Abstract
Neuroblastoma is a malignant paediatric tumour of the sympathetic nervous system1. Roughly half of these tumours regress spontaneously or are cured by limited therapy. By contrast, high-risk neuroblastomas have an unfavourable clinical course despite intensive multimodal treatment, and their molecular basis has remained largely elusive2–4. Here we have performed whole-genome sequencing of 56 neuroblastomas (high-risk, n = 39; low-risk, n = 17) and discovered recurrent genomic rearrangements affecting a chromosomal region at 5p15.33 proximal of the telomerase reverse transcriptase gene (TERT). These rearrangements occurred only in high-risk neuroblastomas (12/39, 31%) in a mutually exclusive fashion with MYCN amplifications and ATRX mutations, which are known genetic events in this tumour type1,2,5. In an extended case series (n = 217), TERT rearrangements defined a subgroup of high-risk tumours with particularly poor outcome. Despite a large structural diversity of these rearrangements, they all induced massive transcriptional upregulation of TERT. In the remaining high-risk tumours, TERT expression was also elevated in MYCN-amplified tumours, whereas alternative lengthening of telomeres was present in neuroblastomas without TERT or MYCN alterations, suggesting that telomere lengthening represents a central mechanism defining this subtype. The 5p15.33 rearrangements juxtapose the TERT coding sequence to strong enhancer elements, resulting in massive chromatin remodelling and DNA methylation of the affected region. Supporting a functional role of TERT, neuroblastoma cell lines bearing rearrangements or amplified MYCN exhibited both upregulated TERT expression and enzymatic telomerase activity. In summary, our findings show that remodelling of the genomic context abrogates transcriptional silencing of TERT in high-risk neuroblastoma and places telomerase activation in the centre of transformation in a large fraction of these tumours.
Several sequencing studies have been performed to uncover genomic alterations underlying the diverse clinical phenotypes of neuroblastoma2–4. Neuroblastoma has a low mutation frequency and a heterogeneous mutation spectrum with few recurrently mutated genes. Besides MYCN, which is amplified in 20% of neuroblastomas1, the most frequently mutated gene is the tyrosine kinase gene ALK, altered in 8–10% of the cases6. Furthermore, inactivating ATRX mutations were found in a small subgroup of neuroblastomas with unfavourable outcome2,5. Beyond these alterations, no mechanisms have so far been identified that define high-risk neuroblastoma at the molecular level.
We hypothesized that additional structural alterations might occur in high-risk neuroblastoma and performed whole-genome sequencing of 56 tumours and matched normal controls (Supplementary Table 1). By searching for breakpoint clusters occurring within 100 kilobase-pair (kb) regions7 we identified four locations exhibiting clustered breakpoints in more than three samples (Fig. 1a). Three of these regions were related to known genetic alterations in neuroblastoma, namely, MYCN amplifications, ATRX deletions, and copy number gains of chromosome 17q. The fourth region was located at chromosome 5p15.33 and affected 12 out of 56 tumours (21%, Fig. 1a and Extended Data Fig. 1). Chromosomal regions translocated to the 5p15.33 breakpoints were scattered across chromosome 5 (seven cases) and other chromosomes (five cases). The types of structural alterations were diverse and included balanced rearrangements, translocations associated with single copy number gain, and focal, high-level amplifications (Fig. 1b). Furthermore, the rearrangement at 5p15.33 was caused by chromothripsis affecting chromosome 5 in two tumours (Fig. 1b and Extended Data Fig. 2a). We noticed that the rearrangements in the 5p15.33 region consistently clustered in a region 50 kb upstream of the TERT transcriptional start site without directly affecting the gene or its core promoter region (Fig. 1b)8,9. We did not observe mutations affecting TERT itself or its promoter10,11.
Figure 1. Genomic rearrangements are clustered at chromosome 5p15.33 in high-risk neuroblastoma.
a, Distribution of genomic rearrangements occurring within regions of 100 kb in 56 primary neuroblastomas. Rearrangements clustering in more than three tumours are highlighted in red. b, Detail of genomic translocations occurring at chromosome 5p15.33 (n = 12) and their corresponding rearrangement partner (right). Levels of genomic copy numbers are colour-coded. c, Prevalence of MYCN amplification, TERT rearrangements, genomic alterations of ATRX and ALK, and chromothripsis in 56 primary neuroblastomas. Samples are ordered from left to the right. The number of somatic mutations per tumour and the clinical risk group assessment are given at the top. Tumour ploidy, telomere length ratio (both estimated from sequencing data), age of patient at diagnosis, and tumour stage are displayed at the bottom. S, stage 4S tumours.
In accordance with previous studies2,3, we found an overall low rate of non-synonymous mutations with 13.3 mutations per genome on average (Fig. 1c and Supplementary Table 2), and a significantly lower mutation rate in low-risk tumours than in the high-risk group (low-risk, 5.9 ± 5.5 mutations; high-risk, 16.6 ± 9.9 mutations; P < 0.001). Four cases exhibited chromothripsis affecting chromosomes 5, 17, and 20 (Extended Data Fig. 2). We detected only three genes that were expressed and altered in more than two samples (MYCN, n = 10; ALK, n = 7; and ATRX, n = 7). Notably, TERT rearrangements occurred exclusively in high-risk tumours in 12 out of 39 of the cases (31%, P = 0.01), similar to ATRX mutations (Fig. 1c). By contrast, ALK mutations and chromothripsis were found across both risk groups (Fig. 1c). Furthermore, TERT rearrangements, ATRX alterations, and MYCN amplifications occurred in a mutually exclusive fashion within the high-risk group (P = 0.008), suggesting that they may converge on similar effector functions.
To further clarify the clinical relevance of TERT rearrangements in a large cohort of patients, we examined 161 additional primary neuroblastomas by fluorescence in situ hybridization (FISH) and targeted sequencing, and found 16 additional TERT-rearranged cases. In the entire set (n = 217, Extended Data Table 1a), we detected 28 TERT rearrangements (13%), 27 of which occurred in high-risk tumours (Fig. 2a and Extended Data Table 1b). The remaining case was classified as intermediate risk (stage 3, 8 years at diagnosis), and the patient is currently in complete remission after surgery and chemotherapy. TERT rearrangements were strongly associated with the unfavourable prognostic markers of stage 4 disease and patient age at diagnosis older than 18 months, but occurred predominantly in tumours without MYCN amplification in the high-risk cohort (Extended Data Table 1b). Together, 27 of 114 high-risk neuroblastomas (24%) and 22 of 65 MYCN-non-amplified high-risk tumours (34%) harboured TERT rearrangements, indicating their frequent occurrence in these groups (Fig. 2a). Patients whose tumours harboured TERT rearrangements had a poor clinical outcome, which was similar to that of patients with MYCN-amplified tumours and significantly worse than that of high-risk patients without MYCN amplification or TERT rearrangements (overall survival, P = 0.056; Fig. 2b; event-free survival, P = 0.038; Extended Data Fig. 3a). The clinical relevance of TERT alterations was substantiated by multivariable analyses, in which TERT rearrangements predicted unfavourable outcome independently of the established variables stage and MYCN (Fig. 2c and Extended Data Fig. 3b).
Figure 2. Genomic TERT rearrangements are associated with poor patient outcome and high TERT mRNA expression.
a, Prevalence of TERT rearrangements in 217 primary neuroblastomas. TERT rearrangements were identified by whole-genome or targeted sequencing and break-apart FISH, exemplarily shown in the lower panel (red, TERT; green, CLPTM1L). b, Overall survival of neuroblastoma patient groups defined by TERT rearrangements (TERT), MYCN amplification (MNA), high-risk disease without these alterations (HR), and low-risk or intermediate-risk disease (non-HR). Patients with tumours bearing both a TERT rearrangement and MYCN amplification (n = 5) were excluded. Overall survival at 5 years: 0.41 ± 0.16 (TERT) versus 0.54 ± 0.10 (MNA) versus 0.79 ± 0.08 (HR) versus 0.98 ± 0.02 (LR).
c,Multivariable Cox regression analysis of the potential prognostic factors stage, MYCN status, and TERT status for overall survival in patients aged >18 months (n = 125). d, Distribution of TERT mRNA levels derived from transcriptome sequencing of the groups defined above: tumours with TERT rearrangements (yellow, n = 10), MYCN amplifications (red, n = 9), high-risk tumours without the aforementioned aberrations (grey, n = 18), among which ATRX-mutated cases are highlighted by blue circles (n = 7), and low-risk tumours (green, n = 17). Error bars, median expression and s.d. e, Comparison of TERT expression in those tumour subgroups defined by TERT copy numbers (bottom: left, two copies; middle, three or four copies; right, more than four copies). TERT expression levels in relation to TERT copy numbers in TERT rearranged cases (top). f, Relative average gene expression levels at the TERT locus. Subgroups of tumours with TERT rearrangements (yellow), MYCN amplification (red), high-risk tumours without the aforementioned aberrations (grey), and low-risk tumours (green) are compared with average expression in a large cohort of neuroblastoma samples (n = 498).
Since MYCN is a known transcriptional activator of TERT12, we compared TERT expression in tumours with 5p15.33 rearrangements with that in MYCN-amplified specimens and with the remaining cases. TERT expression was significantly higher in neuroblastomas with rearrangements than in MYCN-amplified tumours (P = 0.028) and in those without these alterations (P < 0.001, Fig. 2d and Extended Data Fig. 3c). The median TERT expression was 92-fold higher in TERT-rearranged tumours than in low-risk tumours (Extended Data Fig. 3d). In addition, TERT expression was higher in MYCN-amplified tumours than in the remaining samples (P = 0.021, Fig. 2d).
Since the region upstream of TERT consists of condensed chromatin in most somatic cells, which keeps TERT in a silenced state13, we hypothesized that the rearrangements may induce TERT transcriptional upregulation by genomic repositioning, rather than by gain of TERT copy numbers. In support of this hypothesis, TERT expression was massively increased in TERT-rearranged tumours compared with low-risk tumours in both subgroups with and without additional TERT copies (Fig. 2e). In addition, TERT copy numbers and messenger RNA (mRNA) levels did not correlate in TERT-rearranged neuroblastomas (Fig. 2e), indicating that additional TERT copies have little influence on TERT expression in these tumours. In line with that notion, we found evidence for mono-allelic TERT expression in five evaluable tumours, suggesting that only the rearranged allele is expressed in these cases (Extended Data Fig. 3e). Additionally, we found no evidence for the existence of tandem duplications affecting TERT without the presence of complex rearrangements (Fig. 1b). Together, these findings suggest that remodelling of the genomic context by the rearrangement itself—rather than copy number gain of this region14,15—is the major cause of TERT mRNA upregulation in TERT-rearranged neuroblastomas.
We also noticed that the distance of the breakpoints to the transcriptional start site of TERT did not affect TERT expression (Extended Data Fig. 3f). Furthermore, identical rearrangements and elevated TERT expression were detected in four tumours biopsied at initial diagnosis and relapse (Extended Data Fig. 4), supporting the notion that 5p15.33 rearrangements and consecutive upregulation of TERT are stable genetic events during neuroblastoma development and progression.
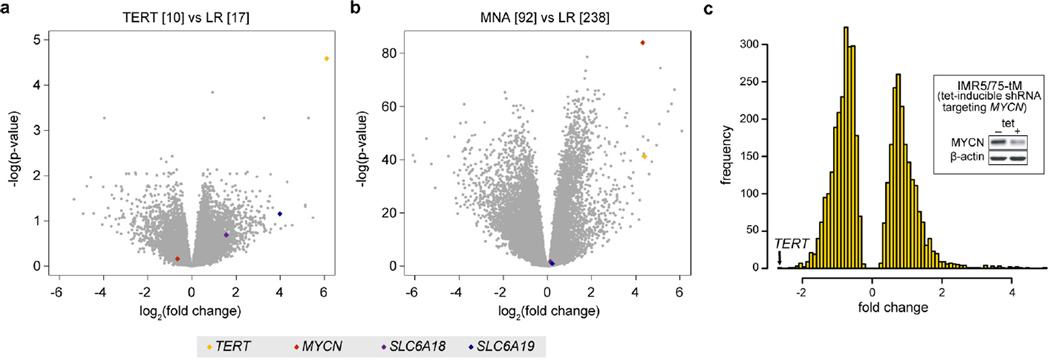
We observed that in TERT-rearranged neuroblastomas three genes (bleyplaby (AceView-annotated human gene; http://www.aceview.org), SLC6A18, and SLC6A19) neighbouring TERT on the distal side of the breakpoint showed markedly increased expression, while expression of suweeby, blorplaby, and CLPTM1L (cleft lip and palate transmembrane protein 1-like protein), located on the proximal side of the breakpoint, did not follow this pattern (Fig. 2f and Extended Data Fig. 5a). By contrast, MYCN-amplified tumours exhibited upregulation of only TERT and bleyplaby, but lacked upregulation of SLC6A18 and SLC6A19, whereas all four genes were silenced in low-risk neuroblastomas and in high-risk tumours without TERT rearrangements or MYCN amplification (Fig. 2f and Extended Data Fig. 5a, b). Our findings were substantiated by analyses of differential gene expression of tumours bearing TERT rearrangements or MYCN amplification versus low-risk tumours: in both comparisons, TERT was among the most highly upregulated genes, while SLC6A18 and SLC6A19 were upregulated in TERT-rearranged tumours only (Extended Data Fig. 6a, b). Thus, genomic rearrangements at 5p15.33 abrogate silencing of gene expression in this chromosomal region, whereas amplified MYCN selectively upregulates TERT by transcriptional activation. In line with this notion, we found that TERT was the most strongly downregulated gene upon short-interfering-RNA-mediated MYCN knockdown in a MYCN-amplified neuroblastoma cell line (Extended Data Fig. 6c). Together, these observations suggest that both 5p15.33 rearrangements and MYCN amplification converge on TERT activation in high-risk neuroblastoma.
The massive upregulation of TERT expression observed in tumours bearing structural rearrangements of the TERT locus suggested that translocation of regulatory elements (for example, enhancers) might be responsible for TERT activation. We therefore examined patterns of H3K27ac and H3K4me1 histone modifications, which are known to mark active enhancers16,17, in three TERT-rearranged tumours using chromatin immunoprecipitation coupled to sequencing (ChIP-seq). In all three tumours, we detected peak signals in the translocated regions that predict the presence of multiple enhancer clusters immediately adjacent to the breakpoints (Fig. 3a). Ranking of H3K27ac peak signals revealed that rearrangements consistently juxtapose TERT to strong enhancer elements, several of which are compatible with the recent definition of super-enhancers (Fig. 3b)18.
Figure 3. Translocation of active enhancers drive TERT expression in TERT-rearranged neuroblastomas.
a, Normalized read counts of H3K27ac and H3K4me1 histone marks derived from ChIP-seq at the TERT rearrangement region in three neuroblastomas. Significant peaks of H3K27ac read counts are displayed by black bars. Enhancer elements identified by stitched peak calls within 12.5 kb regions are shown in pale red. The enhancer element showing highest peak signals within a 0.5 Mb region upstream of the rearrangement breakpoint is highlighted in dark red. b, Enhancer elements were ranked according to cumulated read counts over the stitched peak calls. The strongest element identified within the rearranged region of the respective sample is depicted by red circles. Orange circles indicate the strongest enhancer elements in genomic regions affected by rearrangements in other TERT-rearranged tumours (n = 10; only non-inverted rearrangements are shown). c, Survey of the five histone marks H3K4me3, H3K4me1, H3K27ac, H3K36me3, and H3K9me3 in neuroblastomas harbouring TERT rearrangements (cell line GI-ME-N and primary tumour NBL36; TERT), and neuroblastomas lacking TERT and MYCN alterations (cell line SK-N-FI and primary tumour NBL06; HR). d, DNA methylation of the CpG site cg11625005 proximal to the TERT core promoter in TERT-rearranged (TERT, n = 6) and MYCN-amplified cases (MNA, n = 9) as well as tumours without these alterations (high-risk neuroblastoma, n = 6; low-risk neuroblastoma, n = 18). Error bars, median methylation and s.d.
To examine the chromatin context of the TERT locus further, we analysed additional histone modifications, known to mark active promoters (H3K4me3), transcription elongation (H3K36me3), DNA methylation (H3K9me3), and Polycomb repressive complex 2 (PRC2)-related gene silencing (H3K27me3), in neuroblastoma cells with and without TERT rearrangements (Fig. 3c and Extended Data Figs 7 and 8a). While histone modifications indicative of an active promoter (H3K4me3 and H3K27ac) and transcription elongation (H3K36me3) were spread to the TERT locus in TERT-rearranged cases, these marks were completely lacking in tumours without TERT rearrangements (Fig. 3c). By contrast, the promoter and gene body of TERT were broadly marked by the repressive mark H3K27me3 in cells lacking TERT alterations (Extended Data Fig. 8a).
DNA methylation analyses of 39 primary neuroblastomas revealed a consistent increase in CpG methylation across the TERT locus in rearranged and MYCN-amplified tumours (Extended Data Fig. 8b). The strongest differential methylation was detected in a CpG site located in close proximity to the core promotor of TERT (Fig. 3d and Extended Data Fig. 8b), which has been previously associated with a disabled repressive element and elevated TERT expression19. Thus, structural rearrangements occurring at 5p15.33 lead to juxtaposition of the TERT locus to strong enhancer elements, resulting in a massive epigenetic remodelling of the affected region.
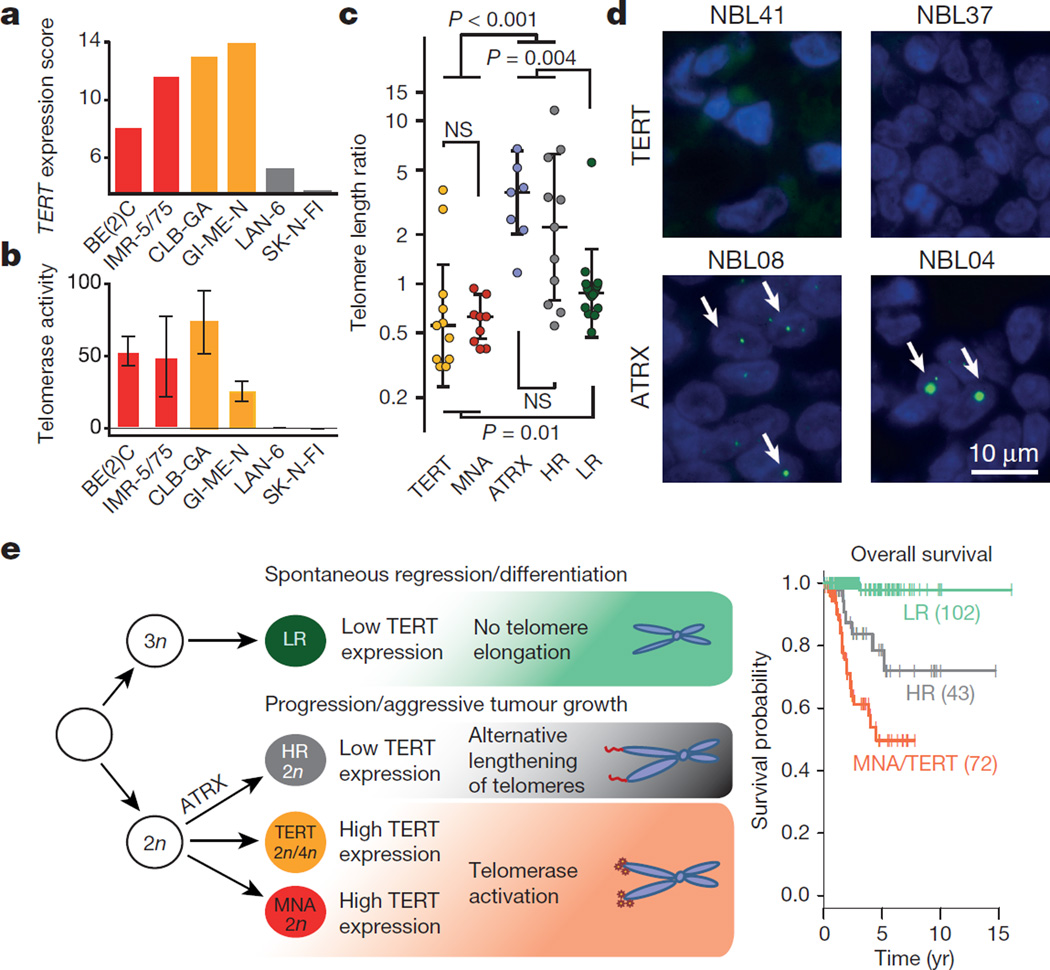
Our findings were further supported by analyses of cell lines derived from high-risk neuroblastoma bearing TERT rearrangements, MYCN amplification, or none of these alterations (Extended Data Fig. 7). Cell lines with TERT translocations or MYCN amplification showed elevated TERT mRNA expression compared with cell lines lacking such alterations (Fig. 4a). High levels of TERT expression were paralleled by a significant increase in enzymatic telomerase activity as assessed by a telomeric repeat amplification assay (Fig. 4b).
Figure 4. Telomerase activity is associated with TERT rearrangements and MYCN amplification, while ALT occurs in high-risk neuroblastoma lacking these alterations.
a, b, TERT expression levels as determined by transcriptome sequencing (a) and telomerase activity (b) as determined by telomeric repeat amplification assay in neuroblastoma cell lines bearing TERT rearrangements (GI-ME-N and CLB-GA), MYCN amplification (SK-N-BE(2)C and IMR-5/75), and cell lines without these alterations (LAN-6 and SK-N-FI). c, Distribution of telomere length ratios between the tumours and matched normals (computed from whole-genome sequencing) in primary neuroblastoma subgroups defined by TERT, MYCN, and ATRX alterations and risk group (HR, high-risk without the aforementioned alterations; LR, low-risk). d, Telomere FISH analyses of two TERT-rearranged (NBL41, NBL37) and two ATRX-mutated (NBL08, NBL04) primary tumours. e, A revised model for neuroblastoma pathogenesis based on recurrent genomic alterations, the presence or absence of telomere maintenance pathways, and clinical courses of the disease (modified after ref. 24). In this model, high-risk tumours are distinguished from low-risk tumours by active mechanisms of telomere lengthening. The most aggressive neuroblastomas are defined by telomerase activation as a result of either TERT rearrangement (TERT) or MYCN amplification (MNA). In addition, near-diploid (2n) or near-tetraploid (4n) karyotypes are preferentially observed in high-risk tumours, while near-triploid karyotypes are mostly found in low-risk tumours24. Overall survival of patient subgroups at 5 years: 0.51 ± 0.08 (MNA/TERT) versus 0.79 ± 0.08 (high-risk tumours) versus 0.98 ± 0.02 (low-risk tumours).
ATRX mutations occurred exclusively in tumours lacking MYCN amplification and TERT rearrangements (Fig. 1c) and have previously been associated with alternative lengthening of telomeres (ALT)5. Consistent with the hypothesis that high-risk neuroblastomas may be associated with ALT in the absence of TERT activation throughMYCN amplification or TERT rearrangement, the ATRX-mutant high-risk neuroblastoma cell line (CHLA-90) has evidence of ALT pathway activation20. Furthermore, we observed activation of the ALT pathway in two high-risk neuroblastoma cell lines without TERT or MYCN alterations (Extended Data Fig. 9)21. In line with these and published observations5, we found that ATRX-mutated primary neuroblastomas and other high-risk tumours lacking TERT or MYCN alterations had abundant telomere repeat sequences, pointing towards activation of ALT22 (Figs 1c and 4c). By contrast, telomeres were short in primary tumours in which telomerase was activated23. We confirmed this result by telomere FISH analysis in primary tumours bearing either ATRX mutations or TERT rearrangements (Fig. 4d). In low-risk tumours, the telomere lengths were similar to matched normal samples, indicating that telomere maintenance mechanisms may be inactive in these tumours (Fig. 4c). Together, these findings support the notion that high TERT expression mediated by 5p15.33 rearrangements or MYCN amplification induces constitutive catalytic telomerase activity, whereas ALT is activated in high-risk neuroblastoma lacking these alterations. Thus, our results suggest that high-risk neuroblastoma is defined molecularly by mechanisms leading to telomere lengthening.
In summary, we discovered recurrent TERT rearrangements in approximately one-quarter of high-risk neuroblastomas. Our results indicate that most high-risk tumours are affected by TERT rearrangements, MYCN amplification, or ATRX mutations, all of which funnel into telomere lengthening, thus providing a molecular, mechanistic definition of this neuroblastoma subtype (Fig. 4e). By contrast, low-risk tumours are characterized by the absence of such alterations and low TERT expression levels, presumably leading to the inability to gain immortal proliferation capacity. In our data set, the most aggressive subtype of neuroblastoma was defined by telomerase activation as a result of either TERT rearrangement or MYCN amplification (Fig. 4e). With the further development of telomerase inhibitors, our finding might point to a novel therapeutic option for the most aggressive subgroup of this deadly paediatric disease.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
METHODS
Patients and tumour samples
Overall, this project comprised tumour samples from 217 German patients with neuroblastoma (Extended Data Table 1a). Patients were diagnosed between 1991 and 2014 and were registered and treated according to several clinical trials of the Gesellschaft für Pädiatrische Onkologie und Hämatologie. The trials were approved by the Ethics Committee of the Medical Faculty, University of Cologne. The Institutional Review Board approved collection and use of all specimens in this study. Informed consent was obtained from all patients. The MYCN gene copy number was determined as a routine diagnostic method using FISH analysis. DNA and total RNA was isolated from tumour samples with at least 60% tumour cell content as evaluated by a pathologist.
TERT rearrangements were established as a novel molecular marker in a discovery cohort of 56 patients. In this set, TERT rearrangements (n = 12) occurred exclusively in high-risk patients (n = 39). We sought to validate this finding in a larger, representative neuroblastoma cohort, comprising approximately 40% high-risk patients. Allowing for a potential occurrence of TERT rearrangements in up to 10% of non-high-risk patients and ensuring a statistical power of 80%, we estimated that at least 75 non-high-risk patients were required for validation. We therefore investigated 161 additional tumours derived from 86 non-high-risk and 75 high-risk patients (including 39 and 36 tumours with and without MYCN amplification, respectively). The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Cell lines
Neuroblastoma cell lines LAN-6, GI-ME-N, as well as SK-N-FI were directly purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany) or American Type Culture Collection (ATCC/LGC Standards, Molsheim Cedex, France), respectively. Furthermore, SK-N-BE(2)C, IMR-5/75, and CLB-GA were provided by the laboratories of H. Deubzer, F. Westermann and J. H. Schulte, respectively. All cell lines not directly purchased from ATCC or DSMZ were authenticated by STR profiling at the DSMZ. IMR-5/75, SK-N-BE(2)C, GI-ME-N, and CLB-GA were grown in RPMI1640 with 10% FBS; LAN-6 and SK-N-FI were cultured in DMEM with 20% FBS. All cell lines were cultured without antibiotics and routinely tested negative for mycoplasma.
Whole-genome sequencing
DNA was extracted from fresh-frozen tumour tissue and the corresponding matched normal of 56 patients using the Puregene Core Kit A (Qiagen) and NucleoSpin Blood DNA extraction kit (Macherey-Nagel), respectively, according to the manufacturers’ instructions. DNA was eluted in 1 × TE buffer, diluted to a working concentration of 100 ng µl−1 and stored at −80 °C. Libraries were prepared with the TruSeq DNA PCR-free sample preparation kit (Illumina) followed by size selection using SPRI beads (Beckman Coulter Genomics). The final libraries were then sequenced on an Illumina HiSeq 2000 instrument with a paired-end read length of 2 × 100 nucleotides.
Whole-genome sequencing data analysis
Raw sequencing reads were aligned to the human genome (NCBI build 37/hg19) by using BWA (version 0.6.1-r104; https://github.com/lh3/bwa). Possible PCR duplicates were then masked in the resulting alignment files by searching for concordant read pairs. Next, somatic mutations were called using a further development of our in-house mutation caller. The major improvements to previous versions25 of the caller were that identified variants were filtered against a library of more than 500 normals (mixed whole-exome and whole-genome) and that the error model contained contributions of human library contamination which were estimated from the sequencing data. With these modifications we were able to gain a larger sensitivity and specificity (data not shown). Rearrangements as well as copy number changes were analysed as described previously25. Patterns of recurrent genomic rearrangements were identified by scanning the genomes for breakpoint clusters that occur within 100 kb regions in a similar approach to that described in ref. 7. To compute the telomere ratio from whole-genome sequencing data, raw sequencing reads containing the telomere repeat sequence (TTAGGG)4 or its reverse complement were counted5, and the ratio between the tumour and matched normal was determined. This ratio was then normalized to the absolute amount of sequenced DNA using the total amount of reads from the tumour and the normal.
Targeted sequencing
In the validation cohort, hybrid-capture-based target enrichment followed by massively parallel sequencing of the genomic region encompassing TERT and CLPTM1L was used to detect TERT rearrangements. Alignment and the detection of genomic rearrangements were performed analogous to whole-genome sequencing. We reported only those TERT rearrangements that had been detected both by FISH analysis and by targeted sequencing.
RNA sequencing
RNA sequencing and gene expression analysis was performed as described previously26. Briefly, a Dynabeads mRNA Purification Kit (Invitrogen) was used to purify mRNA from total RNA. Library construction was performed according to the standard TruSeq protocol. Clusters were generated according to the TruSeq PE cluster Kit version 3 reagent preparation guide (for cBot-HiSeq/HiScanSQ). High-throughput shotgun sequencing was performed on the IlluminaHiSeq 2000 platform. Paired-end reads with lengths of 100 nucleotides were generated. Raw data processing, read mapping, and gene expression quantification were done using the Magic-AceView analysis pipeline as described27. The Magic analysis tool is accessible at ftp://ftp.ncbi.nlm.nih.gov/repository/acedb/Software/Magic; AceView served as primary transcriptome reference (http://www.aceview.org). RNA sequencing was also used to identify MYCN-regulated genes in IMR5/75 cells expressing MYCN shRNA under the control of the tet repressor. Briefly, for IMR5/75 shRNA inducible cells, 1 µg ml−1 tetracycline or the equivalent of volume of 70% ethanol was added to the cells, and cells were incubated for 24 h and then harvested for RNA extraction using RN easy mini kit (Qiagen). Five micrograms of RNA from each sample was processed using the RiboGold kit (Epicentre) to remove rRNA from samples to increase reads from mRNA. The concentration of the resulting RNA was measured using the Qubit RNA assay (Life Technologies). One microgram of RNA was then used to prepare libraries for sequencing using the NEB Ultra directional RNA library prep lit for Illumina (New England Bioscience) according to the manufacturer’s instructions.
Microarray
Single-colour gene expression profiles were generated using customized 4 × 44 K oligonucleotide microarrays produced by Agilent Technologies. Labelling and hybridization was performed following the manufacturer’s protocol. Microarray expression profiles were generated using Agilent’s Feature Extraction software (version 9.5.1). Data were normalized using quantile normalization.
Validation of TERT rearrangements by dideoxy sequencing
Rearrangements of the TERT locus were validated by dideoxy sequencing in both diagnostic and relapsed tumour samples. Dideoxy sequencing was performed by Seqlab.
FISH
BAC clones CTD-2191M2 (to detect the region proximal of CLPTM1L) and CTD-2511M20 (to detect the TERT/SLC6A18/SLC6A19 loci) were labelled with digoxigenin and biotin, respectively (see also Fig. 3a). Cell line cytospin preparations were pre-treated with 2 × SSC solution at 37 °C for 30 min, digested with Digest-All III (dilution 1:2, Invitrogen) at 37 °C for 6 min, fixed in 4% formaldehyde, and subsequently dehydrated in a graded ethanol series. FISH probes and human Cot-1 DNA (Life Technologies) in hybridization buffer (50% formamide, 10% dextran sulfate sodium, in 2 × SSC) were co-denatured at 85 °C for 4 min and hybridized overnight at 37 °C. Post-hybridization washing was done with 0.5 × SSC at 75 °C for 5 min, followed by washes in PBS, a blocking step with CAS-block (Life Technologies, with 10% normal goat serum in PBS) and a 1 h post-incubation with streptavidin–Alexa-555 conjugates (1:500, Life Technologies) and anti-digoxigenin-FITC (1:500, Roche), to enable fluorescence detection. After three subsequent washes in PBS, samples were mounted with VectaShield mounting media containing 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Vectorlabs). Images were acquired using an Olympus Fluoview FV10 scanning confocal microscope system.
Telomeric repeat amplification protocol assay
Telomerase activity in cell lines was determined with a PCR-based telomeric repeat amplification protocol (TRAP) enzyme-linked immunosorbent assay (ELISA) kit (TeloTAGGG Telomerase PCR ELISAPLUS, Roche) according to the manufacturer’s protocol.
ChIP-seq analysis of histone modifications
Formaldehyde cross-linking of cells, cell lysis, sonication, ChIP procedure, and library preparation were performed as described previously28, starting with approximately 4 × 106 cells (1 × 106 cells per individual immunoprecipitation). Direct cell lysis for each sample was achieved by incubation for 30 min in 950 µl RIPA I on ice (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 140 mM NaCl, 0.2% SDS, 0.1% DOC). Tissue disruption, formaldehyde fixation, and sonication of tumour material were done according to a previously published protocol29. Approximately 30 mg of fresh-frozen tumour tissue was used per individual ChIP-seq experiment. All subsequent steps were performed analogous to cell line experiments. The Bioruptor Plus sonication device (Diagenode) was used for high intensity sonication for 30–60 min each with intervals of 30 s on and 30 s off. Library preparation was performed using the NEB Next Ultra DNA Library Prep Kit (New England Biolabs) according to the manufacturer’s protocol. Samples were mixed in equal molar ratios and sequenced on an Illumina sequencing platform.
ChIP-seq data analysis
Single-end reads were aligned to the hg19 genome using Bowtie2 (version 2.1.0). Only uniquely aligned reads were kept. BAM files of aligned reads were further processed using the deepTools suite (https://github.com/fidelram/deepTools). Input files were subtracted from the treatment files using the bamCompare tool, applying the SES method for normalization of signal to noise. Resulting signals were normalized to an average 1 × coverage to produce signal (bigWig) files. Peaks were called using the MACS 1.4 tool using default parameters.
DNA methylation profiling
DNA was isolated from snap-frozen neuroblastoma tissue. Genome-wide DNA methylation was assessed using an Infinium HumanMethylation450 BeadChip (Illumina) according to the manufacturer’s instructions. Probes were removed on the basis of the following criteria: (1) proportion of non-detectable β values >0.3 (n = 379), (2) single nucleotide polymorphism at or near the targeted CpG site according to R-Forge package IMA (https://rforge.net/IMA/, n = 92,600), (3) control probes (n = 65), and (4) mapping to the X or Y chromosome (n = 10,351). Together, 382,182 probes were kept for further analysis. The k-nearest neighbours method was used to impute missing values, and a subset quantile normalization was applied. TERT-related CpGs were annotated using the assignGenomeAnnotation program of the HOMER tool suite (http://homer.salk.edu/homer).
Telomere restriction-fragment analysis and C-circle assay
Both assays have been performed as described previously21. Briefly, genomic DNA was purified and digested with AluI and MboI. For restriction-fragment analysis, 10 µg of digested DNA was electrophoresed on a 0.8% TBE-agarose gel. Subsequently, telomeric DNA was detected by Southern blotting using a [32P]dATP end-labelled (CCCTAA)4 oligonucleotide probe. For the C-circle assay, DNA samples (7.5 ng, 10 µl) diluted in ultraclean water were combined with 10 µl BSA (NEB; 0.2 mg ml−1), 0.1% Tween, 0.2 mM each dATP, dGTP, dTTP, and 1 × Φ29 Buffer (NEB) in the presence or absence of 7.5 U ΦDNA polymerase (NEB), incubated at 30 °C for 8 h and then at 65 °C for 20 min. Reaction products were diluted to 100 µl with 2 × SSC and dot-blotted onto a 2 × SSC-soaked nylon membrane. DNA was ultraviolet cross-linked onto the membrane and hybridized with a 32P-end-labelled (CCCTAA)4 oligonucleotide probe to detect C-circle amplification products. All blots were washed, exposed to PhosphoImager screens, scanned, and quantified using a Typhoon 9400 PhosphoImager (Amersham, GE Healthcare). Genomic DNA from ALT-positive (U2OS) cells served as positive control and reference for the quantification of C-circles detected in other cell lines.
Immunofluorescence-FISH for APBs
ALT-associated promyelocytic leukaemia (PML) bodies were visualized by a combination of immunofluorescence with an anti-PML antibody and FISH using Alexa-488-(TTAGGG)n PNA probes as described previously21.
Telomere FISH on tumour sections
Tumours were sliced into 4 µm sections, paraffin fixed, embedded in formalin, and mounted onto positively charged glass microscope slides. Mounted sections were incubated for 30 min at 55 °C, washed three times for 5 min in xylene, rinsed in successive 100%, 95%, and 70% ethanol baths, and washed in double-distilled H2O and 1% Tween before being placed in antigen unmasking solution in a boiling kitchen steam for 30 min. Next, slides were rinsed in double-distilled H2O and dehydrated in successive ethanol washes of 70%, 95%, and 100%. Slides were incubated at 72 °C for 10 min with an Alexa-488 telomeric-C PNA probe and hybridized overnight in a dark humidity chamber. Slides were washed with PNA wash buffer and PBST and incubated for 10 min in DAPI solution. After washing in double-distilled H2O, slides were mounted with prolong anti-fade mounting medium. Images were taken on a Nikon 90i fluorescent light microscope at × 63 resolution. Full z-stacks were taken at 0.5 µm and projected and focused using Elements software.
Statistical analyses
SPSS (package release 20.0.0, IBM Armonk), R (version 3.1.2), and GraphPad Prism (version 6.05 GraphPad Software) were applied for statistical analyses and data presentation. Overall survival was calculated as the time from diagnosis to death from disease or the last follow-up if the patient survived. Event-free survival was calculated from diagnosis to the time of tumour progression, relapse, or death from disease, or to the last follow-up if no event occurred. Survival curves were estimated according to Kaplan–Meier and compared with the log-rank test (R survival package version 2.15.0). Associations of genomic alterations with clinical risk factors were examined using Fisher’s exact test. Multivariable Cox regression models were used to analyse the simultaneous prognostic impact of TERT rearrangements and established clinical markers (stage (1–3, 4S versus 4), MYCN (non-amplified versus amplified), and age (< 18 months versus >18 months)) on overall survival and event-free survival. Since TERT rearrangements were observed only in patients aged >18 months in this study, multivariable model building was restricted to this cohort (n = 125) and the variables TERT status, stage, and MYCN status. First, the proportional hazard assumption was assessed for each predictor one-at-a-time using the goodness-of-fit test of ref. 30 showing no deviation from the proportional hazard assumption. The proportional hazard assumption was considered valid whenever the P value of the goodness-of-fit test was >0.05. In addition, predicted survival curves under the Cox model were compared with the Kaplan–Meier estimates for each predictor supporting adequateness of model fit. Multivariable models were then built using a backwards selection procedure including the variables TERT status, stage, and MYCN status (inclusion criterion, P value of the score test ≤0.05; exclusion criterion, P value of the likelihood ratio test >0.1). The variables identified at this step formed the model of main effects. Finally, the factors selected in the model of main effects were fitted with all pairwise interactions in a second block by a stepwise forward selection (inclusion criterion, P value of the score test ≤0.05; exclusion criterion, P value of the likelihood ratio test >0.1), resulting in the final model. For the final model, the proportional hazard assumption was assessed using the goodness-of-fit test of ref. 30 as well as by fitting extended Cox models including the prognostic factors from the final model in a first block and the product terms of the prognostic factors with some function of time g(t) in a second block with stepwise forward selection in the second block (inclusion criterion, P value of the score test ≤0.05; exclusion criterion, P value of the likelihood ratio test >0.1). Choices for g(t) were g(t) = t and g(t) = log(t) with t denoting survival time. The proportional hazard assumption was considered as valid if no time-dependent factor was selected in any of the extended Cox models and if, additionally, any P value of goodness-of-fit test was >0.05.
Analyses of TERT expression levels and methylation between subgroups were investigated by Mann–Whitney tests and corrected for multiple hypotheses testing using a Bonferroni correction. To test for mutual exclusivity between TERT rearrangements (TERT), MYCN amplifications (MNA), and ATRX mutations (ATRX) in the high-risk group, Fisher’s exact tests were performed between every alteration and the combination of the remaining two alterations (TERT versus MNA and ATRX; MNA versus TERT and ATRX; ATRX versus TERT and MNA). The largest P value was finally reported.
Extended Data
Extended Data Figure 1. Validation of rearrangements of the TERT locus by dideoxy-sequencing.
Sequencing chromatograms of the breakpoint regions of 5p15.33 rearrangements along with their genomic coordinates (hg19), and the breakpoint-spanning nucleotide sequences. The sequence mapping to the TERT locus is indicated in yellow, the rearrangement partner is indicated in grey; nucleotides inserted at the breakpoint region are indicated in white.
Extended Data Figure 2. Schematic representation of chromothripsis in four primary neuroblastomas.
a, Circos plots showing chromothripsis of chromosome 5 in the tumours NBL39 and NBL37. b, Circos plots showing chromothripsis of chromosomes 17 and 20 in the tumours NBL21 and NBL08, respectively. Regions showing loss of heterozygosity (LOH) are indicated by black segments.
Extended Data Figure 3. TERT rearrangements are associated with poor event-free survival of patients, and with TERT upregulation independent of the breakpoint distance from the TERT transcriptional start site.
a, Kaplan–Meier estimates for event-free survival of neuroblastoma patient groups defined by TERT rearrangements (TERT), MYCN amplification (MNA), high-risk disease without these alterations (HR), and low-risk or intermediate-risk disease (nonHR). Patients with tumours bearing both a TERT rearrangement and MYCN amplification (n = 5) were excluded. Event-free survival at 5 years: 0.17 ± 0.09 (TERT) versus 0.38 ± 0.09 (MNA) versus 0.43 ± 0.09 (HR) versus 0.83 ± 0.05 (nonHR). b, Multivariable Cox regression analysis of the potential prognostic factors stage, MYCN status, and TERT status for event-free survival in patients aged >18 months (n = 125). c, Validation of TERT expression levels by microarrays in the four neuroblastoma subgroups indicated above. Sample numbers are given at the bottom. **P < 0.01, ***P < 0.001. d, Fold-change of median TERT expression levels in TERT-rearranged and MYCN-amplified tumours compared with low-risk (LR) neuroblastomas as measured by transcriptome sequencing. e, Evidence for monoallelic expression of TERT in five tumours bearing TERT rearrangements. The presence of a heterozygous single nucleotide polymorphism and its allelic fraction measured by whole-genome sequencing (WGS) is shown on the left of each panel; monoallelic expression as established by transcriptome sequencing (RNA-seq) is indicated on the right. The genomic position of the single nucleotide polymorphism is indicated at the top, the number of reads available for the analysis is shown at the bottom. f, TERT expression measured by transcriptome sequencing in relation to the distance of the rearrangement breakpoint from the TERT transcriptional start site (TSS). TERT expression levels and breakpoint distances from the TERT transcriptional start site were not correlated (r = 0, P = 0.97; Spearman’s rank correlation test).
Extended Data Figure 4. TERT rearrangements are maintained in relapsed neuroblastoma.
a, Agarose gel electrophoresis of PCR products representing individual TERT rearrangements in four tumours at initial diagnosis (I), and at relapse (R). The non-template controls are indicated by C. b, TERT expression measured by microarrays in relapsed TERT-rearranged tumours (n = 3) compared with TERT-rearranged tumours biopsied at initial diagnosis (n = 10). c, Sequencing chromatograms of the breakpoint regions for the relapse cases.
Extended Data Figure 5. Regional effects of TERT rearrangements on gene expression patterns.
a, Expression levels of genes around TERT measured by transcriptome sequencing are shown for tumours with TERT rearrangements (TERT, yellow), tumours with MYCN amplifications (MNA, red), and tumours without these aberrations classified as either high-risk (HR, grey) or low-risk (LR, green). Five consecutive genes (TERT, bleyplaby, SLC6A18, SLC6A19, and rarjy) located distal of the breakpoint at chromosome 5p15.33, and three genes (suweeby, blorplaby, and CLPTM1L) located proximal of the breakpoint are shown. Genomic positions of the genes are indicated at the top, and sample sizes are indicated at the bottom. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001. b, Averaged expression levels of genes measured by transcriptome sequencing are indicated for an ~500 kb region centring around TERT. Colour codes of neuroblastoma subgroups as indicated above. The breakpoint region is indicated in beige.
Extended Data Figure 6. TERT mRNA levels are massively upregulated in TERT-rearranged and MYCN-amplified neuroblastomas.
a, Differential gene expression between TERT-rearranged (n = 10) and low-risk (n = 17) neuroblastomas and b, between MYCN-amplified (MNA, n = 92) and low-risk tumours (n = 238) measured by transcriptome sequencing26. Negative log(P values) are plotted against log2(fold changes) of the genes expressed in the respective subgroups. Genes of interest are indicated at the bottom. log2(fold changes) correspond to the log difference of established gene expression scores. P values were established using Student’s t-tests and applying a Benjamini and Hochberg false discovery rate multiple testing correction. c, Analysis of differential gene expression of a MYCN-amplified neuroblastoma cell line (IMR-5/75) before and after shRNA-mediated knockdown of MYCN using transcriptome sequencing. The number of genes (frequency) up- or downregulated upon MYCN knockdown is plotted against the fold change of regulation. TERT is the most strongly downregulated gene upon shRNA-mediated MYCN knockdown. Only differentially expressed genes (t-test, false discovery rate controlled with P < 0.01) are shown. The insert shows reduced MYCN protein levels after induction of a MYCN-specific shRNA in IMR-5/75 cells as well as β-actin protein levels as control analysed by immunoblotting.
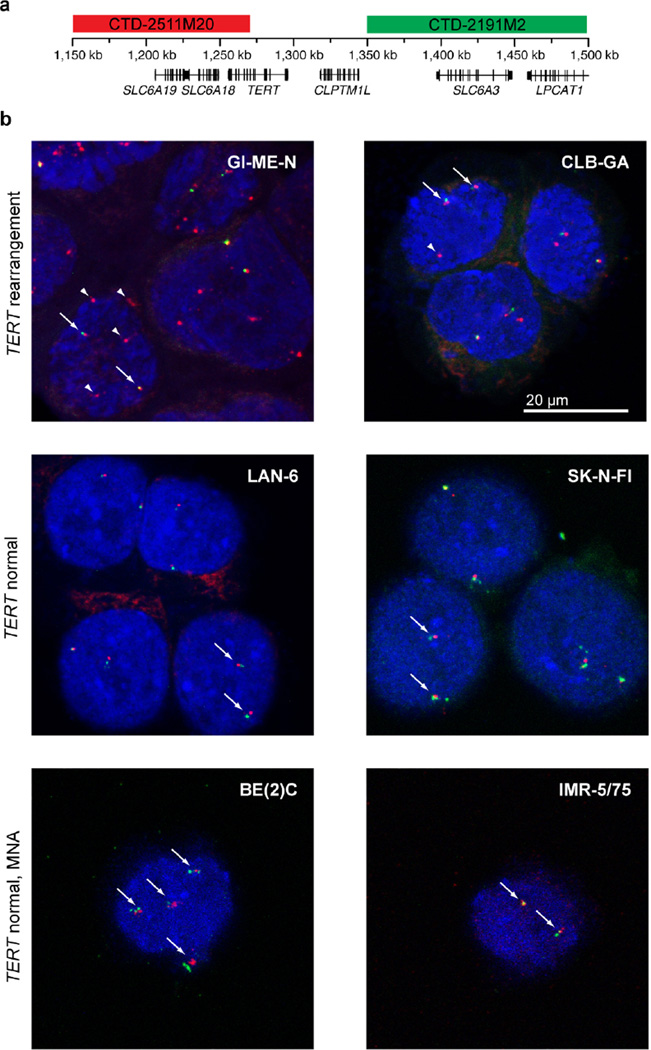
Extended Data Figure 7. Detection of TERT rearrangements in neuroblastoma cell lines.
a, Schematic representation of binding sites of probes used for FISH at the TERT locus. b, FISH analysis of neuroblastoma cell lines with 5p15.33 rearrangements and without these rearrangements; MNA, MYCN-amplified. Arrows indicate the unaltered status of chromosome 5p15.33 (red signals in close proximity to green signals); arrowheads indicate a rearrangement of 5p15.33 (red signals without adjacent green signals).
Extended Data Figure 8. Patterns of H3K27me3 histone modification and DNA methylation at the TERT locus.
a, Survey of the histone mark H3K27me3 in neuroblastomas harbouring TERT rearrangements (cell line GI-ME-N and primary tumour NBL36) and a neuroblastoma cell line lacking these alterations (SK-N-FI). b, DNA methylation patterns of CpG sites at the TERT locus in TERT-rearranged (TERT, n = 6) and MYCN-amplified (MNA, n = 9) primary neuroblastomas, as well as tumours lacking these alterations (others, n = 24) using HumanMethylation450 microrrays. Samples are ordered from top to bottom. CpG sites are indicated relative to their position to the TERT locus. Average methylation levels of each CpG site in the three subgroups are shown at the top. Similar to the highlighted CpG site cg11625005 upstream of the TERT transcriptional start site (Fig. 3d), CpG sites scattered over the TERT gene body were methylated significantly higher in TERT-rearranged and MYCN-amplified cases than in tumours without these alterations (P < 0.001 each).
Extended Data Figure 9. ALT activity in neuroblastoma cell lines lacking TERT rearrangements and MYCN amplifications.
a, Detection of extrachromosomal telomeric repeat DNA by C-circle assays with genomic AluI/MboI digested DNA from the indicated neuroblastoma cell lines. The ALT-positive control cell line, U2OS, is at the left. b, Detection of ALT-associated PML bodies in the indicated cell lines. Telomeric TTAGGG FISH (green) and immunofluorescence for PML (red) were combined and DNA was counterstained with DAPI (blue). c, Telomere restriction-fragment analysis of telomeric DNA from LAN-6, SK-N-FI, and the ALT-positive control U2OS. Telomeric DNA was detected by Southern blot with a [32P]dATP end-labelled (CCCTAA)4 oligonucleotide.
Extended Data Table 1.
Prognostic characteristics of the neuroblastoma cohort and prevalence of TERT rearrangements in patient subgroups
| a | ||
|---|---|---|
| Prognostic characteristics of the neuroblastoma cohort | ||
| n | % of total | |
| INSS stage | ||
| 1 | 31 | 14.3 % |
| 2 | 28 | 12.9 % |
| 3 | 25 | 11.5 % |
| 4 | 104 | 47.9 % |
| 4S | 29 | 13.4 % |
| Age at diagnosis | ||
| < 18 months | 92 | 42.4 % |
| > 18 months | 125 | 57.6 % |
| MYCN status | ||
| Normal | 168 | 77.4 % |
| Amplified (MNA) | 49 | 22.6 % |
| Risk groups | ||
| High -risk [HR] patients | 114 | 52.5 % |
| - HR w/o MNA | 65 | 30.0 % |
| Non -HR patients | 103 | 47.5 % |
| b | ||||
|---|---|---|---|---|
| Prevalence of TERT rearrangements in prognostic neuroblastoma subgroups | ||||
| nALL | nTERTr | odds ratio | P-value | |
| INSS stage | ||||
| 1–3, 4S | 113 | 1 | ||
| 4 | 104 | 27 | 38.81 | <0.001 |
| Age at diagnosis | ||||
| < 18 months | 92 | 0 | ||
| > 18 months | 125 | 28 | ∞ | <0.001 |
| Risk groups | ||||
| Non-HR patients | 103 | 1 | ||
| High-risk [HR] patients | 114 | 27 | 31.33 | <0.001 |
| MYCN status | ||||
| Normal | 168 | 23 | ||
| Amplified (MNA) | 49 | 5 | [0.72] | 0.633 |
| HR w/o MNA | 65 | 22 | ||
| HR w/ MNA | 49 | 5 | 0.22 | 0.004 |
Supplementary Material
Acknowledgments
We are indebted to the patients and their parents for making available the tumour specimens analysed in this study. We thank the German Neuroblastoma Biobank for providing samples from patients. The Institutional Review Board approved collection and use of all specimens in this study. We also thank our colleagues N. Hemstedt, H. Düren, E. Hess, J. Kreth, and J. Gopalakrishnan; and our collaboration partners I. Amit and F. Paul at the Weizmann Institute of Science for technical assistance. We further acknowledge the Center for Molecular Medicine Cologne light microscope facility for helping us to obtain high-quality data of FISH analyses, and S. Wolf and the Next Generation Sequencing Unit at the German Cancer Research Center (DKFZ) for sequencing. This work was supported by the German Cancer Aid (grant 110122) to M.F., F.W., J.H.S., A.S., and S.A.; the German Ministry of Science and Education (BMBF) as part of the e:Med initiative (grant 01ZX1303A to M.P., M.F., J.H.S., R.B., U.L., and R.K.T., grant 01ZX1406 to M.P., and grant 01ZX1307D to M.F. and F.W.); the BMBF (grant 0316076A to F.W.); the European Union (grant 259348 to F.W.); the Fördergesellschaft Kinderkrebs-Neuroblastom-Forschung e.V. (to M.F.); the German-Israeli Helmholtz Research School in Cancer Biology (to M.G. and F.W.); the Volkswagenstiftung (Lichtenberg Program) (to M.R.S.); and the Center for Molecular Medicine Cologne.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions Conception and design: M.P., F.H., F.R., D.D., M.G., F.W., R.K.T., and M.F. Administrative support, provision of study materials and patients: M.P., J.H., I.L., T.H., P.N., V.A., U.L., A.Eg., F.B., Z.P., C.Z., L.Sh., R.B., S.P., B.H., A.S., J.H.S., F.W., R.J.O., R.K.T., and M.F. Conduct of the experiments, data analysis, and interpretation: M.P., F.H., F.R., D.D., M.G., R.M., A.K., J.L.R., F.S., J.H., F.I., R.S., S.A., A.En., Y.K., W.V., J.A., J.T.-M., D.T.-M., A.M., S.H., E.M., K.-O.H., C.G., G.B., M.-R.S., L.Sa., S.C.W., C.S., E.B., J.T., R.V., M.S., B.D., M.O., B.H., C.H., R.J.O., F.W., R.K.T., and M.F. Manuscript writing: M.P., F.H., F.R., F.W., R.K.T., and M.F. All authors read and approved the final manuscript.
All high-throughput data have been deposited at the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/) under accession number EGAS00001001308.
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 3.Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nature Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sausen M, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the child hood cancer neuroblastoma. Nature Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung NK, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. J. Am. Med. Assoc. 2012;307:1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mossé YP, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Northcott PA, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagel I, et al. Deregulation of the telomerase reverse transcriptase (TERT) gene by chromosomal translocations in B-cell malignancies. Blood. 2010;116:1317–1320. doi: 10.1182/blood-2009-09-240440. [DOI] [PubMed] [Google Scholar]
- 9.Davis CF, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 11.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mac SM, D’Cunha CA, Farnham PJ. Direct recruitment of N-myc to target gene promoters. Mol. Carcinog. 2000;29:76–86. [PubMed] [Google Scholar]
- 13.Zhao Y, Wang S, Popova EY, Grigoryev SA, Zhu J. Rearrangement of upstream sequences of the hTERT gene during cellular immortalization. Genes Chromosom. Cancer. 2009;48:963–974. doi: 10.1002/gcc.20698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumps C, et al. Focal DNA copy number changes in neuroblastoma target MYCN regulated genes. PLoS One. 2013;8:e52321. doi: 10.1371/journal.pone.0052321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobrinik D, et al. Recurrent pre-existing and acquired DNA copy number alterations, including focal TERT gains, in neuroblastoma central nervous system metastases. Genes Chromosom. Cancer. 2013;52:1150–1166. doi: 10.1002/gcc.22110. [DOI] [PubMed] [Google Scholar]
- 16.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 17.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castelo-Branco P, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol. 2013;14:534–542. doi: 10.1016/S1470-2045(13)70110-4. [DOI] [PubMed] [Google Scholar]
- 20.Farooqi AS, et al. Alternative lengthening of telomeres in neuroblastoma cell lines is associated with a lack of MYCN genomic amplification and with p53 pathway aberrations. J. Neurooncol. 2014;119:17–26. doi: 10.1007/s11060-014-1456-8. [DOI] [PubMed] [Google Scholar]
- 21.O’Sullivan RJ, et al. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nature Struct. Mol. Biol. 2014;21:167–174. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nature Rev. Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 23.Martínez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nature Rev. Cancer. 2011;11:161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- 24.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nature Rev. Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Cuesta L, et al. Frequent mutations in chromatin-remodelling genes in pulmonary carcinoids. Nature Commun. 2014;5:3518. doi: 10.1038/ncomms4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, et al. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 2015;16:133. doi: 10.1186/s13059-015-0694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng X, et al. Tissue-specific transcriptome sequencing analysis expands the nonhuman primate reference transcriptome resource (NHPRTR) Nucleic Acids Res. 2015;43:D737–D742. doi: 10.1093/nar/gku1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blecher-Gonen R, et al. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nature Protocols. 2013;8:539–554. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- 29.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nature Protocols. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 30.Harrel F, Lee K. Proc. Eleventh Annual SAS User’s Group International. SAS Institute; 1986. pp. 823–838. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.