Abstract
Chromosomal translocations typically impair cell differentiation and often require secondary mutations for malignant transformation. However, the role of a primary translocation in the development of collaborating mutations is debatable. To delineate the role of leukemic translocation NUP98-HOXD13 (NHD13) in secondary mutagenesis, DNA break and repair mechanisms in stimulated mouse B lymphocytes expressing NHD13 were analyzed. Our results showed significantly reduced expression of non-homologous end joining (NHEJ)-mediated DNA repair genes, DNA Pkcs, DNA ligase4, and Xrcc4 leading to cell cycle arrest at G2/M phase. Our results showed that expression of NHD13 fusion gene resulted in impaired NHEJ-mediated DNA break repair.
Keywords: Chromosomal translocation, NUP98, HOXD13, DNA double strand break, Non-homologous end joining
1. Introduction
Chromosomal translocations (CT) are the hallmark features associated with many hematological malignancies [1]. The majority of CT are considered class II mutations resulting in impaired hematopoietic cell differentiation but require collaborating secondary mutations for complete malignant transformation [1–3]. Class II mutations target transcription factors and function predominately to impair hematopoietic differentiation and subsequently apoptosis [3,4]. However, the mechanisms by which the cell acquires secondary mutations are not completely understood. Increased DNA damage and/or impaired DNA repair mechanisms are widely accepted as mechanisms for genomic mutations [5,6]. An emerging concept is that CT might increase the chance for further DNA damage or that the fusion gene can somehow impair the DNA repair mechanisms [7–9]. Of the different types of DNA damage to occur, one of the most important types is the double strand breaks (DSB), which occurs in each dividing cell at an estimated rate of 10 breaks per day [10]. These breaks are typically repaired by non-homologous end joining (NHEJ) which is a tightly controlled process involving multiple factors [11]. Ironically, impaired NHEJ can lead to mutations including the formation of CT [12,13]. Delineating DNA break induction and repair pathways in cells with primary CT during leukemic progression will identify the potential mechanism by which CT induce secondary mutations. These mechanisms may be through direct DNA–DNA interactions, fusion protein–DNA interactions, or fusion protein–protein interactions. By doing so, identification of these mechanisms could reveal potential therapeutic targets.
Transgenic mice expressing the leukemic fusion gene NUP98-HOXD13 (NHD13) progress through myelodysplastic syndrome (MDS) and develop acute leukemia with the occurrence of collaborating mutations [14]. We have recently shown that expression of NHD13 in stimulated B-lymphocytes results in: (1) impaired B-cell differentiation at stages in the bone marrow that are dependent on VDJ recombination and (2) impaired class switch recombination (CSR) and antibody production [15]. During CSR naïve B lymphocytes undergo DNA breaks on immunoglobulin heavy chain gene and NHEJ-mediated break repair when stimulated with appropriate antigens and cofactors to produce different immunoglobulin isotypes [16]. DNA DSB results in the phosphorylation of histone H2AX by Ataxia telangiectasia mutated (ATM), which in turn results in the stabilization of break ends [17,18]. Proteins Ku70 and Ku80 bind to the break ends to form a heterodimer and recruits DNAP-Kcs [19]. DNAPKcs acts as a scaffold and DNA repair factors DNA ligase 4 and XRCC4 complex [20]. As CSR involves physiological DNA breaks and NHEJ mediated repair, expression of the NHD13 fusion in B lymphocytes is a suitable system for delineating the mechanism by which CT can induce DNA instability and result in secondary mutations. Based on these observations, we hypothesized that NHD13 enhances double strand break induction and impairs the DNA repair mechanism as an underlying reason for secondary mutations. Here we used CSR as a tool to induce DNA DSB repair in the presence of NHD13. Our results show that NHD13 fusion gene impairs expression of critical NHEJ repair factors, and impairs DNA repair mechanism resulting in cell cycle arrest.
2. Materials and methods
2.1. Animals
Five young (8–12 weeks) NHD13 mice and five wild type (WT) littermates on an FVB background were used for each experiment. Mice were bred and maintained at the AAALAC accredited animal facility at Virginia Maryland Regional College of Veterinary Medicine, Virginia Tech. All experiments were carried out as per NIH guidelines with prior approval from the Virginia Tech Institutional Animal Care and Use Committee.
2.2. In vitro class switch recombination
To induce in vitro class switch recombination, splenic B lymphocytes were harvested using anti-mouse IgM magnetic beads and magnetic assisted cell sorting (Milteny Biotec, Auburn, CA). A total of 2 × 105 cells were treated with 5 μM CFSE and cultured with media containing E. coli Lipopolysaccharide (LPS, Sigma–Aldrich, St. Louis, MO) (25 μg/ml) and IL-4 (PeproTech, Rocky Hill, NJ) (25 ng/ml). To verify that CSR had occurred, cells were harvested at 72 h, labeled with anti-mouse IgG1 and IgE antibodies and analyzed by flow cytometry as previously published [15].
2.3. DNA break analysis by confocal microscopy
Splenic B lymphocytes were harvested and cultured to induce CSR as described above. Cells were harvested at 0, 24, 48 and 72 h and cytospun onto charged slides. Cells were fixed using 4% paraformaldehyde (Thermo Scientific, Rockford, IL) and incubated with rabbit anti-mouse γH2AX (Cell Signaling, Danvers, MA) at 1:500 dilution in TBS + 1% FBS followed by labeling with Alexafluor-488 conjugated anti-rabbit antibody (Cell Signaling, Danvers, MA). After labeling with DAPI (Cell Signaling), cells were visualized using LSM700 Carl Zeiss confocal microscope (Carl Zeiss Microimaging, Thornwood, NY) and LSM900® software (Carl Zeiss).
2.4. DNA break analysis by flow cytometry
Splenic B lymphocytes cultured in the presence of LPS (Sigma–Aldrich) and IL-4 (Sigma–Aldrich) were harvested at 0, 24, 48 and 72 h. Cells were fixed with 70% ethanol and incubated with rabbit anti-mouse phosphoH2AX antibody (Cell Signaling) followed by incubation with Alexa-fluor 488 conjugated anti-rabbit antibody (Cell Signaling). Cells were analyzed using a FACScan flow cytometer (BD Biosciences).
2.5. Cell cycle analysis
B lymphocytes from NHD13 and WT mice were harvested and cultured in the presence of LPS and IL-4 as described above. Cells were harvested at 24 h intervals, fixed in 70% ethanol and labeled with propidium iodide (PI) (Sigma–Aldrich). Cells were analyzed by flow cytometry.
2.6. Gene expression analysis
Lymphocytes stimulated with LPS + IL-4 were harvested at 72 h of culture, total RNA was extracted and cDNA was synthesized. Gene expression was determined by reverse quantitative PCR using gene specific primers and Sybrgreen mastermix (Applied Biosystems). Gapdh was used as the internal control.
2.7. Data and statistical analysis
Flow cytometric data were analyzed using FlowJo software 7.6 (FlowJo, Ashland, OR). Confocal images were analyzed using Zen 2009® software (Carl Zeiss Microimaging). Data was analyzed with GraphPad Prism 5.0® (GraphPad Software, La Jolla, CA), using either two tailed t-test or ANOVA and Bonferroni post test; a p value < 0.05 was considered significant.
3. Results
3.1. B cells from NHD13 mice had reduced DNA double strand break repairs
Class switch recombination essentially involves DNA double strand breaks and NHEJ-mediated DNA recombination. To delineate the effect of NHD13 on the pattern of DNA DSB during CSR, we analyzed stimulated B cells from NHD13 and WT mice for DNA breaks using flow cytometry and confocal microscopy. Lymphocyte stimulation using E. coli lipopolysaccharide (LPS) and IL-4 results in expression of activation induced deaminase (Aid), which will initiate DNA DSB. These DNA DSB result in phosphorylation of histone H2AX (γH2AX), and can be employed as a marker for DNA DSB. Flow cytometric analysis for γH2AX revealed a comparable percentage of cells with DNA breaks in NHD13 and WT mice at 0, 24 and 48 h (Fig. 1A and B). However, at 72 h, NHD13 mice had increased DNA DSB as evidenced by flow cytometric analysis (Fig. 1A and C). We also analyzed the pattern of breaks at the single cell level using confocal microscopy (Fig. 1B). The DNA repair efficiency was estimated and expressed as the percentage reduction in break positive cells between 48 and 72 h. Our results showed a significantly lower DNA repair efficiency in NHD13 B cells (Fig. 1D). These results indicate that NHD13 B cells have comparable DNA double strand break induction following stimulation, but have reduced efficiency for break repair during physiological class switch recombination in B cells.
Fig. 1.

B cells from NHD13 mice have impaired DNA double strand break repair following stimulation. LPS and IL-4 stimulated splenic B cells from NHD13 and WT mice were harvested at 24 h intervals and labeled for γH2AX, a marker for DNA double strand breaks. (A) Representative flow cytometric pattern of DNA double strand breaks at 24 h intervals. (B) Confocal analysis of DNA double strand breaks showing DNA breaks in NHD13 and WT B-cells at 72 h. (C) Analysis of the percentage of DNA breaks using flow data showed significantly high levels in NHD13 B-cells at 72 h following stimulation. (D) DNA repair efficiency calculated based on the percentage of breaks repaired between 48 and 72 h showed significantly lower repair efficiency in NHD13 B-cells. n = 5, p < 0.05.
3.2. NHD13 B cells had a G2/M cell cycle arrest following stimulation
To better understand the impact of NHD13 on DNA repair, we determined the cell cycle kinetics of stimulated B cells from NHD13 mice at 24 h intervals. B cells were stimulated with LPS and IL-4, harvested at 24 h intervals and labeled with PI for flow cytometric analysis. Our results showed a comparable cell cycle pattern in NHD13 and WT cells at 0, 24 and 48 h following stimulation as indicated by a comparable percentage of apoptotic cells and cells in G1, S and G2/M phases (Fig. 2A and B). However, at 72 h of stimulation, NHD13 mice had a significantly higher percentage of cells in the G2/M stage (Fig. 2A and B) indicating cell cycle arrest at this stage. Cells in other stages, apoptotic, G1 and S phase were altered in NHD13 mice at 72 h, but not significant (Fig. 2B). We assayed expression of the cell cycle regulator gene p53 and found that its expression was higher at 72 h in NHD13 B cells (not statistically significant, Fig. 2C). Our results suggest that the NHD13 fusion gene impairs the DNA break repair mechanism and results in a G2/M check point arrest during the cell cycle in stimulated B-cells.
Fig. 2.
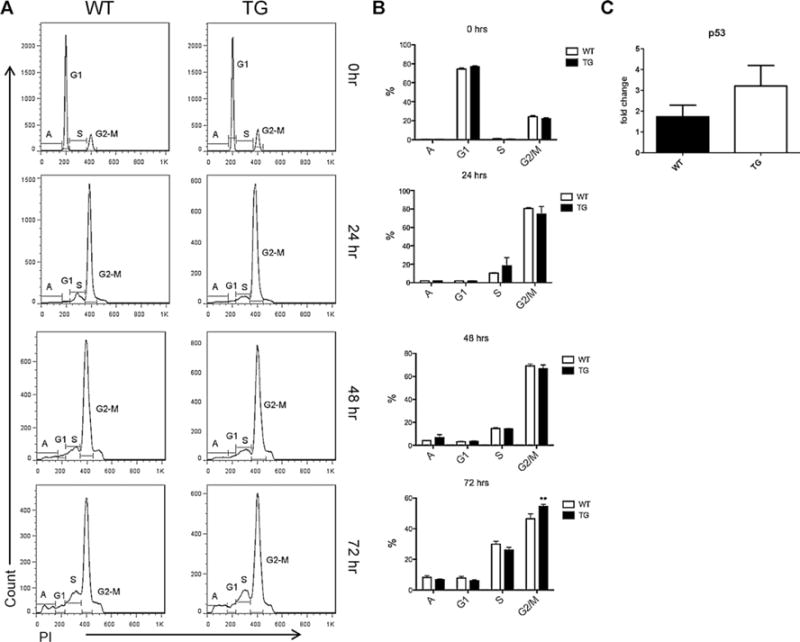
Cell cycle is arrested at G2/M phase in B-cells from NHD13 mice following stimulation. Isolated splenic B cells were stimulated with LPS and IL-4 to induce CSR. Cells were harvested at 24 h intervals and stained with propidium iodide to analyze cell cycle pattern. (A) Representative cell cycle plots from NHD13 and WT cells at 24 h intervals, 0 indicating prior to stimulation. (B) Statistical analysis of different phases of cell cycle pattern showed comparable kinetics at 0, 24 and 48 h. However, at 72 h, cells in G2 phase were significantly high in NHD13 mice suggesting a cell cycle arrest. (C) Expression analysis of p53 showed elevated levels in NHD13 lymphocytes 72 h following stimulation. n = 5, **p < 0.01.
3.3. DNA ligase 4, XRCC4, and DNAPKcs were downregulated in NHD13 B lymphocytes
Considering the essential role of NHEJ during DNA double strand repair, we hypothesized that NHD13 fusion gene might interfere with the NHEJ mechanism. To test this assumption, we harvested RNA from stimulated B cells at 72 h of culture and assayed the gene expression of critical genes involved in NHEJ mechanism. Our results showed comparable levels of Ku70 and Ku80 in cells from both NHD13 and WT mice (Fig. 3A). However, DNA ligase 4, Xrcc4, and DNAPKcs were significantly lower in NHD13 B lymphocytes. We also considered the possibility that Alternative End Joining (AEJ) could play a role during the DNA repair process. We evaluated expression of possible AEJ factors including Ligase 1 and Ligase 3α (Fig. 3B) and found that there was no statistical significance between transgenic and wild type cells. Our results indicate that expression of NHD13 fusion gene results in reduced expression of DNA ligase 4, Xrcc4, and DNAPKcs, leading to reduced DNA repair.
Fig. 3.

DNA ligase 4 and DNAPKcs expression is downregulated in NHD13 B cells following stimulation. LPS and IL-4 stimulated splenic B cells were harvested at 72 h following stimulation and, RNA was harvested for RQPCR. (A) Expression analysis of critical NHEJ genesf showed that Ku70, and Ku80 were comparable between NHD13 and WT B cells. However, DNA ligase 4, Xrcc4 and DNAPKcs were significantly low in NHD13 mice following stimulation. (B) Expression analysis of AEJ factors Ligase 1 and Ligase 3 showed comparable levels in both WT and NHD13 TG B lymphocytes. n = 5, p < 0.05.
4. Discussion
Chromosomal translocations are hallmark features of hematopoietic malignancies, which generally require collaborating mutations for malignant transformation [4]. Recent studies have proposed that CT can induce secondary mutations before malignant transformation [21,22]. A wide variety of factors can cause DNA damage, promote misrepairs and lead to the development of mutations [23]. Of these etiologies, DNA DSB [24] are frequent in mammalian cells as a result of physiological and pathological factors including reactive oxygen species, ionizing radiations or chemicals [25,26], and are repaired mainly by NHEJ [5]. Effective repair is essential for the maintenance of genomic stability and cell viability, whereas defective break repair results in cell cycle arrest, apoptosis, mutagenesis and malignant transformation [7,27]. Here, we analyzed the effects of a primary leukemic fusion gene, NHD13, on the DNA DSB repair pathways. Previous studies in transgenic mice expressing NHD13 have shown that leukemic progression is accompanied by random mutations in a wide variety of collaborating genes [14]. Taken together, it is reasonable to believe that the presence of a primary leukemic fusion gene can induce DNA instability by impairing the NHEJ pathway and lead to the formation of secondary mutations.
Analysis of lymphocyte development and gene recombination pattern in NHD13 mice suggests that the VDJ recombination mechanism is perturbed in NHD13 lymphocytes [28]. Furthermore our previous studies have shown that B lymphocyte differentiation and CSR are altered in NHD13 mice, with partial blocks occurring during gene recombination events [15]. Based on these findings, we used CSR as a tool to determine the role of NHD13 to induce DNA breaks and thereby analyze the NHEJ-mediated break repair pattern. As CSR involves DNA double strand break induction and NHEJ-mediated repair mechanisms, delineating the underlying molecular pathways of impaired CSR can have wider applications [29]. Our current study showed a comparable percentage of cells with DNA breaks prior to stimulation and at 24 and 48 h following stimulation (Fig. 1A and B). However, the percentage of cells with DNA breaks was significantly higher at 72 h of culture, suggesting an impaired DNA repair mechanism. Single cell analysis for DNA breaks using confocal microscopy showed a comparable amount of breaks in both WT and TG B cells, suggesting that the breaks are induced in a similar pattern. DNA break repair efficiency calculated based on the percentage of repair in a 24 h time frame (Puthiyaveetil et al., 2013, Mol Immunol, in press) showed a significantly lower repair in stimulated cells from NHD13 mice between 48 and 72 h (Fig. 1D), suggesting that DNA repair is impaired in NHD13 mice rather than break induction. Analysis of cell cycle pattern showed a G2/M stage cell cycle arrest in NHD13 B lymphocytes at 72 h of stimulation. Consistent with previous studies that have shown DNA repair defects will result in cell cycle arrest at G2/M checkpoint [30,31], our results also indicate that cell cycle arrest can be mediated by NHD13 leading to, or as a consequence of, defective or inefficient DNA repair.
Our analysis of expression of DNA repair factors showed comparable levels of Ku70, Ku80 and, but significantly lower levels of DNA Ligase 4, Xrcc4, and DNAPKcs at 72 h following stimulation. Reduced expression of repair factors has been shown to impair classical NHEJ pathway, leading to more error prone repair mechanisms, increasing the chance for secondary mutations [9]. During NHEJ-mediated DNA repair, Ligase 4 expression is often associated with increased AEJ activity. However, in our model, the C-NHEJ factors DNA Ligase 4, Xrcc4, and DNAPKcs are down regulated without any substantial increase in expression of AEJ factors, i.e., Ligase 1 and Ligase 3α. Considering the phosphorylation of H2AX, an early break detection process, is intact but that NHEJ factors are not up-regulated, we propose that NHD13 impairs DNA break repair induction. In conclusion, our results showed that the myeloid leukemic fusion gene NHD13 suppressed the expression of DNA Ligase 4, Xrcc4, and DNAPKcs, DNA double strand break accumulation and cell cycle arrest as probable underlying mechanisms for secondary mutations. Additional studies are warranted to more specifically determine the molecular mechanisms by which expression of NHD13 regulates the activity of DNA Ligase 4, XRCC4, and DNAPKcs. Doing so could reveal novel pathways or therapeutic targets useful in the treatment of leukemia and other types of cancer.
Acknowledgments
We thank Dr. Peter Aplan for providing the NHD13 mice. We thank our current lab members, Benjamin Okyere and Bettina Heid for comments and suggestions. We also thank the animal facility staff for animal management and care and Melissa Makris for flow cytometry. The study was funded by an internal competition grant from Virginia Maryland Regional College of Veterinary Medicine, Virginia Tech, USA.
Footnotes
Conflict of interest statement
The authors declare that they have no competing interests.
Contributors: AG designed and performed experiments and prepared the first draft. CR and TP designed experiments and helped in experiments and manuscripts preparation. DC conceived and designed the study and prepared the final draft of manuscript.
References
- 1.Caudell D, Aplan PD. Leukemia. Vol. 22. Official Journal of the Leukemia Society of America, Leukemia Research Fund; UK: 2008. The role of CALM-AF10 gene fusion in acute leukemia; pp. 678–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caudell D, Harper DP, Novak RL, Pierce RM, Slape C, Wolff L, et al. Retroviral insertional mutagenesis identifies Zeb2 activation as a novel leukemogenic collaborating event in CALM-AF10 transgenic mice. Blood. 2010;115:1194–203. doi: 10.1182/blood-2009-04-216184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilliland DG, Tallman MS. Focus on acute leukemias. Cancer Cell. 2002;1:417–20. doi: 10.1016/s1535-6108(02)00081-8. [DOI] [PubMed] [Google Scholar]
- 4.Gilliland DG. Molecular genetics of human leukemias: new insights into therapy. Semin Hematol. 2002;39:6–11. doi: 10.1053/shem.2002.36921. [DOI] [PubMed] [Google Scholar]
- 5.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–60. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sallmyr A, Fan J, Rassool FV. Genomic instability in myeloid malignancies: increased reactive oxygen species (ROS), DNA double strand breaks (DSBs) and error-prone repair. Cancer Lett. 2008;270:1–9. doi: 10.1016/j.canlet.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 8.Skorski T. Genomic instability: the cause and effect of BCR/ABL tyrosine kinase. Curr Hematol Malig Rep. 2007;2:69–74. doi: 10.1007/s11899-007-0010-6. [DOI] [PubMed] [Google Scholar]
- 9.Fan J, Li L, Small D, Rassool F. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: implications for genomic instability and therapy. Blood. 2010;116:5298–305. doi: 10.1182/blood-2010-03-272591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–20. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 11.Kenter AL. Class switch recombination: an emerging mechanism. Curr Top Microbiol Immunol. 2005;290:171–99. doi: 10.1007/3-540-26363-2_8. [DOI] [PubMed] [Google Scholar]
- 12.Lieber MR, Gu J, Lu H, Shimazaki N, Tsai AG. Nonhomologous DNA end joining (NHEJ) and chromosomal translocations in humans. Subcell Biochem. 2010;50:279–96. doi: 10.1007/978-90-481-3471-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nijnik A, Dawson S, Crockford TL, Woodbine L, Visetnoi S, Bennett S, et al. Impaired lymphocyte development and antibody class switching and increased malignancy in a murine model of DNA ligase IV syndrome. J Clin Invest. 2009;119:1696–705. doi: 10.1172/JCI32743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slape C, Liu LY, Beachy S, Aplan PD. Leukemic transformation in mice expressing a NUP98-HOXD13 transgene is accompanied by spontaneous mutations in Nras, Kras, and Cbl. Blood. 2008;112:2017–9. doi: 10.1182/blood-2008-01-135186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puthiyaveetil AG, Heid B, Reilly CM, Hogenesch H, Caudell DL. A NUP98-HOXD13 leukemic fusion gene leads to impaired class switch recombination and antibody production. Exp Hematol. 2012;40:622–33. doi: 10.1016/j.exphem.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–92. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–7. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin B, Savic V, Juntilla MM, Bredemeyer AL, Yang-Iott KS, Helmink BA, et al. Histone H2AX stabilizes broken DNA strands to suppress chromosome breaks and translocations during V(D)J recombination. J Exp Med. 2009;206:2625–39. doi: 10.1084/jem.20091320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. 2006;22:511–9. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Calsou P, Delteil C, Frit P, Drouet J, Salles B. Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment. J Mol Biol. 2003;326:93–103. doi: 10.1016/s0022-2836(02)01328-1. [DOI] [PubMed] [Google Scholar]
- 21.Sallmyr A, Fan J, Datta K, Kim KT, Grosu D, Shapiro P, et al. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: implications for poor prognosis in AML. Blood. 2008;111:3173–82. doi: 10.1182/blood-2007-05-092510. [DOI] [PubMed] [Google Scholar]
- 22.Fernandes MS, Reddy MM, Gonneville JR, DeRoo SC, Podar K, Griffin JD, et al. BCR-ABL promotes the frequency of mutagenic single-strand annealing DNA repair. Blood. 2009;114:1813–9. doi: 10.1182/blood-2008-07-172148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin C, Yang L, Rosenfeld MG. Molecular logic underlying chromosomal translocations, random or non-random? Adv Cancer Res. 2012;113:241–79. doi: 10.1016/B978-0-12-394280-7.00015-4. [DOI] [PubMed] [Google Scholar]
- 24.Kasparek TR, Humphrey TC. DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Semin Cell Dev Biol. 2011;22:886–97. doi: 10.1016/j.semcdb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Vilenchik MM, Knudson AG., Jr Inverse radiation dose-rate effects on somatic and germ-line mutations and DNA damage rates. Proc Natl Acad Sci U S A. 2000;97:5381–6. doi: 10.1073/pnas.090099497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci U S A. 2003;100:12871–6. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng RC, Hsieh FJ, Shih CM, Hsu HS, Chen CY, Wang YC. Lung cancer susceptibility and prognosis associated with polymorphisms in the nonhomologous end-joining pathway genes: a multiple genotype–phenotype study. Cancer. 2009;115:2939–48. doi: 10.1002/cncr.24327. [DOI] [PubMed] [Google Scholar]
- 28.Choi CW, Chung YJ, Slape C, Aplan PD. A NUP98-HOXD13 fusion gene impairs differentiation of B and T lymphocytes and leads to expansion of thymocytes with partial TCRB gene rearrangement. J Immunol. 2009;183:6227–35. doi: 10.4049/jimmunol.0901121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42:319–29. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata A, Barton O, Noon AT, Dahm K, Deckbar D, Goodarzi AA, et al. Role of ATM and the damage response mediator proteins 53BP1 and MDC1 in the maintenance of G(2)/M checkpoint arrest. Mol Cell Biol. 2010;30:3371–83. doi: 10.1128/MCB.01644-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturgeon CM, Knight ZA, Shokat KM, Roberge M. Effect of combined DNA repair inhibition and G2 checkpoint inhibition on cell cycle progression after DNA damage. Mol Cancer Ther. 2006;5:885–92. doi: 10.1158/1535-7163.MCT-05-0358. [DOI] [PubMed] [Google Scholar]


