Abstract
Purpose
We undertook this analysis of KRAS mutation in four trials of adjuvant chemotherapy (ACT) versus observation (OBS) to clarify the prognostic/predictive roles of KRAS in non–small-cell lung cancer (NSCLC).
Methods
KRAS mutation was determined in blinded fashion. Exploratory analyses were performed to characterize relationships between mutation status and subtype and survival outcomes using a multivariable Cox model.
Results
Among 1,543 patients (763 OBS, 780 ACT), 300 had KRAS mutations (codon 12, n = 275; codon 13, n = 24; codon 14, n = 1). In OBS patients, there was no prognostic difference for overall survival for codon-12 (mutation v wild type [WT] hazard ratio [HR] = 1.04; 95% CI, 0.77 to 1.40) or codon-13 (HR = 1.01; 95% CI, 0.47 to 2.17) mutations. No significant benefit from ACT was observed for WT-KRAS (ACT v OBS HR = 0.89; 95% CI, 0.76 to 1.04; P = .15) or codon-12 mutations (HR = 0.95; 95% CI, 0.67 to 1.35; P = .77); with codon-13 mutations, ACT was deleterious (HR = 5.78; 95% CI, 2.06 to 16.2; P < .001; interaction P = .002). There was no prognostic effect for specific codon-12 amino acid substitution. The effect of ACT was variable among patients with codon-12 mutations: G12A or G12R (HR = 0.66; P = .48), G12C or G12V (HR = 0.94; P = .77) and G12D or G12S (HR = 1.39; P = .48; comparison of four HRs, including WT, interaction P = .76). OBS patients with KRAS-mutated tumors were more likely to develop second primary cancers (HR = 2.76, 95% CI, 1.34 to 5.70; P = .005) but not ACT patients (HR = 0.66; 95% CI, 0.25 to 1.75; P = .40; interaction, P = .02).
Conclusion
KRAS mutation status is not significantly prognostic. The potential interaction in patients with codon-13 mutations requires validation. At this time, KRAS status cannot be recommended to select patients with NSCLC for ACT.
INTRODUCTION
KRAS, a member of the RAS oncogene family, encodes a protein that binds guanine nucleotides, has GTPase activity, and is involved in signal transduction.1 In non–small-cell lung cancer (NSCLC), KRAS mutations occur most frequently in codons 12 and 13, usually are found in cancers of smokers, and are most common in nonsquamous NSCLC. Mutations have been reported in ∼30% of lung adenocarcinomas.2,3 Slebos et al4 were among the first to suggest that KRAS mutation was prognostic of poorer outcome. Since then, several meta-analyses have reported similar results, although there has been considerable variability among studies with respect to the magnitude of the prognostic effect.5,6
On the basis of early in vitro studies, it was postulated that RAS mutations might be associated with resistance to therapy.7,8 However, subsequent clinical studies could not always confirm the preclinical effects.9–11 The North American Intergroup trial JBR.10 randomly assigned patients with completely resected stage IB-II NSCLC to receive adjuvant chemotherapy (ACT) with cisplatin/vinorelbine or observation (OBS) alone.10,11 No differential overall survival (OS) benefit could be demonstrated in patients with wild-type (WT) RAS compared with those with mutations. However, disease-specific survival appeared to be prolonged with ACT in patients with WT-RAS (hazard ratio [HR] = 0.72; P = .06), with no apparent benefit in patients with mutations (HR= 1.07; P = .82). However, the study lacked power to demonstrate significant interaction.
In an attempt to clarify the impact of KRAS mutation on survival benefit from platinum-based ACT, we performed this pooled analysis of four randomized trials of ACT or OBS.
METHODS
Clinical Trials
This study used the LACE (Lung Adjuvant Cisplatin Evaluation) database of 3,533 patients from three LACE cisplatin-based ACT trials (IALT [International Adjuvant Lung Cancer Trial],12,13 ANITA [Adjuvant Navelbine International Trialist Association],14 JBR.1010,11)15 and Cancer and Leukemia Group B (CALGB) 9633, which used carboplatin-based ACT.16 Scientists, clinicians, and statisticians from these trials represent the LACE-Bio Collaborative Group.
KRAS Mutation Analyses
KRAS mutation analyses were performed in one laboratory for ANITA and JBR.10 (M.-S.T.) and in the laboratories of P.A.J. and P.H. for CALGB-9633 and IALT, respectively. Scientists were blinded regarding study arm and outcome. Laboratory methods are provided (Data Supplement).
Statistical Methods
Analyses included only patients from the previously mentioned trials with KRAS mutation results. OS, the primary end point, was defined as the time from randomization to death from any cause or last follow-up in surviving patients. Disease-free survival (DFS) was defined as the time from randomization to recurrence or death from any cause or last follow-up in surviving patients. Second primaries were analyzed as a time-to-event end point from randomization to occurrence of a second primary.
A logistic model stratified by trial was used to study correlations between KRAS status and covariates. A Cox model stratified by trial and including the covariates of treatment, sex, age (< 55, 55 to 64, > 64 years), performance status (PS 1/2, PS 0), tumor stage (T1, T2, T3/4), nodal stage (N0, N1, N2), and histology (squamous, adenocarcinoma, other) was used to examine the prognostic value of KRAS status on OS, DFS, and second primary rate (Data Supplement). Prognostic analyses were performed in the OBS and ACT arms together, and in OBS patients alone when a potential treatment and KRAS interaction was identified. The interaction between treatment and KRAS status was assessed to determine the predictive value of mutation status. Prognostic and predictive effects of KRAS mutation subtypes were examined using the same methods. KRAS subtypes were grouped prospectively into three subgroups based on their potential association with tobacco carcinogens. To assess the effect of KRAS status by histologic subgroup, an interaction with histology was introduced in the models.
A test for heterogeneity was used to compare HRs among trials. Within pooled analyses, statistical significance was set at P < .01, and HRs were reported with 95% CIs. Survival curves were based on Kaplan-Meier methods and represented with adjusted/stratified HRs and P values. Statistical analyses were performed using SAS Software, version 9.2 (SAS Institute, Cary, NC). All P values are two-sided.
RESULTS
Patients
Of 3,532 patients randomly assigned (840 ANITA, 482 JBR.10, 1,867 IALT, 343 CALGB-9633; Data Supplement), 1,718 underwent KRAS testing, with mutation results for 1,543 (44% of randomized cases). Baseline demographics for patients with and without KRAS results and for patients with KRAS results by trial are shown in the Data Supplement.
KRAS Mutation Results
KRAS mutations (MUT) were identified in 300 specimens (19%). Mutations were more frequent in female patients (27% v 17% male; P = .001), patients with adenocarcinoma (34% v 6% squamous, 23% other; P < .001), and younger patients (trend P = .0003; Table 1). In the multivariable model, only age (P = .044) and histology (P < .001) remained significant.
Table 1.
Patient and Tumor Characteristics for Patients With KRAS Mutated and Wild-Type Tumors
| Characteristic |
KRAS Mutated (n = 300) |
KRAS Wild Type (n = 1,243) |
P* | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Sex | |||||
| Male | 195 | 17 | 957 | 83 | .001 |
| Female | 105 | 27 | 286 | 73 | |
| Age, years | |||||
| < 55 | 107 | 25 | 325 | 75 | .001 (.0003) |
| 55-64 | 115 | 18 | 513 | 82 | |
| > 64 | 78 | 16 | 405 | 84 | |
| WHO performance status | |||||
| 0 | 162 | 20 | 648 | 80 | |
| 1/2 | 137 | 19 | 592 | 81 | .44 |
| Unknown | 1 | 25 | 3 | 75 | |
| Smoking status | |||||
| No | 10 | 17 | 48 | 83 | |
| Yes | 123 | 25 | 379 | 75 | .20 |
| Unknown† | 167 | 17 | 816 | 83 | |
| Stage‡ | |||||
| I | 175 | 23 | 590 | 77 | .15 (.02) |
| II | 96 | 18 | 430 | 82 | |
| III | 29 | 12 | 218 | 88 | |
| Unknown | 0 | 0 | 5 | 100 | |
| N stage‡ | |||||
| 0 | 184 | 21 | 6,752 | 79 | .50 (.21) |
| 1 | 87 | 18 | 390 | 82 | |
| 2 | 28 | 14 | 171 | 86 | |
| Unknown | 1 | 13 | 7 | 87 | |
| T stage‡ | |||||
| 1 | 33 | 19 | 141 | 81 | .009 (.01) |
| 2 | 252 | 22 | 912 | 78 | |
| 3/4 | 15 | 8 | 185 | 92 | |
| Unknown | 0 | 0 | 5 | 100 | |
| Histology | |||||
| Squamous cell | 43 | 6 | 664 | 94 | < .001 |
| Adenocarcinoma | 204 | 34 | 401 | 66 | |
| Other NSCLC | 53 | 23 | 178 | 77 | |
| Type of surgery | |||||
| Lobectomy/other | 240 | 23 | 836 | 77 | .001 |
| Pneumonectomy | 59 | 13 | 405 | 87 | |
| Unknown | 1 | 33 | 2 | 67 | |
Abbreviations: IALT, International Adjuvant Lung Cancer Trial; NSCLC, non–small-cell lung cancer.
P values are calculated from univariate logistic regression stratified by trial in patients with no missing values. Test for trend in parentheses.
No tobacco-related data collected in IALT trial (718 patients); this variable was not included in the multivariable models.
From 6th edition TNM staging classification.
Mutations were found at codon 12 in 275 patients (three double codon 12 mutations), codon 13 in 24, and codon 14 in one patient. The most frequent nucleotide change was a guanine>thymidine (G>T) transversion in 223 patients (Data Supplement). Amino acid substitutions are described in the Data Supplement. Mutations were found in 11 (1.6%) of 68 patients who were lifetime nonsmokers (codon 12, n = 10; codon 13, n = 1; Data Supplement). G>T transversions, considered the characteristic mutation from tobacco smoke, represented 78% (75 of 134) of codon 12 mutations in smokers and 50% (five of 10) in nonsmokers.
Prognostic Effect of KRAS Mutations
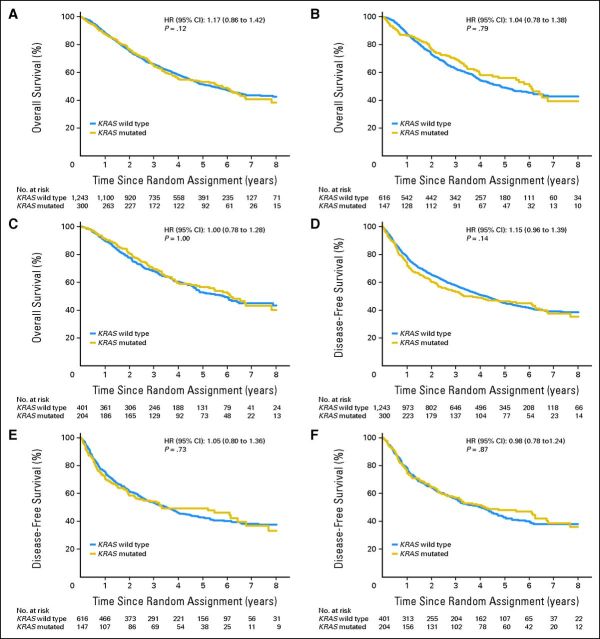
With 5.5 (95% CI, 5.3 to 5.7) years of median follow-up and 754 deaths (48.9%) among all 1,543 patients (Fig 1A), after accounting for the prognostic effect of baseline characteristics in multivariable analyses (Data Supplement), there was no significant difference in OS based on KRAS (multivariable HR MUT v WT, 1.17; 95% CI, 0.96 to 1.42; P = .12), with no heterogeneity among trials (P = .47). There was no prognostic effect in OBS patients (Fig 1B; HR = 1.04; 95% CI, 0.78 to 1.38; P = .79) or patients with adenocarcinoma (Fig 1C; HR = 1.00; 95% CI, 0.78 to 1.29; P = .97). Trends toward worse outcome were seen for MUT-KRAS in patients with nonadenocarcinoma (squamous HR = 1.41 [95% CI, 0.89 to 2.23]; other nonadenocarcinoma NSCLC HR = 1.86 [95% CI, 1.22 to 2.82], Data Supplement).
Fig 1.
Overall survival (A, B, C) and disease-free survival (D, E, F) for patients with KRAS wild-type tumors compared with patients with KRAS mutated tumors. (A) All patients; (B) patients in observation arm; (C) all patients with adenocarcinoma; (D) all patients; (E) patients in observation arm; (F) all patients with adenocarcinoma. HR, hazard ratio.
For DFS (Figs 1D, 1E, and 1F), similar results were found for all patients (HR = 1.15; 95% CI, 0.96 to 1.39; P = .14), OBS (HR = 1.05; 95% CI, 0.80 to 1.36; P = .73), and adenocarcinoma (HR = 0.98; 95% CI, 0.78 to 1.24; P = .87).
Because of potential treatment interactions, prognostic analyses for KRAS subtypes were performed in OBS patients alone. There was no significant prognostic effect on OS (Table 2) for codon 12 (HR v WT = 1.04; 95% CI, 0.77 to 1.40) or 13 mutations (HR v WT = 1.01; 95% CI, 0.47 to 2.17; P = .96). There was no significant difference in prognosis for codon 12 subgroups (Table 2) for OS (P = .99) or DFS (P = .98).
Table 2.
Summary of the Prognostic and Predictive Effects of Codon 12 and 13 KRAS Mutations and of Codon 12 Amino Acid Substitutions
| Variable | Survival Measure | Effect | KRAS | Comparison | HR | 95% CI | P |
|---|---|---|---|---|---|---|---|
| Codon-specific KRAS mutation | OS | Prognostic* | .96 | ||||
| Codon12 mutation v WT | 1.04 | 0.77 to 1.40 | .79 | ||||
| Codon13 mutation v WT | 1.01 | 0.47 to 2.17 | .98 | ||||
| Predictive | WT | ACT v OBS | 0.89 | 0.76 to 1.04 | .15 | ||
| Codon12 mutation | ACT v OBS | 0.95 | 0.67 to 1.35 | .77 | |||
| Codon13 mutation | ACT v OBS | 5.78 | 2.06 to 16.22 | < .001 | |||
| Interaction test | .002 | ||||||
| DFS | Prognostic* | .93 | |||||
| Codon12 mutation v WT | 1.05 | 0.80 to 1.39 | .71 | ||||
| Codon13 mutation v WT | 1.00 | 0.49 to 2.04 | 1.0 | ||||
| Predictive | WT | ACT v OBS | 0.86 | 0.73 to 1.00 | .04 | ||
| Codon12 mutation | ACT v OBS | 0.86 | 0.62 to 1.19 | .36 | |||
| Codon13 mutation | ACT v OBS | 3.88 | 1.44 to 10.46 | .007 | |||
| Interaction test | .01 | ||||||
| Specific amino acid† substitutions on KRAS codon 12 | OS | Prognostic* | .99 | ||||
| G12C or G12V v WT | 1.04 | 0.74 to 1.46 | .81 | ||||
| G12D or G12S v WT | 0.95 | 0.50 to 1.81 | .87 | ||||
| G12A or G12R v WT | 1.08 | 0.49 to 2.37 | .86 | ||||
| Predictive | WT | ACT v OBS | 0.89 | 0.76 to 1.05 | .16 | ||
| G12C or G12V | ACT v OBS | 0.94 | 0.63 to 1.41 | .77 | |||
| G12D or G12S | ACT v OBS | 1.39 | 0.55 to 3.54 | .49 | |||
| G12A or G12R | ACT v OBS | 0.66 | 0.21 to 2.10 | .48 | |||
| Interaction test | .76 | ||||||
| DFS | Prognostic* | .98 | |||||
| G12C or G12V v WT | 1.04 | 0.76 to 1.42 | .82 | ||||
| G12D or G12S v WT | 1.03 | 0.57 to 1.85 | .94 | ||||
| G12A or G12R v WT | 1.15 | 0.55 to 2.39 | .72 | ||||
| Predictive | WT | ACT v OBS | 0.86 | 0.74 to 1.00 | .04 | ||
| G12C or G12V | ACT v OBS | 0.89 | 0.61 to 1.30 | .55 | |||
| G12D or G12S | ACT v OBS | 1.11 | 1.46 to 2.64 | .82 | |||
| G12A or G12R | ACT v OBS | 0.48 | 0.15 to 1.47 | .20 | |||
| Interaction test | .70 |
Abbreviations: ACT, adjuvant chemotherapy; DFS, disease-free survival; HR, hazard ratio; OBS, observation; OS, overall survival; WT, wild type.
In OBS arm only.
Amino acids are designated as follows: A, alanine; C, cystine; D, aspartic acid; G, glycine; R, arginine; S, serine; V, valine.
Predictive Effect of KRAS Mutation Status
The HR for death comparing ACT with OBS for patients with WT-KRAS tumors (Fig 2A) was 0.89 (95% CI, 0.76 to 1.05; P = .15) and 1.05 (95% CI, 0.76 to 1.46; P = .77; interaction P = .37) for patients with MUT-KRAS (Fig 2C). Results were not significantly different among trials P = .52 (Data Supplement). Results were similar for DFS (WT-KRAS HR = 0.86, 95% CI, 0.74 to 1.00, P = .04; MUT-KRAS HR = 0.93, 95% CI, 0.68 to 1.27, P = .65, interaction P = .63) and in patients with adenocarcinoma (interaction P = .86; Figs 2B and 2D, Data Supplement).
Fig 2.
Overall survival benefit from adjuvant chemotherapy (ACT) compared with observation (OBS) in (A) patients with KRAS wild-type tumors and (C) patients with KRAS-mutated tumors, for patients with (B) adenocarcinoma and KRAS wild-type tumors and (D) adenocarcinoma with KRAS-mutated tumors, and for patients with (E) KRAS codon 12 mutations and (F) codon 13 mutations. (A) KRAS wild type; (B) adenocarcinoma KRAS wild type; (C) KRAS mutated; (D) adenocarcinoma KRAS mutated; (E) KRAS codon 12 mutated; (F) KRAS codon 13 mutated. HR, hazard ratio.
The predictive effects of codon 12 or 13 mutations are summarized in Table 2 (Figs 2E and 2F). Patients with tumors harboring codon 12 mutations seemed to derive no benefit from ACT (HR = 0.95; 95% CI, 0.67 to 1.35; P = .77). In contrast, the presence of codon 13 mutations was associated with significantly worse OS with ACT (HR = 5.78; 95% CI, 2.06 to 16.22; P < .001; interaction P = .002). Results were similar for DFS.
The predictive effects of specific codon 12 amino acid substitutions are summarized in Table 2. The effect of ACT was variable among patients with codon-12 mutations: G12A or G12R OS HR = 0.66 (95% CI, 0.21 to 2.10; P = .48), G12C or G12A HR = 0.94 (95% CI, 0.63 to 1.41; P = .77) and G12D or G12S HR = 1.39 (95% CI, 0.55 to 3.54; P = .49); the differences were not significant (comparison of four HRs, including WT, P = .76). Results were similar for DFS (interaction P = .70).
Second Primary Cancers
Second primary cancers were identified in 86 (6%) of 1,543 patients (not reported in ANITA). The analysis was therefore performed in 1,433 patients from the other trials. In OBS patients, the risk of developing a second primary was higher for patients with KRAS-mutated primary tumors (HR = 2.76; 95% CI, 1.34 to 5.70; P = .005; Table 3; Fig 3). In contrast, patients with KRAS mutations receiving ACT seemed to have a lower risk of second primaries (HR = 0.66; 95% CI, 0.25 to 1.75; P = .40; interaction P = .02).
Table 3.
Risk of Developing a Second Primary Malignancy in Patients With KRAS Mutated and KRAS Wild-Type Tumors in CALGB-9633, JBR.10, and IALT Trials
| ACT Arm |
OBS Arm |
ACT v OBS |
|||||
|---|---|---|---|---|---|---|---|
| No. of Second Primaries | No. of Patients | No. of Second Primaries | No. of Patients | HR | 95% CI | P | |
| KRAS wild type, n = 1,146 | 38 | 581 | 25 | 565 | 1.34 | 0.81 to 2.22 | .26 |
| KRAS mutated, n = 276 | 5 | 143 | 13 | 133 | 0.32 | 0.11 to 0.91 | .03 |
| Mutated v wild type | |||||||
| HR | 0.66 | 2.76 | 0.24* | 0.08 to 0.76 | |||
| 95% CI | 0.25 to 1.75 | 1.34 to 5.70 | |||||
| P | .40 | .005 | |||||
NOTE. Eleven patients were excluded because of missing covariates. HR of interaction KRAS*treatment P = .02.
Abbreviations: ACT, adjuvant chemotherapy; CALGB, Cancer and Leukemia Group B; HR, hazard ratio; IALT, International Adjuvant Lung Cancer Trial; OBS, observation.
Fig 3.
The risk of developing a second primary malignancy in observation patients with KRAS-mutated tumors. HR, hazard ratio.
DISCUSSION
More than 20 years have passed since the first reports that KRAS mutation was associated with poorer outcome in patients with lung adenocarcinoma.2,4 Since then, most but not all studies have confirmed a prognostic effect.6,17–20 However, there has been considerable heterogeneity among studies with respect to tumor type (some included only adenocarcinoma), stage, and treatment. Furthermore, many investigators reported only univariate comparisons without correcting for other prognostic variables.6
With more than 1,500 patients (300 with mutations), our study is the largest to report on the prognostic effect of KRAS. Furthermore, our study examines a homogeneous group of surgically staged patients with complete follow-up of more than 5 years duration. We could demonstrate only modest prognostic effects of KRAS mutation that were not significant in either univariate or multivariate adjusted analyses. Furthermore, when examining the adenocarcinoma subset (accounting for almost 70% of mutations), there was absolutely no prognostic effect.
In NSCLC, most KRAS mutations occur at codon 12, with many consisting of G>T transversions, the preferential mutation type induced by tobacco.20 Our results agree, with more than 90% of mutations on codon 12, including mainly G>T transversions. In contrast, codon 13 is not known as a preferential site for tobacco carcinogen mutagenesis, and so KRAS codon 13 mutations might be expected to be more prevalent in cancers of nonsmokers. However, in our small nonsmoking subgroup, 10 of 11 mutations were on codon 12 and only one on codon 13. Riely et al20 reported similar results with 10 of 12 mutations in nonsmokers being G12D mutations, with no codon13 mutations. They also reported that mutations in nonsmokers usually were transition G>A mutations rather than G>T or G>C transversions. G>A mutations were seen in only four of our nonsmoking population, with the rest being G>T or G> C.
Although codon 12 and 13 mutations differ in their structure, cellular, and molecular effects, they also have been shown to have quite different oncogenic effects, as reported by Prior et al.3 In our study, the first to assess the potential for a differential prognostic effect in NSCLC, we could identify no difference in prognosis based on codon. In colorectal cancer, Abubaker et al21 reported significantly poorer survival (P = .0009) in patients with KRAS mutations, with codon 12 mutation associated with the worst 5-year survival (64.4%; P = .0025) compared with codon 13 (75.8%) or WT-KRAS (78.2%). Among 195 patients with advanced colorectal cancer with KRAS results treated on the OBS arm of National Cancer Institute of Canada Clinical Trials Group CO.17, which randomly assigned patients to single-agent cetuximab or supportive care alone, 82 (42%) had mutations (13 G13D, 69 other). In contrast to the results of Abubaker et al,21 patients with G13D mutations had significantly shorter survival compared with those with other KRAS mutations.22 However, on multivariable analysis, the differences were not significant. These colorectal cancer studies with conflicting results emphasize the potential pitfalls of drawing conclusions from small sample sets (only 28 codon 13 mutations from the two studies and 24 in our study). Furthermore, although KRAS mutations in lung and colorectal cancer are seen predominately on codons 12 and 13, the amino acid substitutions vary considerably between the two cancers, and so comparisons of prognostic effects across cancer types may not be appropriate.
This study is the first to examine the prognostic effect of different KRAS codon 12 amino acid substitutions in lung cancer. As expected, the majority (81%) were smoking-related G12C and G12V mutations. We could identify no prognostic effect for any mutation subtype. In colorectal cancer, the international RASCAL-II (Kirsten Ras in Colorectal Cancer Collaborative Group) study examined the effect of KRAS codon 12 and 13 mutations in 4,268 patients.23 Only G12V mutations (in 8.6% of patients), showed a significant prognostic effect on failure-free survival (HR = 1.3; P = .004) and OS (HR = 2.9; P = .008).
With respect to the predictive effects of KRAS, patients with WT tumors demonstrated a trend to greater survival benefit from ACT as compared with those with mutations. However, even in this relatively well-powered study, the test for interaction was not significant. Furthermore, in the adenocarcinoma subset, modest but nonsignificant effects in favor of ACT were seen in patients with both mutated and WT tumors.
Our observations concerning the predictive effects of codon 12 and 13 mutations are intriguing. Despite a total lack of prognostic effect, we found that, even after accounting for other prognostic variables, patients with codon 13 mutations seemed to derive significant harm from ACT (HR = 5.78; P < .001; interaction P = .002). Only 24 patients had codon 13 mutations, and so our results, although statistically significant, must be interpreted with caution and require validation. Although in vitro and clinical studies suggest that KRAS predicts for poorer response to chemotherapy and radiation,7–9 we could identify no other reports of predictive studies for chemotherapy based on KRAS codon in NSCLC. Tejpar et al24 reported shorter survival in patients treated with chemotherapy alone for colorectal cancer if their tumors had G13D mutations compared with WT-KRAS or other MUT-KRAS. This study did not examine untreated patients, and so the predictive effect of mutation subtype for chemotherapy could not be confirmed; however, the poorer outcome associated with chemotherapy and G13D mutation may support our observations.
The predictive effect of KRAS for response to epidermal growth factor receptor (EGFR) inhibitors has been studied more extensively, with several reports suggesting poorer outcomes in response to tyrosine kinase inhibitors (TKIs) in NSCLC.25–31 Most studies did not differentiate between codon 12 and 13 mutations. Recently, however, Metro et al32 reported significantly shorter progression-free survival and OS in response to EGFR TKIs for patients with NSCLC whose tumors had KRAS codon 13 mutations compared with those with codon 12 mutations, a result that is similar to our observation with chemotherapy.
In colorectal cancer,22,24,33,34 EGFR monoclonal antibody therapy is restricted to patients with WT- KRAS tumors. Recently however, some but not all studies suggest that patients with G13D-mutated tumors may respond to EGFR monoclonal antibodies.22,24,33 In colorectal cancer, the situation also may be complicated further by mutations in other genes, such as BRAF, which may have a significant effect on pathways downstream from RAS.32,34 BRAF mutations are rare in NSCLC, and the interaction of BRAF or other potential markers and KRAS has not been studied. In contrast, KRAS was not shown to be a predictor of a differential outcome in patients with NSCLC treated with chemotherapy and cetuximab.35,36
Our results concerning codon 12 amino acid substitutions are inconclusive, with HRs varying from 0.66 to 1.39. The lack of statistical significance may, in part, be due to low numbers in some of the subsets. Clearly, a large international collaboration (akin to RASCAL-II) would be required to provide sufficient numbers for statistical power. This may not be possible, though, because prospective ACT studies in NSCLC will no longer have a no-treatment control arm, and many of the older trials did not collect samples for correlative studies.
There is evidence that structural differences between various codon 12 and 13 mutations may affect GTPase activity and binding of effector proteins.37,38 Certainly, different amino acid substitutions have been shown in lung and other cancers to be associated with greater or lesser carcinogenic potential and downstream signaling effects,3 and so the nonsignificant differences we observed potentially can be explained. Recently Ihle et al36 reported that patients whose tumors had G12C or G12V mutations had worse survival than patients with other MUT-KRAS or WT-KRAS. Their in vitro NSCLC cell line studies showed that mutant KRAS G12A cell lines had activated phosphatidyl-inositol 3-kinase and mitogen-activated protein/extracellular signal-regulated kinase signaling. In contrast, cell lines with mutant KRAS G12C or G12V had activated Ral signaling and decreased growth factor-dependent Akt activation. These different downstream effects may result in a differential response to therapy.
Second primary cancers have been reported in approximately 15% of patients with NSCLC who undergo curative resection.39,40 The most frequent malignancy is a second lung cancer, with two-fold higher risk in patients who continue to smoke. Other smoking-related cancers also are seen frequently, including head and neck cancers and malignancies of the esophagus, stomach, and bladder.39 Because smoking is so strongly associated with KRAS mutation in NSCLC, and because of the field effect of this carcinogen in the upper aero-digestive tract, we postulated that KRAS mutation, an early event in lung carcinogenesis, might correlate with the development of second primaries. Indeed, in our observation cohort, the risk of developing a second primary cancer was almost three-fold greater in patients with KRAS-mutant tumors. To our knowledge, this is the first report of such an association, and our observation may have implications for follow-up in this high-risk group. Our finding of a reduced risk for second cancers in the ACT group was unexpected and unexplained. We theorize that ACT may have had a therapeutic effect on subclinical deposits of KRAS mutant cells in lung or other organs, and that these cells with KRAS-activated growth pathways may have been sensitive to the effects of and, potentially, were even eradicated by the ACT. Our observation requires validation in other NSCLC cohorts.
In summary, KRAS is not a significant prognostic marker in patients with resected NSCLC. Overall, it is not significantly predictive of a differential benefit from ACT, particularly in patients with adenocarcinoma, and at this time, it cannot be recommended as a tool to select patients for ACT. Although our results suggest a potentially detrimental effect from chemotherapy in patients with codon 13 mutations, validation studies will be critical. However, large international collaborations will be necessary to confirm these results and also to explore the potential for a differential treatment effect in patients with various amino acid substitutions. Postoperative follow-up of patients with lung cancer is variable, with no clear guidelines, and only low level evidence concerning the use of computed tomography or positron emission tomography scans. KRAS mutation status may identify a cohort of patients who, in the absence of ACT, may be at particularly high risk of developing second primary cancers and who potentially might benefit from more intense follow-up.
Supplementary Material
Acknowledgment
We thank Ni Liu (Princess Margaret Hospital) for technical assistance in sample analysis.
Footnotes
Written on behalf of the LACE-Bio Collaborative Group.
Supported by la Ligue Nationale Contre le Cancer (France), le Programme National d'Excellence Spécialisé cancer du poumon de l'Institut National du Cancer (INCa) (France), the National Cancer Institute (United States), the Canadian Cancer Society Research Institute (Canada), and an unrestricted grant from Sanofi.
P.H., J.-C.S., and M.-S.T. are co-senior scientists for this work.
Presented in part at the European Society for Medical Oncology Annual Meeting, September 29, 2012, Milano, Italy, and at the 48th Annual Meeting of the American Society of Clinical Oncology, June 4, 2012, Chicago, IL.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Frances A. Shepherd, Pierre Hainaut, Jean-Pierre Pignon, Jean-Yves Douillard, Elizabeth Brambilla, Thierry Le Chevalier, Lesley Seymour, Robert Pirker, Jean-Charles Soria, Ming-Sound Tsao
Financial support: Jean-Pierre Pignon
Administrative support: Caroline Domerg, Jean-Pierre Pignon, Lesley Seymour, Abderrahmane Bourredjem, Gwénaël Le Teuff
Provision of study materials or patients: Frances A. Shepherd, Pasi A. Jänne, Stephen Graziano, Jean-Yves Douillard, Elizabeth Brambilla, Thierry Le Chevalier, Robert Pirker, Martin Filipits, Rafael Rosell, Robert Kratzke, Bizhan Bandarchi, Xiaoli Ma, Marzia Capelletti, Jean-Charles Soria, Ming-Sound Tsao
Collection and assembly of data: Frances A. Shepherd, Pierre Hainaut, Pasi A. Jänne, Jean-Pierre Pignon, Stephen Graziano, Elizabeth Brambilla, Thierry Le Chevalier, Lesley Seymour, Gwénaël Le Teuff, Martin Filipits, Rafael Rosell, Robert Kratzke, Bizhan Bandarchi, Xiaoli Ma, Marzia Capelletti, Jean-Charles Soria, Ming-Sound Tsao
Data analysis and interpretation: Frances A. Shepherd, Caroline Domerg, Pierre Hainaut, Jean-Pierre Pignon, Stephen Graziano, Jean-Yves Douillard, Elizabeth Brambilla, Thierry Le Chevalier, Lesley Seymour, Abderrahmane Bourredjem, Gwénaël Le Teuff, Robert Pirker, Jean-Charles Soria, Ming-Sound Tsao
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Harvey JJ. An unidentified virus which causes the rapid production of tumours in mice. Nature. 1964;204:1104–1105. doi: 10.1038/2041104b0. [DOI] [PubMed] [Google Scholar]
- 2.Rodenhuis S, van De Wetering ML, Mooi WJ, et al. Mutational activation of the K-ras oncogene: A possible pathogenic factor in adenocarcinoma of the lung. N Engl J Med. 1987;317:929–935. doi: 10.1056/NEJM198710083171504. [DOI] [PubMed] [Google Scholar]
- 3.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323:561–565. doi: 10.1056/NEJM199008303230902. [DOI] [PubMed] [Google Scholar]
- 5.Huncharek M, Muscat J, Geschwind JF. K-ras oncogene mutation as a prognostic marker in non-small cell lung cancer: A combined analysis of 881 cases. Carcinogenesis. 1999;20:1507–1510. doi: 10.1093/carcin/20.8.1507. [DOI] [PubMed] [Google Scholar]
- 6.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: A systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sklar MD. The ras oncogenes increase the intrinsic resistance of NIH 3T3 cells to ionizing radiation. Science. 1988;239:645–647. doi: 10.1126/science.3277276. [DOI] [PubMed] [Google Scholar]
- 8.Sklar MD. Increased resistance to cis-diammine-dichloroplatinum(II) in NIH 3T3 cells transformed by ras oncogenes. Cancer Res. 1998;48:793–797. [PubMed] [Google Scholar]
- 9.Rodenhuis S, Boerrigter L, Top B, et al. Mutational activation of the K-ras oncogene and the effect of chemotherapy in advanced adenocarcinoma of the lung: A prospective study. J Clin Oncol. 1997;15:285–291. doi: 10.1200/JCO.1997.15.1.285. [DOI] [PubMed] [Google Scholar]
- 10.Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. Observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 11.Butts CA, Ding K, Seymour L, et al. Randomized phase III trial of vinorelbine plus cisplatin compared with observation in completely resected stage IB and II non-small cell lung cancer: Updated survival analysis of JBR-10. J Clin Oncol. 2009;28:29–34. doi: 10.1200/JCO.2009.24.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arriagada R, Bergman B, Dunant A, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 13.Arriagada R, Dunant A, Pignon JP, et al. Long-term results of the international adjuvant lung trial evaluating adjuvant cisplatin-based adjuvant chemotherapy in resected lung cancer. J Clin Oncol. 2010;28:35–42. doi: 10.1200/JCO.2009.23.2272. [DOI] [PubMed] [Google Scholar]
- 14.Douillard JY, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): A randomised controlled trial. Lancet Oncol. 2006;7:719–727. doi: 10.1016/S1470-2045(06)70804-X. [DOI] [PubMed] [Google Scholar]
- 15.Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: A pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–3559. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 16.Strauss GM, Herndon JE, 2nd, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol. 2008;26:5043–5051. doi: 10.1200/JCO.2008.16.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodenhuis S, Slebos RJ. The ras oncogenes in human lung cancer. Am Rev Respir Dis. 1990;142:S27–S30. doi: 10.1164/ajrccm/142.6_Pt_2.S27. [DOI] [PubMed] [Google Scholar]
- 18.Rodenhuis S, Slebos RJ, Boot AJ, et al. Incidence and possible clinical significance of K-ras oncogene activation in adenocarcinoma of the human lung. Cancer Res. 1998;48:5738–5741. [PubMed] [Google Scholar]
- 19.Feng Z, Hu W, Chen JX, et al. Preferential DNA damage and poor repair determine ras gene mutational hotspot in human cancer. J Natl Cancer Inst. 2002;94:1527–1536. doi: 10.1093/jnci/94.20.1527. [DOI] [PubMed] [Google Scholar]
- 20.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abubaker J, Bavi P, Al-Haqawi W, et al. Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal cancer. J Pathol. 2009;219:435–445. doi: 10.1002/path.2625. [DOI] [PubMed] [Google Scholar]
- 22.De Roock W, Jonker DJ, Di Nicolantonio F, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2008;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 23.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: The ‘RASCAL II’ study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tejpar S, Celik I, Schlichting M, et al. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol. 2012;30:3570–3577. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 25.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 26.Zhu CQ, da Cunha Santos G, Ding K, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol. 2008;26:4268–4275. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 28.Roberts PJ, Stinchcombe TE, Der CJ, et al. Personalized medicine in non-small cell lung cancer: Is KRAS a useful biomarker in selecting patients for epidermal growth factor receptor-targeted therapy? J Clin Oncol. 2010;28:4769–4777. doi: 10.1200/JCO.2009.27.4365. [DOI] [PubMed] [Google Scholar]
- 29.Cadranel J, Mauguen A, Faller M, et al. Impact of systematic EGFR and KRAS mutation evaluation on progression-free survival and overall survival in patients with advanced non-small-cell lung cancer treated by erlotinib in a French prospective cohort (ERMETIC project–part 2) J Thorac Oncol. 2012;7:1490–1502. doi: 10.1097/JTO.0b013e318265b2b5. [DOI] [PubMed] [Google Scholar]
- 30.Califano R, Landi L, Cappuzzo F. Prognostic and predictive value of K-RAS mutations in non-small cell lung cancer. Drugs. 2012;19(suppl 1):28–36. doi: 10.2165/1163012-S0-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 31.Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: A systematic review and meta-analysis of studies in advanced non-small-cell lung cancer colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 32.Metro G, Duranti S, Chiari R, et al. KRAS mutational status and sensitivity to a reversible epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) in EGFR wild type (WT) advanced non-small cell lung cancer (NSCLC) patients (pts) J Clin Oncol. 2012:30. abstr e18023. [Google Scholar]
- 33.Rizzo S, Bronte G, Fanale D, et al. Prognostic vs predictive molecular biomarkers for colorectal cancer: Is KRAS and BRAF wild type status required for anti-EGFR therapy? Cancer Treat Rev. 2010;36(suppl 3):S56–S61. doi: 10.1016/S0305-7372(10)70021-9. [DOI] [PubMed] [Google Scholar]
- 34.Imamura Y, Morikawa T, Liao X, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorrectal cancers. Clin Cancer Res. 2012;18:4753–4763. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khambata-Ford S, Harbison CT, Hart LL, et al. Analysis of potential predictive markers of cetuximab benefit in BMS099, a phase III study of cetuximab and first-line taxane/carboplatin in advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:918–927. doi: 10.1200/JCO.2009.25.2890. [DOI] [PubMed] [Google Scholar]
- 36.O'Byrne KJ, Gatzemeier U, Bondarenko I, et al. Molecular biomarkers in non-small-cell lung cancer: A retrospective analysis of data from the phase 3 FLEX study. Lancet Oncol. 2011;12:795–805. doi: 10.1016/S1470-2045(11)70189-9. [DOI] [PubMed] [Google Scholar]
- 37.Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: Implications for signalling and clinical outcome. J Natl Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spoerner M, Hozsa C, Poetzl JA, et al. Conformational states of human rat sarcoma (Ras) protein complexed with its natural ligand GTP and their role for effector interaction and GTP hydrolysis. J Biol Chem. 2010;285:39768–39778. doi: 10.1074/jbc.M110.145235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice D, Kim HW, Sabichi A, et al. The risk of secondary primary tumors after resection of stage I nonsmall cell lung cancer. Ann Thorac Surg. 2003;76:1001–1008. doi: 10.1016/s0003-4975(03)00821-x. [DOI] [PubMed] [Google Scholar]
- 40.Bhaskaria A, Tang PC, Mashtare T, et al. Analysis of second primary lung cancers in the SEER database. J Surg Res. 2010;162:1–6. doi: 10.1016/j.jss.2009.12.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.