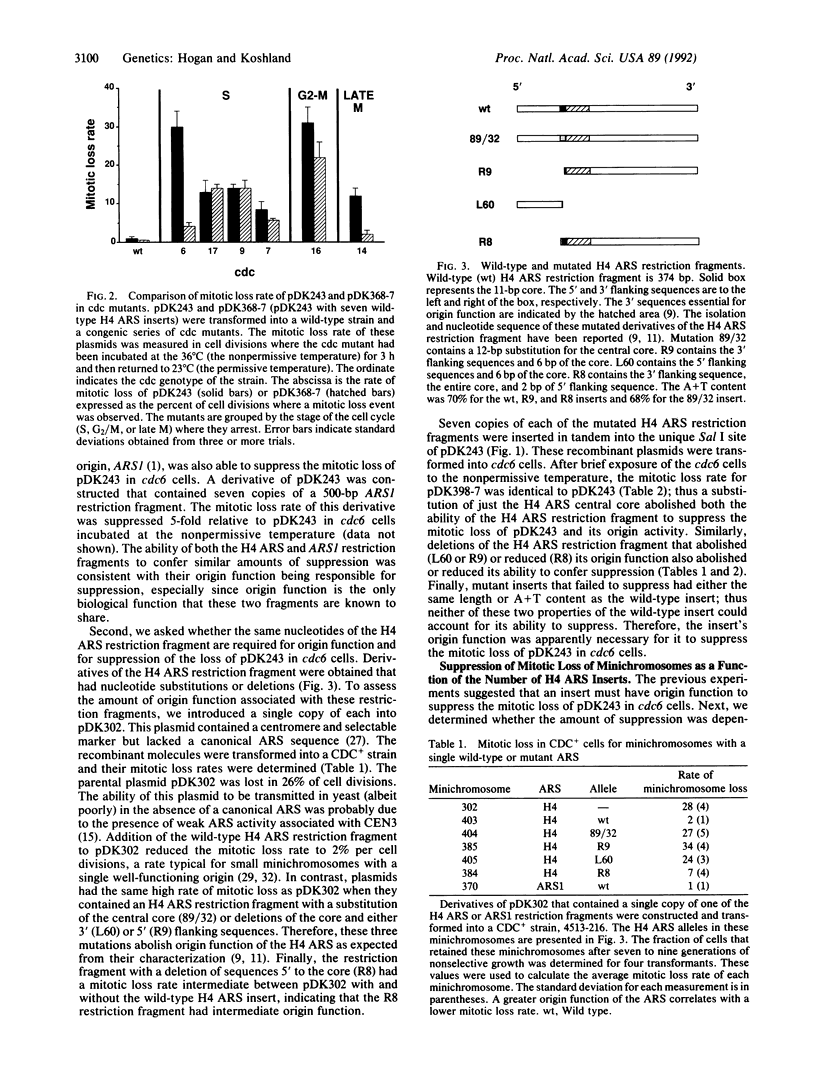
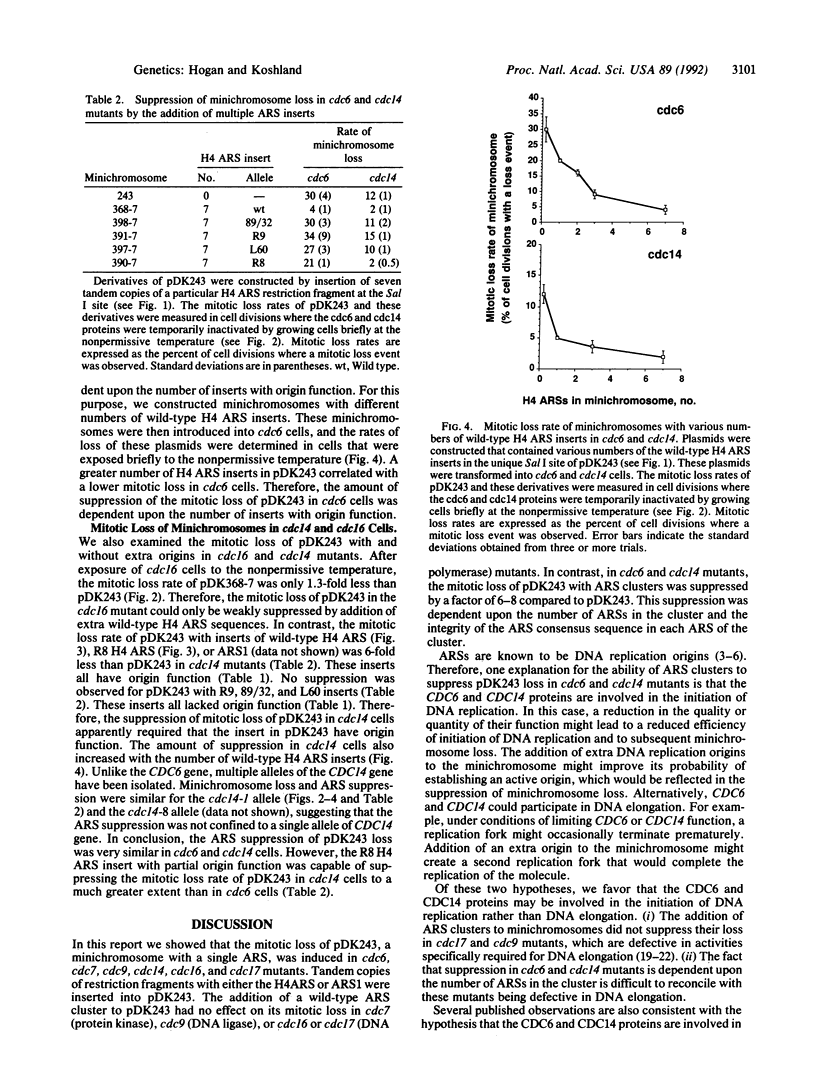
Many cell division cycle (cdc) mutants of Saccharomyces cerevisiae exhibit elevated mitotic loss of pDK243, a 14-kilobase minichromosome with a centromere and one autonomous replicating sequence (ARS). Tandem copies of different ARSs were added to pDK243. The addition of these ARS clusters to pDK243 had no effect on its mitotic loss in cdc7 (protein kinase), cdc9 (DNA ligase), or cdc16 or cdc17 (DNA polymerase) mutants. However, in cdc6 and cdc14 mutants, the mitotic loss of pDK243 with an ARS cluster was suppressed by a factor of 6-8 compared to pDK243 without the cluster. This suppression was dependent upon the number of ARSs in the cluster and the integrity of the ARS consensus sequence in each ARS of the cluster. ARSs are known to be DNA replication origins. Therefore, the suppression of mini-chromosome loss by ARSs in cdc6 and cdc14 mutants suggests that these mutants are defective in the initiation of DNA replication. Since the CDC6 protein appears to act at the G1/S phase transition, the CDC6 protein may be a factor required at the beginning of S phase to initiate DNA replication at origins. In contrast, the CDC14 protein acts after mitosis. We suggest that the CDC14 protein performs a function late in the cell cycle that may be required for efficient initiation of DNA replication during S phase of the next cell cycle.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker D. G., Johnston L. H. Saccharomyces cerevisiae cdc9, a structural gene for yeast DNA ligase which complements Schizosaccharomyces pombe cdc17. Eur J Biochem. 1983 Aug 1;134(2):315–319. doi: 10.1111/j.1432-1033.1983.tb07568.x. [DOI] [PubMed] [Google Scholar]
- Blow J. J., Laskey R. A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988 Apr 7;332(6164):546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Boulet A., Simon M., Faye G., Bauer G. A., Burgers P. M. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989 Jun;8(6):1849–1854. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton A. H., Smith M. M. Fine-structure analysis of the DNA sequence requirements for autonomous replication of Saccharomyces cerevisiae plasmids. Mol Cell Biol. 1986 Jul;6(7):2354–2363. doi: 10.1128/mcb.6.7.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Li Y. Y., Feldman J., Jayaram M., Abraham J., Nasmyth K. A., Hicks J. B. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Sweder K., Srienc F., Bailey J. E., Campbell J. L. Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B. M., Brewer B. J., Reynolds A. E., Fangman W. L. A yeast origin of replication is activated late in S phase. Cell. 1991 May 3;65(3):507–515. doi: 10.1016/0092-8674(91)90468-e. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976 Jul 15;104(4):803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy K. M., Clark C. D., Botstein D. Subcellular localization of yeast CDC46 varies with the cell cycle. Genes Dev. 1990 Dec;4(12B):2252–2263. doi: 10.1101/gad.4.12b.2252. [DOI] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hofmann J. F., Gasser S. M. Identification and purification of a protein that binds the yeast ARS consensus sequence. Cell. 1991 Mar 8;64(5):951–960. doi: 10.1016/0092-8674(91)90319-t. [DOI] [PubMed] [Google Scholar]
- Hollingsworth R. E., Jr, Sclafani R. A. DNA metabolism gene CDC7 from yeast encodes a serine (threonine) protein kinase. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6272–6276. doi: 10.1073/pnas.87.16.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes S. G., Smith M. M. Interaction of the H4 autonomously replicating sequence core consensus sequence and its 3'-flanking domain. Mol Cell Biol. 1989 Dec;9(12):5464–5472. doi: 10.1128/mcb.9.12.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C. L., Carbon J. High-frequency transformation of yeast by plasmids containing the cloned yeast ARG4 gene. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3829–3833. doi: 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Spotila L. D., Nawotka K. A., el-Assouli S. M., Davis L. R. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987 Nov 6;51(3):473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., Zhu J. G., Davis L. R., Newlon C. S. Close association of a DNA replication origin and an ARS element on chromosome III of the yeast, Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 25;16(14A):6373–6384. doi: 10.1093/nar/16.14.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Hieter P. Visual assay for chromosome ploidy. Methods Enzymol. 1987;155:351–372. doi: 10.1016/0076-6879(87)55024-8. [DOI] [PubMed] [Google Scholar]
- Koshland D., Kent J. C., Hartwell L. H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985 Feb;40(2):393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- Koshland D., Rutledge L., Fitzgerald-Hayes M., Hartwell L. H. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell. 1987 Mar 13;48(5):801–812. doi: 10.1016/0092-8674(87)90077-8. [DOI] [PubMed] [Google Scholar]
- Lisziewicz J., Godany A., Agoston D. V., Küntzel H. Cloning and characterization of the Saccharomyces cerevisiae CDC6 gene. Nucleic Acids Res. 1988 Dec 23;16(24):11507–11520. doi: 10.1093/nar/16.24.11507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J. L., Erfle M., Wykes E. J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984 Dec;32(3):481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- Palmer R. E., Hogan E., Koshland D. Mitotic transmission of artificial chromosomes in cdc mutants of the yeast, Saccharomyces cerevisiae. Genetics. 1990 Aug;125(4):763–774. doi: 10.1093/genetics/125.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A. M., Herterich S. U., Krauss G. A single-stranded DNA binding protein from S. cerevisiae specifically recognizes the T-rich strand of the core sequence of ARS elements and discriminates against mutant sequences. EMBO J. 1991 Apr;10(4):981–985. doi: 10.1002/j.1460-2075.1991.tb08032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitney K. C., Budd M. E., Campbell J. L. DNA polymerase III, a second essential DNA polymerase, is encoded by the S. cerevisiae CDC2 gene. Cell. 1989 Feb 24;56(4):599–605. doi: 10.1016/0092-8674(89)90582-5. [DOI] [PubMed] [Google Scholar]
- Srienc F., Bailey J. E., Campbell J. L. Effect of ARS1 mutations on chromosome stability in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Jul;5(7):1676–1684. doi: 10.1128/mcb.5.7.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D. T., Struhl K., Davis R. W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979 Nov 1;282(5734):39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Linskens M. H., Kowalski D., Huberman J. A. New beginnings in studies of eukaryotic DNA replication origins. Biochim Biophys Acta. 1989 Jan 23;1007(1):1–14. doi: 10.1016/0167-4781(89)90123-1. [DOI] [PubMed] [Google Scholar]
- Wood J. S., Hartwell L. H. A dependent pathway of gene functions leading to chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1982 Sep;94(3):718–726. doi: 10.1083/jcb.94.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. J., Campbell J. L. The CDC7 protein of Saccharomyces cerevisiae is a phosphoprotein that contains protein kinase activity. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3574–3578. doi: 10.1073/pnas.88.9.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Huang S. H., Jong A. Y. Molecular cloning of Saccharomyces cerevisiae CDC6 gene. Isolation, identification, and sequence analysis. J Biol Chem. 1989 May 25;264(15):9022–9029. [PubMed] [Google Scholar]