Abstract
Purpose
The aim of this study was to use massively parallel DNA sequencing to identify GNAQ/11, BAP1 and SF3B1 mutations in ophthalmic melanocytoma.
Procedures
Six ophthalmic melanocytoma specimens (1 iridociliary and 5 optic nerve) were profiled for genomic alterations in GNAQ/11, BAP1 and SF3B1 using a custom deep sequencing assay. This assay uses solution phase hybridization-based exon capture and deep-coverage massively parallel DNA sequencing to interrogate all protein-coding exons and select introns.
Results
The only iridociliary melanocytoma showed a mutation in GNAQ but not in BAP1. Of the 2 optic-nerve melanocytomas that developed into melanoma, one had a GNAQ mutation and both a BAP1 mutation and monosomy 3. The remaining 3 optic-nerve melanocytomas did not reveal mutations in GNAQ/11 or BAP1. SF3B1 mutations were not detected in any specimen.
Conclusions
The presence of GNAQ mutation in some iridociliary and optic-nerve melanocytomas suggests a possible relationship between ophthalmic melanocytoma and other ophthalmic melanocytic neoplasms. BAP1 mutation may accompany the transformation of ophthalmic melanocytoma to melanoma.
Key Words: Melanoma, Genetics, Ocular tumors, Melanocytoma, GNAQ/11, SF3B1, BAP1
Introduction
In the 1960s, Dr. Lorenz E. Zimmerman made some important observations regarding ophthalmic melanocytoma, asserting that due to its nonmetastasizing nature, this benign lesion could be observed and the eye saved from enucleation [1]. He also postulated a histopathologic similarity to ocular melanocytosis and suggested that melanocytoma may have a clinical presentation analogous to cutaneous blue nevi [2]. Previous work has revealed mutations in GNAQ/11 as a genetic basis shared between a case of melanosis oculi and blue nevi [3], and recent work by Mudhar et al. [4] demonstrates the same genetic alteration in ciliochoroidal melanocytoma, thereby suggesting a genetic link between these three benign, pigmented ophthalmic entities.
Our published knowledge of genetic mutations in ophthalmic melanocytoma consists of 2 cases, both of which were ciliochoroidal in location and established by mutational analysis of a single exon in three target genes (GNAQ, GNA11 and BRAF)[4]. The unavailability of enucleated melanocytoma specimens (which may ironically be a result of Dr. Zimmerman's original work) contributes to the absence of information about the genetic events underlying the lesional pathogenesis.
Here, we present our findings describing the genetic analysis of 6 ophthalmic melanocytoma samples (1 iridociliary and 5 optic nerve) using targeted exome sequencing for GNAQ/11, BAP1 and SF3B1. This method is unique in that it sequences the entire gene (exons) and can identify both common and rare mutations in addition to other alterations such as copy number gains and losses [5]. This methodology, developed at the Memorial Sloan-Kettering Cancer Center, has become the standard technique by which all tumors are evaluated at our institution.
Methods
At the participating institutions, melanocytoma specimens were retrieved from the archives of the department of pathology. The specimens were obtained through a search of the respective database, patient and tissue records. A diagnosis of melanocytoma was confirmed by each institution's respective ophthalmic pathologist prior to inclusion in the study. Cytogenetic analysis by fluorescence in situ hybridization was performed for the specimens (No. 4 and 6) that had malignantly transformed to melanoma, by a method that has previously been reported [6]. Chromosome number variation was also evaluated with the IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets) assay for all specimens. The iridociliary specimen was obtained through a needle biopsy, and the reason for enucleation of the remaining specimens is depicted in table 1.
Table 1.
Mutational analysis results of 6 ophthalmic melanocytoma specimens
| Specimen | Source | Fixation status | GNAQ/GNA11 mutation | SF3B1 | BAP1 | Notes |
|---|---|---|---|---|---|---|
| 1 | Iridociliary | FFPE | GNAQ: missense mutation: exon 5 Q209L | – | – | Needle biopsy |
| 2 | ON | FFPE | – | – | – | Enucleated due to coincidental closed-angle glaucoma |
| 3 | ON | FFPE | – | – | – | Enucleated due to being blind and painful |
| 4 | ON | FFPE | – | – | Frameshift insertion: D663fs | Transformed to melanoma |
| 5 | ON | FFPE | – | – | – | Enucleated due to patient preference |
| 6 | ON | Frozen | GNAQ: missense mutation: exon 5 p.Q209L | – | Nonsense mutation: R385* | Transformed to melanoma |
ON = Optic nerve.
Isolation and Purification of DNA
Microdissection was performed on the 5 formalin-fixed and paraffin-embedded (FFPE) samples on 10-µm-thick unstained sections, employing hematoxylin and eosin-stained (HE) sections when needed as a guide. This included a malignantly transformed melanocytoma (No. 4), which was submitted entirely, without distinction between benign- and malignant-appearing areas. The remaining specimen (No. 6) was biopsied from the original lesion and snap frozen in liquid nitrogen at the time of enucleation and stored at −80°C. Prior to freezing, this specimen was dissected into melanoma and melanocytoma with the assistance of a microscope. However, without the guidance of HE sections, it is difficult to know whether these were true representations of benign and malignant tumors. These two parts of the specimens had an identical genetic signature and were therefore reported together. The DNeasy Tissue Kit (Qiagen) was used for DNA extraction according to the manufacturer's recommendations. The Nano-Drop 8000 (Thermo Scientific) and Qubit (Life Technologies) devices were employed to quantify the extracted DNA. The minimum concentration of FFPE DNA is 250 ng.
Exon Capture Sequencing
Genetic alterations in GNAQ/11, BAP1 and SF3B1 were profiled using the IMPACT assay. As previously described in detail [5], this assay employs solution-phase hybridization-based exon capture and massively parallel DNA sequencing to interrogate all protein-coding exons and select introns of 279 oncogenes, tumor suppressor genes and members of pathways considered actionable by targeted therapies. The concentration of genomic DNA and DNA sequence library are both tested at the beginning of the assay before and after exon capture. If the yield is too low at any of these steps, the sample is failed. None of the specimens in this study failed these steps.
In brief, barcoded sequence libraries (New England Biolabs, Kapa Biosystems) underwent exon capture by hybridization (Nimblegen SeqCap) using a custom probe design, with approximately 112-250 ng of DNA used as input for library construction and 100 ng of the library used as input for exon capture [7]. To prevent off-target hybridization, a pool of blocker oligonucleotides, complementary to the full sequences of all barcoded adaptors, was spiked into a final total concentration of 10 mmol/l. An Illumina HiSeq 2000 device was used to sequence DNA and generate paired-end 75-bp reads. The reads were aligned to the reference human genome (hg19) using the Burrows-Wheeler alignment tool [8], and local realignment and quality score recalibration were completed using the Genome Analysis Toolkit (GATK) according to GATK best practices [9]. A mean unique-sequence coverage of ×281 was achieved. Single-nucleotide variants were identified using muTect [10]. For specimens without matched normal DNA, all silent variants and all single-nucleotide polymorphisms validated by the 1,000 Genomes Project were filtered out. Indels were detected using the SomaticIndelDetector tool in GATK, and the Integrative Genomics Viewer was used to manually review all candidate mutations and indels [11].
Results
In this collection of 6 (1 iridociliary and 5 optic-nerve) ophthalmic melanocytoma specimens, two mutations were detected in GNAQ in codon 209 (Gln209Leu). They occurred in 1 iridociliary melanocytoma (fig. 1) and 1 of the optic-nerve melanocytomas (fig. 2). The 2 specimens in which the melanocytoma had clinically and histopathologically transformed to a melanoma contained a mutation in BAP1. The mutational analysis of the 6 specimens is summarized in table 1. No specimen had a mutation in SF3B1.
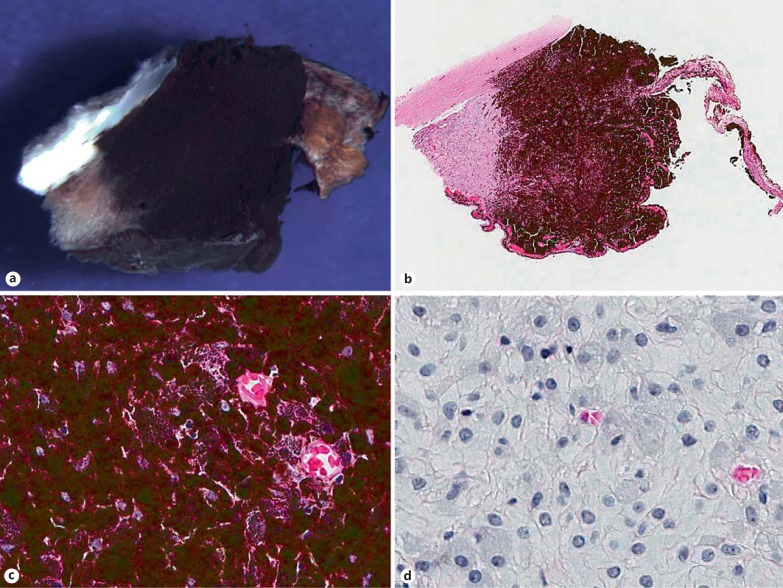
Fig. 1.
Iridociliary melanocytoma (specimen 1). Gross photograph (a) and low-power photomicrograph (b) of an iridocyclectomy specimen, demonstrating a darkly pigmented tumor, which involves the iris root and ciliary body stroma and extends into the anterior chamber angle (b HE stain; original magnification ×5). c The neoplasm is composed of polyhedral cells with dense intracytoplasmic pigments obscuring nuclear details (HE stain; original magnification ×100). d Bleached preparations highlight the bland nuclei with inconspicuous nucleoli, low nucleus-to-cytoplasm ratios and the absence of appreciable pleomorphism or mitotic figures (bleach-hematoxylin stain; original magnification ×100).
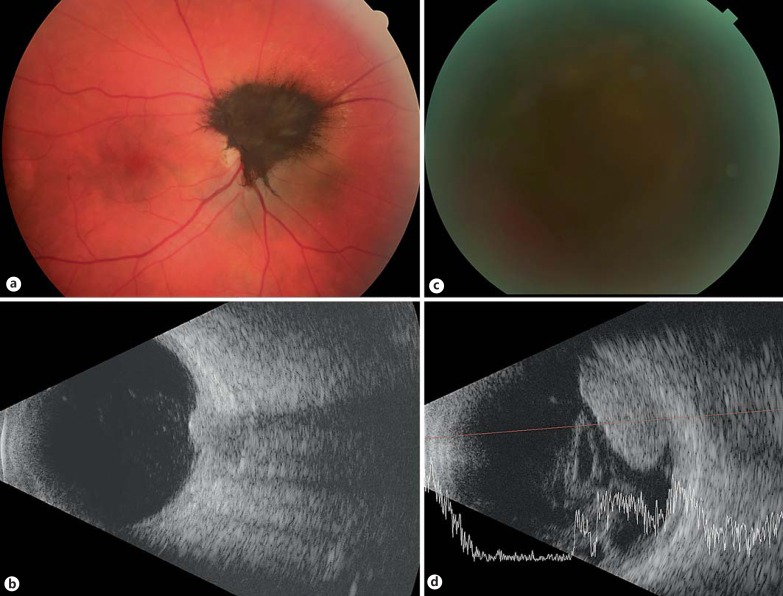
Fig. 2.
Optic nerve melanocytoma transformed to melanoma (specimen 6). Color fundus photograph of an optic-nerve melanocytoma at presentation (a) and the corresponding ultrasound image, demonstrating a 2-mm-thick tumor (b). The patient had a vision of 20/25 at that time. The color photograph at 3 years of follow-up demonstrates that the fundus is obscured by vitreous hemorrhage (c), at which point the patient's vision had decreased to light perception. The corresponding ultrasound (d) at that point demonstrates growth of the lesion to a height of 5.5 mm and overlying vitreous hemorrhage.
Copy number variations were identified in specimen 6 and revealed a loss of chromosome 1 and chromosome 3 and a gain of chromosome 8. For the remaining 5 specimens, the analysis did not reveal any significant amplifications or deletions. Specimens 4 and 6 were monosomy 3 by fluorescence in situ hybridization.
No patient developed metastases over a median follow-up of 5.8 years, and both patients with malignantly transformed melanocytomas are still alive.
Specimen 6 was grossly dissected into melanoma and melanocytoma and snap frozen at the time of enucleation. Both the melanocytoma and melanoma specimens were found to have a mutation in GNAQ and BAP1, but the patient-matched normal tissue had neither, confirming that these mutations were likely somatic.
Discussion
Despite the clinical and histopathological differences, molecular studies show an overlap with morphologically distinct lesions, namely blue nevi, melanocytoma, ophthalmic melanocytosis and uveal nevi/melanomas. The IMPACT assay employed in this study has the advantage of providing a combination approach for the detection of multiple categories of genetic alterations [7]. It gives us the advantage of investigating the entire exons of GNAQ/11, BAP1 and SF3B1 for both the common and rare mutations as well as identifying additional genomic alterations such as copy number losses and gains [5].
Using this method, we discovered the presence of a Q209L exon 5 GNAQ mutation in the iridociliary melanocytoma and in 1 of 5 of the optic-nerve melanocytomas. It is unclear whether these findings are representative of all melanocytomas or just of those that come to enucleation due to aggressive clinical features. The reasons for detecting mutations in only 2 out of 6 melanocytomas include a lower prevalence of mutation in melanocytoma, the small sample size of this study or, less likely, the presence of low allele frequency mutations that could have been missed due to low tumor purity or subclonality.
Somatic mutations are typically mutually exclusive in either GNAQ or GNA11. They have been revealed in a number of melanocytic neoplasms, including 83% of all uveal melanomas (including 22% of the iris melanomas), 90% of the uveal melanoma metastases, choroidal nevi, blue nevi (skin and oral cavity), oculodermal melanocytosis, and central nervous system and ciliochoroidal melanocytomas [3,12,13,14,15,16,17,18,19] (table 2). The present study indicates that ophthalmic melanocytomas of the iridociliary body and optic nerve can be added to this list.
Table 2.
Summary of the literature on the anatomic location and frequency of GNAQ/11 mutations
| First author [Ref.] | Year | Site | GNAQ, exon 4 | GNAQ, exon 5 | GNA11, exon 4 | GNA11, exon 5 |
|---|---|---|---|---|---|---|
| Melanocytoma | ||||||
| Küsters-Vandevelde [12] | 2010 | CNS | 3/4 | |||
| Küsters-Vandevelde [13] | 2010 | CNS | 6/12 | |||
| Murali [14] | 2012 | CNS | 1/4 | 1/4 | ||
| Mudhar [4] | 2013 | Ciliochoroid | 2/2 | |||
| Küsters-Vandevelde [19] | 2015 | CNS | 9/16 | 1/4 | ||
| Blue nevus | ||||||
| Van Raamsdonk [3] | 2009 | Skin | 24/29 | |||
| Lamba [25] | 2009 | Skin | 6/13 | |||
| Van Raamsdonk [17] | 2010 | Skin | 1/96 | 76/139 | 1/96 | 9/139 |
| Küsters-Vandevelde [12] | 2010 | CNS | 1/1 | |||
| Cohen [16] | 2012 | Oral cavity | 1/4 | |||
| Nevus of Ota | ||||||
| Van Raamsdonk [3] | 2009 | Skin | 1/17 | |||
| Van Raamsdonk [17] | 2010 | Skin | 1/11 | 2/20 | 0/11 | 1/20 |
| Nevus | ||||||
| Van Raamsdonk [17] | 2010 | Uvea | 0/1 | 1/1 | 0/1 | 0/1 |
| Cohen [16] | 2012 | Oral cavity | 0/6 | |||
| Melanoma | ||||||
| Van Raamsdonk [17] | 2010 | Uvea | 4/145 | 73/163 | 3/145 | 52/163 |
| Van Raamsdonk [17] | 2010 | Uvea-metastatic | 1/17 | 5/23 | 1/17 | 13/23 |
| Van Raamsdonk [3] | 2009 | Uvea | 22/48 | |||
| Cohen [16] | 2012 | Oral cavity | 0/4 | |||
| Küsters-Vandevelde [12] | 2010 | CNS | 1/4 | |||
| Küsters-Vandevelde [19] | 2015 | CNS | 2/4 | 1/4 | ||
CNS = Central nervous system.
It has been proposed that lesions with GNAQ/11 mutations may have two common traits: (1) they occur in extraepithelial melanocytes such as the leptomeninges, uvea and dermis, and (2) they appear to arise preferentially in melanocytes derived from cranial (rather than truncal) neural crests [15]. Our findings of mutations in iridociliary and optic-nerve melanocytomas could be consistent with both characteristics, since both are extraepithelial. Furthermore, the iris stroma and melanocytes of the optic nerve originate from melanocytes derived from cranial neural crest precursors (either the leptomeninges, lamina fusca or choroid).
GNAQ/11 mutations may be detected in uveal melanoma development but are not considered sufficient for malignant transformation [3]. Because 84% of all metastasizing uveal melanoma tumors have inactivating somatic mutations in BAP1 (BRCA1-associated protein 1), BAP1 is implicated as a later aberration and an associated event in uveal melanoma metastasis [20]. Malignant transformation is the most likely indication for removal of a melanocytoma-containing eye. In this study, 2 optic nerve melanocytomas with clinically suspected and histopathologically confirmed transformation to melanoma revealed a BAP1 mutation. These mutations consisted of a nonsense mutation (R385*) and a frameshift insertion (D663fs). A germline nonsense mutation has been found at the same location in a family with uveal melanoma; and while this specific frameshift mutation has not been previously reported, alterations at the nuclear localization signal domain have been previously found [21].
This suggests that mutations in BAP1 may accompany the malignant transformation of ophthalmic melanocytoma to melanoma. In this study, it is unclear whether the BAP1 mutation existed exclusively in the malignantly transformed portion of the melanocytoma, or whether it was also present in the benign-appearing melanocytoma. Despite our small sample size, future confirmation of this discovery could suggest the need for aggressive management of ophthalmic melanocytomas (and any benign-appearing melanocytic lesion), for instance, in patients carrying a known germline mutation in BAP1. If confirmed in larger numbers of samples, it may be an option to biopsy suspicious melanocytomas for the presence of a somatic BAP1 mutation.
Mutations in SF3B1 have recently been identified in approximately 15-18.6% of all uveal melanomas; they are associated with good prognosis and rarely occur coincidentally with BAP1 mutations [22,23,24]. None of the melanocytoma specimens in this study, including those without BAP1 mutations, had an SF3B1 mutation.
A collection of melanocytic neoplasms, mainly extradermal and of neural-crest derivation, possesses alterations in GNAQ/11, and this study demonstrates that ophthalmic melanocytomas of the iridociliary body and optic nerve can be added to this list.
Statement of Ethics
This study was approved by the Institutional Review Board of the Memorial Sloan-Kettering Cancer Center, New York Eye and Ear Infirmary and University of Washington School of Medicine and Public Health, and adhered to the Declaration of Helsinki. All patients gave informed consent.
Disclosure Statement
None of the authors have conflicts of interests to disclose.
Acknowledgments
This study was supported by The Fund for Ophthalmic Knowledge, The New York Community Trust, and Research to Prevent Blindness.
References
- 1.Zimmerman LE, Garron LK. Melanocytoma of the optic disc. Int Ophthalmol Clin. 1962;2:431–440. [Google Scholar]
- 2.Zimmerman LE. Melanocytes, melanocytic nevi, and melanocytomas. Invest Ophthalmol. 1965;4:11–41. [PubMed] [Google Scholar]
- 3.Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mudhar HS, Doherty R, Salawu A, et al. Immunohistochemical and molecular pathology of ocular uveal melanocytoma: evidence for somatic GNAQ mutations. Br J Ophthalmol. 2013;97:924–928. doi: 10.1136/bjophthalmol-2013-303291. [DOI] [PubMed] [Google Scholar]
- 5.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Y, Wang X, Dusza S, et al. Use of fluorescence in situ hybridization to distinguish metastatic uveal from cutaneous melanoma. Int J Surg Pathol. 2012;20:246–251. doi: 10.1177/1066896912438589. [DOI] [PubMed] [Google Scholar]
- 7.Wagle N, F BMI, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Küsters-Vandevelde HVN, Klaasen A, Küsters B, et al. Activating mutations of the GNAQ gene: a frequent event in primary melanocytic neoplasms of the central nervous system. Acta Neuropathol. 2010;119:317–323. doi: 10.1007/s00401-009-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Küsters-Vandevelde HV, van Engen-van Grunsven IA, Küsters B, et al. Improved discrimination of melanotic schwannoma from melanocytic lesions by combined morphological and GNAQ mutational analysis. Acta Neuropathol. 2010;120:755–764. doi: 10.1007/s00401-010-0749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murali R, Wiesner T, Rosenblum MK, et al. GNAQ and GNA11 mutations in melanocytomas of the central nervous system. Acta Neuropathol. 2012;123:457–459. doi: 10.1007/s00401-012-0948-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Raamsdonk CD, Fitch KR, Fuchs H, et al. Effects of G-protein mutations on skin color. Nat Genet. 2004;36:961–968. doi: 10.1038/ng1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen Y, Goldenberg-Cohen N, Akrish S, et al. BRAF and GNAQ mutations in melanocytic tumors of the oral cavity. Oral Surg Oral Med Oral Pathol Oral Radiol. 2012;114:778–784. doi: 10.1016/j.oooo.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirley MD, Tang H, Gallione CJ, et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013;368:1971–1979. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Küsters-Vandevelde HV, van Engen-van Grunsven IA, Coupland SE, et al. Mutations in G protein encoding genes and chromosomal alterations in primary leptomeningeal melanocytic neoplasms. Pathol Oncol Res. 2015;21:439–447. doi: 10.1007/s12253-014-9841-3. [DOI] [PubMed] [Google Scholar]
- 20.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Njauw CN, Kim I, Piris A, et al. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PLoS One. 2012;7:e35295. doi: 10.1371/journal.pone.0035295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin M, Maßhöfer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harbour JW, Roberson ED, Anbunathan H, et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furney SJ, Pedersen M, Gentien D, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamba S, Felicioni L, Buttitta F, et al. Mutational profile of GNAQQ209 in human tumors. PLoS One. 2009;4:e6833. doi: 10.1371/journal.pone.0006833. [DOI] [PMC free article] [PubMed] [Google Scholar]