Abstract
Integration of the reverse-transcribed viral DNA into the host genome is an essential step in the lifecycle of retroviruses. Retrovirus integrase (IN) catalyzes insertions of both ends of the linear viral DNA into a host chromosome 1. IN from HIV-1 and closely related retroviruses share the three-domain organization, consisting of a catalytic core domain flanked by N- and C-terminal domains essential for the concerted integration reaction. Although structures of the tetrameric IN-DNA complexes have been reported for IN from prototype foamy virus (PFV) featuring an additional DNA-binding domain and longer interdomain linkers 2–5, the architecture of a canonical three-domain IN bound to DNA remained elusive. Here we report a crystal structure of the three-domain IN from Rous sarcoma virus (RSV) in complex with viral and target DNAs. The structure shows an octameric assembly of IN, in which a pair of IN dimers engage viral DNA ends for catalysis while another pair of non-catalytic IN dimers bridge between the two viral DNA molecules and help capture target DNA. The individual domains of the eight IN molecules play varying roles to hold the complex together, making an extensive network of protein-DNA and protein-protein contacts that show both conserved and distinct features compared to those observed for PFV IN. Our work highlights diversity of retrovirus intasome assembly and provides insights into the mechanisms of integration by HIV-1 and related retroviruses.
INs from lentiviruses including HIV-1 and the phylogenetically closely related alpharetroviruses including avian Rous sarcoma virus (RSV) share the conserved three-domain organization consisting of the Zn++-coordinating N-terminal domain (NTD), the catalytic core domain (CCD), and the β-strand-rich C-terminal domain (CTD) (Fig. 1a, Extended Data Fig. 1). We used the three-domain RSV IN construct biochemically fully active in concerted integration 6 and a branched DNA substrate mimicking the product of the concerted integration reaction (Fig. 1b) 7 to assemble and crystallize the stable RSV intasome complex (Extended Data Figs. 2, 3). The crystallized RSV intasome showed in solution an apparent molecular mass of 255 kDa by size-exclusion chromatography (SEC) and 240 (+/−10) kDa by SEC-multi-angle light scattering analysis, larger than the expected mass of 168 kDa for a tetramer of RSV IN(1–270) bound to the branched DNA (Extended Data Fig. 4a–c). The structure of the RSV intasome was determined by molecular replacement phasing and refined to 3.8 Å resolution (Extended Data Table 1).
Figure 1. Overall structure of the RSV intasome.
a, Comparison of the domain organization between RSV, HIV-1, and PFV INs. b, Branched DNA structure mimicking the product of the concerted integration reaction used in assembling the RSV intasome. c, Structure of the RSV intasome, viewed along its pseudo two-fold axis from the viral DNA side. The three structural domains of IN are color-coded as in (a). The grey spheres represent zinc ions in NTD. Two subunits within each IN dimer are colored slightly different from each other. d–f, Structure of the RSV intasome with the eight IN molecules colored individually, shown in three orthogonal orientations. The protein surface is shown in (d) and (e). The DNA strands are color-coded as in (b).
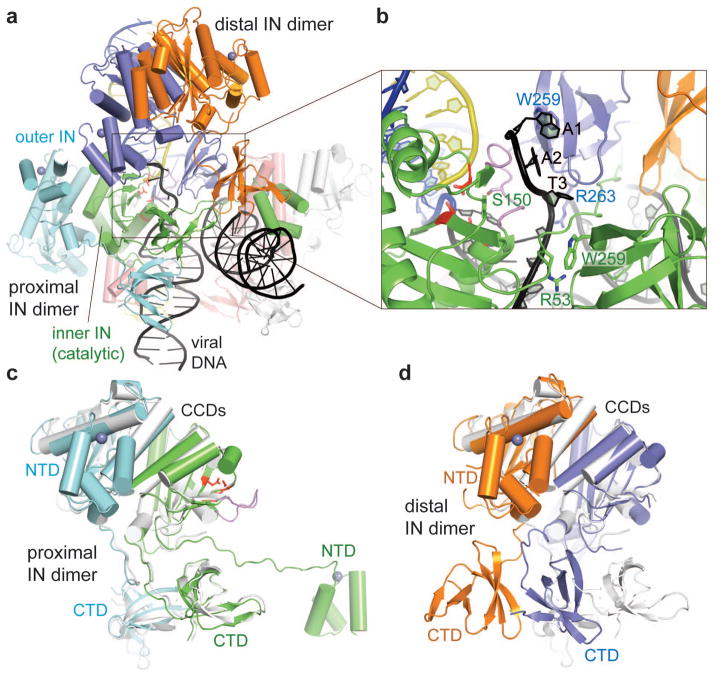
In contrast to the general assumption that retrovirus intasomes consist of an IN tetramer, the RSV intasome structure shows that it contains eight IN molecules (Fig. 1c–f and Supplementary Video), clearly supported by the selenium anomalous difference Fourier peaks for a crystal grown with selenomethionine-labeled IN (Extended Data Fig. 5). The observed octameric assembly (288 kDa) is consistent with the larger apparent molecular mass of RSV intasome in vitro and chemical cross-linking analysis (Extended Data Fig. 4d). The RSV IN octamer contains a core tetramer, which consists of two sets of ‘proximal’ IN dimers, and two additional sets of ‘distal’ IN dimers. The CCDs of the four IN dimers are positioned in an approximately two-fold symmetrical arrangement that resembles a parallelogram (Fig. 1c,d). The proximal IN dimer consists of ‘inner’ and ‘outer’ IN subunits, where the active-site of the inner subunit accommodates the viral/target DNA junction (Fig. 2a–c). The NTD of each inner IN subunit interacts in trans with the viral DNA engaged by the opposing proximal IN dimer, with the extended linker between NTD and CCD traversing the grooves of both viral DNA molecules to make additional contacts (Figs. 2a, 3a–c). The domain-swapped arrangement of NTD and CCD of the proximal IN dimers is analogous to that observed in the tetrameric PFV intasome 2.
Figure 2. Proximal and distal IN dimers.
a, A view highlighting interactions between a proximal (green/cyan) and a distal (slate/orange) IN dimers and their interactions with DNA. The IN and DNA strands are colored as in Fig. 1d–f. b, A close-up view around the viral DNA terminus. The CCD loop (residues 144–153) centered around Ser150 of the catalytic IN subunit is colored in violet. The catalytic triad residues (D64, D121, E157) are shown in red sticks in (a–c). c,d, Superposition of the proximal IN dimer (c; green/cyan) or the distal IN dimer (d; slate/orange) on the RSV IN CCD-CTD dimer in its native conformation (PDB 4FW1, light grey) 6. The grey spheres represent zinc ions in NTD.
Figure 3. Comparison between the RSV and PFV intasomes.
a, b, The octameric RSV intasome. The eight IN molecules and the DNA strands are colored as in Fig. 1d–f. The catalytic triad (DDE) of the inner subunit of the proximal IN dimers are shown in red in (b). c, Conformation of the inner catalytic IN subunit in the RSV intasome. The protein chain is colored in a gradient of blue to red from N to C-terminus, respectively. The NTDs and CCDs of the distal IN dimer are omitted. d,e, The tetrameric PFV intasome 3. The color scheme follows that used for the proximal IN dimers of RSV intasome in (a, b). The direction of DNA helical axis between the two integration sites on opposing strands (6 bp for RSV and 4 bp for PFV) differs significantly between the RSV (b) and PFV (e) intasomes. f, The inner catalytic IN subunit in the PFV intasome, colored in a gradient of blue to red as in (c). Note the presence of an extra domain on the N-terminus (NED) and different positioning of CTD compared to RSV IN in (c).
However, despite the shared structural features that support the basic chemistry of integration, the RSV and PFV intasomes have very different architectures (Fig. 3, Extended Data Fig. 6). The PFV intasome consists of four IN molecules, corresponding to only the core tetramer (two proximal IN dimers) of the RSV intasome. In the PFV intasome the inner IN subunits make all DNA interactions, while only the CCD is ordered for the outer subunits (Fig. 3d–f). In the RSV intasome, CTDs of the inner and outer subunits of the proximal IN dimer take unique conformations and tightly associate with each other (Fig. 2c), and this CTD dimer makes viral DNA contacts both in cis and trans (Fig. 2a). Moreover, the octameric RSV intasome contains two additional IN dimers; the CTDs from the distal IN dimers bridge between the proximal IN dimers, making additional contacts with both viral DNAs analogously to the CTD of the catalytic IN subunit in the PFV intasome (Fig. 3b,e). The CCDs of the distal IN dimers, anchored to the core of the RSV intasome through these CTD interactions (Fig. 2a), are positioned at the outer corners of the parallelogram, loosely associated with the distal regions of target DNA through non-specific interactions (Figs. 1d–f, 2a). The six remaining NTDs from the outer subunits of the proximal IN dimers and both subunits of the distal IN dimers are bound intra-molecularly to CCD (Extended Data Fig. 7c,d). In total, over 10,000 Å2 of molecular surface is buried in IN-IN interfaces within the RSV intasome, approximately half of which is accounted for by the conserved CCD dimerization interface, and ~6,000 Å2 in IN-DNA interfaces.
Although the CCDs and CTDs from all four IN dimers within the RSV intasome individually self-dimerize in the same fashions, the relative positioning of the CTDs with respect to the CCDs differs between the proximal and distal IN dimers, corresponding to their different roles. The CCD-CTD configuration for the proximal IN dimers is very similar to that previously observed for DNA-free RSV INs 6,8 (Fig. 2c), suggesting that RSV IN dimer in its native conformation is poised for viral DNA binding and catalysis, as noted earlier 6. The CTD of the inner catalytic subunit of the proximal dimer binds to viral DNA near the viral/target junction but on the opposite face of the double-stranded DNA, while it also makes contacts in trans with the other viral DNA (Fig. 2a–b). The CTD of the outer subunit of the proximal dimer binds to the distal region of the viral DNA in cis. Unlike in the proximal IN dimer, the CCD-CTD configuration for the distal IN dimers shows deviation from the canonical conformation, which can be described by a swing of the CTDs relative to the CCDs and disruption of the parallel β–sheet-like conformation of the CCD-CTD linkers 8 (Fig. 2d). This alternative CCD-CTD orientation allows the distal IN dimers to fit in the intasome without making steric clashes with the proximal INs or the 5’ overhang of the viral DNA strand (Fig. 2a). The CCDs of the distal IN dimers may be positioned similarly in the absence of target DNA, serving as a platform for target DNA capturing.
The asymmetrically associated CTDs of the proximal and distal IN dimers further interact with each other and with the NTD of the catalytic IN subunit to crosslink between the two viral DNAs (Figs. 2a, 4a,b). Each CTD dimer interacts with both of the viral DNA molecules at different positions, resulting in four distinct DNA-binding modes of individual CTDs. The basic amino acids Arg227, Arg244, Arg263, and Lys266 from various CTD monomers make contacts with the viral DNA molecules, and each viral DNA is sandwiched between separate CTD dimers (Fig. 4c). Arg244 of the inner catalytic subunit of the proximal IN dimer is positioned in the major groove of viral DNA, closest to G7 of the non-transferred strand. The GC pair at this position is critical for concerted integration by RSV IN 9. The corresponding residue Glu246 of HIV-1 IN was shown by disulfide cross-linking studies to interact with A7 of the non-transferred strand 10, suggesting similar modes of viral DNA sequence recognition by CTD between RSV and HIV-1 INs. Both RSV R244A/C and HIV-1 E246A IN mutants show reduced 3’ processing and strand-transfer activities 6,11,12, possibly reflecting the importance of these residues from various IN subunits. Mutation of conserved Trp233, which is stacked between the Arg227 and Lys266 side chains, to Glu or Ala but not Phe, abolishes binding to the viral DNA LTR sequence and concerted integration by RSV IN 13. The corresponding HIV-1 IN mutations W235E/A/F have parallel effects on concerted integration activity and virus replication, suggesting the importance of an aromatic residue at this position in orienting the basic side chains 14,15. Similarly, mutation of Trp259 buried in the CTD dimer interface (Fig. 4b) 6,8 as well as involved in multiple interactions near the viral DNA 5′ end (Fig. 2b) abolishes all enzymatic activities of RSV IN 6,16, reflecting the important roles of this residue in engaging viral DNA.
Figure 4. Viral DNA contacts.
a, Surface representation of the RSV intasome similar to Fig. 3a. b, A crown-like structure formed by the CTD of all eight INs and NTD and CCD of the catalytic INs, which encircles the two viral DNA molecules in black (boxed region in a). Trp259 side chains buried in the interface of each CTD dimer are shown. c, Viral DNA contacts by the CTDs. The side chains of Arg227, Arg244, Arg263, Lys266 are shown. NTD and CCD are omitted. d, A close-up view around the viral DNA end, showing interactions by CCD of a catalytic subunit, NTD of the opposing catalytic subunit contributed in trans, and the CTDs of a distal IN dimer. The catalytic triad residues are shown in red sticks. The grey sphere represents zinc ion in NTD.
The CCD of the catalytic RSV IN molecules engages viral and target DNAs primarily through interactions in their minor grooves, as observed for PFV IN 2,3. The long α-helix (α7; residues 154–174) that harbors one of the catalytic triad of metal-coordinating residues, Glu157, inserts into a significantly widened minor groove near the viral DNA terminus, where Arg158, Arg161, and Lys164 side chains make DNA base or backbone contacts (Fig. 4d, Extended Data Fig. 7h,i). The preceding loop centered around Ser150, which is highly flexible in the DNA-free IN 6,8,17, is positioned between the viral DNA 5′-overhang and the 3′-end of the cleaved target DNA, displacing T3 base opposite the terminal adenine (A20) of the transferred strand (Fig. 2b). DNA contacts in this minor groove also include hydrogen-bonding between Thr66 and the backbone phosphate group of the terminal nucleotide (A20) of the transferred viral DNA strand (Fig. 4d). Mutations of the corresponding HIV-1 IN residue T66A/I/K confer resistance against the IN strand-transfer inhibitors (INSTIs) 18, likely as a result of subtle changes of the IN-viral DNA interface. The NTD of the opposing catalytic subunit, contributed in trans, binds in the adjacent major groove and places Arg17 and Arg31 side chains for potential base-specific contacts (Fig. 4d). The hydrophobic NTD-CCD interface is centered on Phe199 from CCD, which explains why the F199K mutation selectively abolishes concerted integration by RSV IN 6,16. The CTDs of the distal IN dimer further extend the viral DNA interactions in this region (Fig. 4d).
The three flipped-out terminal nucleotides at the 5′ end of the non-transferred viral DNA strand, including the two overhanged bases and the displaced T3, bridge between CCD of the catalytic IN subunit and a CTD from the distal IN dimer, rather than between CCD and CTD of the same molecule as observed in the PFV intasome (Fig. 2b, 4b, 5a). Arg263 from this CTD points toward the non-transferred strand G4 opposite the CA dinucleotide at the viral DNA terminus (Fig. 2b), which could be relevant to the effect of mutation of the corresponding HIV-1 residue Arg263 on catalysis and drug resistance 19. The two viral DNAs in the RSV intasome branch out with their helical axes skew and at an angle of ~60°, which is smaller than the viral DNA split of ~80° in the PFV intasome (Extended Data Fig. 8). Accordingly, the viral DNA molecules in the RSV intasome are positioned closer to each other than those in the PFV intasome, with the backbone phosphate oxygen atoms at the closest point ~5 Å apart. The viral DNA molecules in the RSV intasome are surrounded by a highly positively charged surface formed by a network of CTDs and NTDs, which may alleviate potential electrostatic repulsions between DNA strands and help hold the complex together (Fig. 4a–c).
Figure 5. Target DNA.
a, The sharply bent target DNA spanning the CCDs of the catalytic IN subunits. Ser124 and Glu229 side chains potentially involved in target DNA contacts are shown. b, The central 6 bp region viewed from the viral DNA side, showing distorted DNA conformation with severely compromised base-stacking. c, The target DNA viewed from the viral DNA side, highlighting a large shift in the helical axis.
The target DNA in the RSV intasome shows a strong overall bending of ~90° away from the core of the complex (Fig. 5a, Extended Data Fig. 7b), which may help prevent the reversal of integration as noted for the PFV intasome and related transpososome structures 3,20. The bent conformation is stabilized by the minor groove contacts made by CCD of the catalytic subunits near the viral/target DNA junction, which include insertion of a short helix (α5) harboring Ser124, a residue important in target DNA capturing 21. Localized kinks at the viral/target DNA junctions cause the DNA trajectory to also zigzag in the plane perpendicular to the direction of the primary bending, creating a ~20 Å shift in the helical axis with an overall positive writhe (Fig. 5c). As a result, the target DNA conformation in the RSV intasome deviates significantly from that in the PFV intasome 3 (Extended Data Fig. 8c) or the DNA structure in nucleosomes22. This suggests that RSV integration into nucleosomes would require a large conformational change in the target DNA, likely more extensive than that observed in the PFV intasome-nucleosome complex 4.
The active-sites of the catalytic IN subunits in the RSV intasome are separated by ~27 Å, larger than the distance of ~21 Å across the major groove between the backbone phosphates 6 bp apart in a B-form DNA. Accordingly, the 6 bp spacer region of target DNA is underwound and shows severely compromised base-stacking in addition to widening of the major groove (Fig. 5b, Extended Data Fig. 9). In particular, the central dinucleotide step shows a distorted conformation where the unstacked nucleobases stack on the deoxyribose moiety of the adjacent nucleotide; the unique conformation may explain the modest target sequence preference in RSV integration 23,24. A CTD from each distal IN dimer has a loop (between β6 and β7) positioned over the major groove of the target DNA in this region, with Glu229 side chain positioned for potential base contacts (Fig. 5a). This loop from various CTDs are involved in viral DNA contacts, interactions between NTD and CTDs, and those linking the CTDs from the proximal and distal IN dimers in the RSV intasome (Fig. 4b, Extended Data Fig. 7e–g).
The RSV intasome structure shows a novel architecture of IN-DNA complex and highlights a remarkable diversity among retrovirus intasome assemblies. In particular, the structurally conserved CTD plays critical roles in the octameric RSV intasome distinct from those played by CTD in the tetrameric PFV intasome 2,3,5. PFV IN features a unique ~30 amino-acid insertion between CCD and CTD (Fig. 1a, Extended Data Fig. 1), and the resulting long interdomain linker not only makes direct viral DNA contacts but also allows CTD of the catalytic IN subunit to fit between the NTD and CCD from the same molecule to interact with both viral DNAs (Fig. 3f). In case of RSV IN with a much shorter CCD–CTD linker, the NTD and CCD of the catalytic IN molecule is instead bridged by the CTDs contributed in trans from the distal IN dimer (Fig. 3c). Analogously, the extra ~50 amino acids on the N-terminus of PFV IN constitute an independent DNA-binding domain (NED), and its interaction with distal region of the viral DNA substitutes for viral DNA interactions made by the multiple copies of CTDs as observed in the RSV intasome (Fig. 3a, d). The larger (408 amino acids) IN from a gammaretrovirus murine leukemia virus (MLV) also has both the NED and a long CCD-CTD linker 25, consistent with the idea that these two structural features have complementary functions. The different modes of viral DNA engagement by either tetrameric or octameric IN, using similar sets of structurally conserved domains, suggest divergent evolution of the integration machineries. Of note, an octameric intasome assembly similar to that reported here has been recently observed by cryo-EM for another three-domain IN from mouse mammary tumor virus 26.
The integration of retroviral DNA into the host genome was first postulated based on the observation of RSV-infected cells 27, and identification and characterization of IN from retroviruses including RSV have provided a basic understanding of retrovirus integration and foundation for studying HIV-1 IN 28–31. Our findings on the RSV intasome structure shed light on the previously unappreciated diversity in retroviral DNA integration and define a new framework for future studies of retrovirus IN. Availability of the intasome structures from multiple retrovirus systems will also allow more accurate modeling of the HIV-1 IN-DNA interactions to help development of novel antiretroviral drugs, including IN inhibitors that target outside the most conserved active-site features.
METHODS
RSV intasome preparation
RSV IN and its mutant forms were overexpressed in Escherichia coli BL21 (DE3) and purified as previously described 6. The proteins were stored in aliquots at −80°C in 20 mM HEPES-NaOH (pH 7.5), 1.0 M NaCl, 20μM ZnCl2, 5 mM 2-mercaptoethanol and 10% (w/v) glycerol. The branched DNA substrate mimicking the product of the concerted integration reaction was obtained by annealing three synthetic oligonucleotides (Integrated DNA Technologies). A similar strategy had been used previously to prepare a PFV IN-DNA complex, which demonstrated that the IN-DNA complex assembled on the designed integration product has essentially an identical structure to the equivalent complex formed via the forward integration reaction 7. The viral DNA branches carrying the high-affinity gain-of-function mutant RSV U3 long terminal repeat (LTR) sequence (GU3) 32 were attached to the target DNA duplex with a palindromic 6 bp spacer to generate a fully symmetrized structure. To prepare the RSV IN-DNA complexes for crystallization, 30 μM half-site DNA substrate was mixed with 120 μM RSV IN in 20 mM HEPES-NaOH (pH 7.5), 500 mM NaCl, 20% (w/v) glycerol, and 1 mM tris-(2-carboxyethyl)phosphine (TCEP). The mixture was dialyzed against low salt buffer (20 mM HEPES-NaOH pH 7.5, 125 mM NaCl, 20% (w/v) glycerol, 1 mM TCEP) at 25°C for overnight. At the end of this first dialysis, essentially all IN and IN-DNA complex precipitated. The mixture was subsequently dialyzed against high salt buffer (20 mM MES-NaOH pH 6.0, 1.2 M NaCl, 20% (v/v) dimethyl sulfoxide (DMSO), 5% (w/v) glycerol, 1 mM TCEP) at 25°C for 2–4 hours. At the end of this second dialysis, the reaction mixture became clear. The solubilized RSV IN-DNA complex (intasome) was purified through size-exclusion chromatography (Superdex 200 10/300 GL, GE Healthcare) running with 20 mM MES-NaOH pH 6.0, 1.2 M NaCl, 20% (v/v) DMSO, 5% (w/v) glycerol, and 1 mM TCEP. The isolated intasome remains stable in the high-salt condition containing 1.2 M NaCl that precludes complex formation, suggesting that once the intasome forms, it is kinetically trapped. The solubility-enhancing RSV IN mutation F199K completely abolished intasome formation. For the SEC-MALS analysis, a modified condition was used for the intasome re-solubilization and isolation for better baseline stability (Extended Data Fig. 4c).
Crystallization
For crystallization, the RSV intasome assembled through dialysis and purified by size-exclusion chromatography was concentrated to 4–6 mg/ml using centrifugal concentrator (Amicon). Various combinations of the lengths of the viral DNA (ranging from 16 bp to 25 bp) and flanking target DNA (ranging from 14 bp to 22 bp, corresponding to the full target DNA lengths of 34 bp to 50 bp) were screened in crystallization trials. The extensive screening yielded only one crystal form with a specific combination of 22-base (length of the non-transferred strand) viral DNA branches and 16 base-pair (bp) target DNA flanks on either side of the central six bp spacer (Fig. 1b, Extended Data Fig. 2). DNA substrates with two slightly different target sequences were used (5′-aatgttgtcttatgcaatactc-3′/5′-gagtattgcataagacaacagtgcacgaatcttgaagacact-3′/5′-agtgtcttcaagattc-3′, or 5′-aatgttgtcttatgcaatactc-3′/5′-gagtattgcataagacaacagtcgaccaaccttcaacttagc-3′/5′-gctaagttgaaggttg-3′), and they produced essentially the same crystals. The RSV intasome crystals were grown through reverse vapor diffusion in hanging drops at 22°C, by mixing 1.5 μL IN-DNA complex solution with 1.5 μL reservoir solution (3.2 M sodium formate). Crystals appeared within 3–5 days and reached a size of ~150–300 μm in 3~5 days. Even though the RSV intasome crystals initially diffracted X-ray poorly (~ 10 Å), soaking the crystals with a metatungstate cluster compound dramatically improved the resolution (Extended Data Fig. 3b, c). The tungsten cluster was later found to bind between IN dimers from separate intasome complexes to mitigate crystal lattice disorder. The crystals were soaked overnight with 0.15 mM metatungstate cluster Na6[H2W12O40] in a stabilization buffer consisting of 3.2 M sodium formate, 16 mM MES-NaOH pH 6.0, 0.8 M NaCl, 16% (v/v) DMSO, 4% (w/v) glycerol, and 1 mM TCEP. After soaking, the crystals were cryo-protected in 3.2 M sodium formate, 16 mM MES pH 6.0, 0.8 M NaCl, 16% (v/v) DMSO, 12% (w/v) glycerol, and 1 mM TCEP, and were frozen by rapid immersion in liquid nitrogen. The full-length wild-type RSV IN(1–286), the C-terminally truncated RSV IN(1–270), and its various mutant forms tested produced essentially the same crystals with indistinguishable X-ray diffraction properties.
Structure determination
X-ray diffraction data were collected at the Advanced Photon Source Northeastern Collaborative Access Team beamlines (24-ID-C/E) and the Advanced Light Source Molecular Biology Consortium (4.2.2) beamline and processed using HKL2000 33 or XDS 34. The RSV intasome crystals showed varied degrees of pseudo-merohedral twinning with twin operator [l, −k, h], owing to the very similar a and c unit cell dimensions of the primitive monoclinic lattice. Thus, we screened a large number of crystals to identify ones that diffract to higher resolution and have smaller twin fractions. The structure of RSV intasome was determined by molecular replacement with PHASER 35, using the RSV IN CCD, CTD (PDB ID: 4FW1) 6, and a 16 bp B-form DNA as search models. 8 copies of CCD, one copy of CTD and three copies of DNA molecules were located. Refinement of the partial model revealed electron density for two copies of the metatungstate clusters. The metatungstate clusters were placed into the electron density by molecular replacement using MOLREP 36. Subsequent iterative model building using COOT 37 and refinement with PHENIX suite 38 allowed placement of the remaining 7 copies of CTD, 8 copies of NTD generated using MODELLER 39 based on HIV-1 NTD (PDB ID: 1K6Y) 40, and building of the inter-domain linkers as well as the remaining parts of the DNA molecule guided by the difference electron density maps. A third metatungstate cluster, which is more weakly bound compared to the first two, was positioned manually into residual density. The DNA base-pairs and base-stacking restraints were used throughout the refinement. The geometry restraints for protein included the reference-model restraints for CCD and CTD based on the higher resolution RSV IN structure (4FW1) 6, and the secondary structure and zinc-coordination restraints for NTD. Atomic displacement parameters were refined with grouped B-factors per residue for protein and DNA, and a total of 53 TLS groups assigned by PHENIX 38. Twelve tungsten atoms representing each cluster were refined as a rigid body. The asymmetric unit of the crystal contains one complete RSV intasome, which includes eight IN molecules and two viral DNA branches emanating from a strongly bent 38 bp target DNA. The dataset used for the final refinement was from an RSV intasome crystal grown using selenomethionine-labeled RSV IN (1–270) with the following amino acid substitutions; C23S, L112M, L135M, L162M, L163M, L188M, and L189M, which we confirmed to be active in concerted integration and inhibited by INSTI similarly to the wild-type RSV IN, and a DNA substrate carrying a nick at the middle of each target DNA branch (the 16-base DNA strand shown in olive in Fig. 1b has a nick 8 bases from either end). The nick occasionally facilitated crystal growth, but it was not necessary for crystallization and did not change the space group or the unit cell parameters compared to crystals grown without the nick. Because this nick in the target DNA is not biologically relevant, it is not shown in Figs. 1b, 1d–f, 3b, 5a,c to avoid confusion. Twin refinement protocol was not used as the dataset used for the final refinement had a low (less than 10%) twin fraction. The summary of data collection and refinement statistics is shown in Extended Data Table 1. The paired-refinement procedure 41 was performed in steps of 0.1 Å to determine the high-resolution limit (Extended Data Fig. 3f). The register of amino acids in the final model was verified by the selenium anomalous difference Fourier peaks (Extended Data Fig. 5). Ramachandran analysis shows that 96.0, 3.9, and 0.1 % of the protein residues are in the most favored, allowed, and disallowed region, respectively. The NTD-CCD linker in some of the non-catalytic IN molecules and the last four bp at the distal end of one of the viral DNA molecules were not built due to poor electron density. The molecular graphics images were produced using PYMOL (www.pymol.org).
Extended Data
Extended Data Figure 1. Amino acid sequence alignment of RSV, HIV-1, and PFV INs.
The secondary structure elements for RSV IN are color-coded based on the three IN domains similarly to Fig. 1a. The residue numbering at the top is for RSV IN. For each IN, the black dots mark every ten residues. This figure was made using ESPript 42.
Extended Data Figure 2. DNA substrate used for assembling the RSV intasome.
a, The half-site (gaped duplex) substrate prepared by annealing three oligonucleotides dimerizes via the self-complementary 6-base spacer sequence (underlined) to form a branched structure mimicking the product of the concerted integration reaction (Fig. 1b). b, DNA structure in the RSV intasome. Viral DNA nucleotides are numbered, and some of the structural elements of RSV IN involved in the viral DNA interactions are shown.
Extended Data Figure 3. RSV intasome crystal.
a, A crystal of the RSV intasome. b, X-ray diffraction pattern from a crystal not treated with metatungstate. c, X-ray diffraction pattern after the metatungstate-soaking (see method for details). d,e, Lattice contacts within the RSV intasome crystal. DNA strands are colored as in Fig. 1b, while IN subunits from only one intasome are colored. The unit cell is shown in green. The small blue spheres represent tungsten atoms. The view in (e) is perpendicular to the two-fold screw (b) axis of the monoclinic lattice, which lies horizontally. f, Paired-refinement analysis 41 to assess the resolution limit of the RSV intasome diffraction data. For each pairwise comparison, model refinements were run at two different resolution limits and the R-factors calculated for a common (lower) resolution cutoff were compared. Inclusion of data beyond 3.7 Å in the refinement compromised model quality.
Extended Data Figure 4. Biochemical characterization of RSV intasome.
a, A representative size-exclusion chromatography (SEC) profile for RSV intasome, overlaid with that for a mixture of molecular weight markers. The buffer condition was as mentioned in the methods. b, SEC profiles for RSV intasomes formed with IN of varying C-termini. c, SEC-MALS (multi-angle light scattering) analysis of RSV intasome. The intasome formed with RSV IN (1–269 aa) was separated by SEC in a modified condition containing 20 mM HEPES pH 7.5, 1.0 M NaCl, 5 % glycerol, and 1.0 mM TCEP. The absolute molecular mass was determined by light scattering using in-line detectors described previously 43. The mass profile for the intasome is shown in red across the peak. The molecular mass of RSV intasome was 240 ±10 kDa (n=4). A similar SEC-MALS analysis of the intasome formed with wt full-length RSV IN (1–286 aa) yielded a molecular mass of 268 kDa (n=2, data not shown). The calculated mass of an intasome containing eight RSV IN (1–269 or 270) molecules is ~288 kDa. d, Chemical cross-linking analysis of RSV intasome. The RSV intasome and free IN (1–269 aa) were purified by size-exclusion chromatography in the running buffer: 20 mM HEPES (pH 7.5), 1.0 M NaCl, 5 % glycerol, and 1.0 mM TCEP. The peak fractions of the intasome and IN were cross-linked with the indicated amount of ethylene glycol bis-succinimidylsuccinate (EGS) as described previously 43 and analyzed by SDS-PAGE. The majority of cross-linked species within the intasome were larger than a tetramer. The highest oligomeric species observed is consistent with an octamer migrating at ~220 kDa. The molecular weight markers are in the far right lane. The NuPAGE 4–12% gradient gel with a MES-based SDS-PAGE running buffer was used.
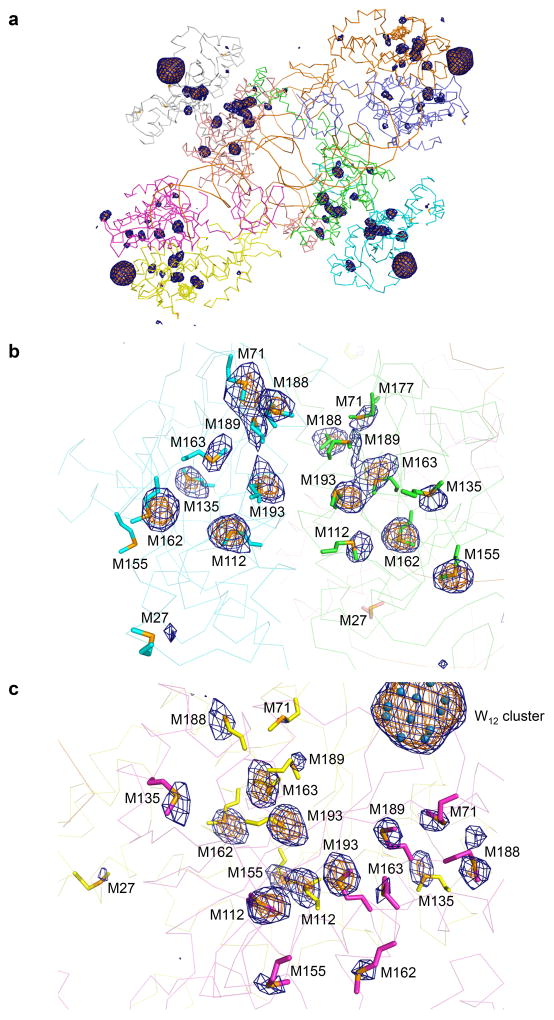
Extended Data Figure 5. Selenium anomalous difference Fourier peaks confirming the model.
Anomalous difference Fourier maps calculated using the data collected on selemomethionine-labeled RSV intasome, contoured at 3.5σ (blue mesh) or 5.0 σ (orange mesh). Methionine side chains are shown in sticks. a, A view covering the octameric RSV intasome. b, c, Close-up view of a proximal (b) and distal (c) IN dimer, respectively.
Extended Data Figure 6. Comparison between RSV and PFV intasomes.
a, b, Protein arrangement in the octameric RSV intasome. The inner and outer subunit of one proximal IN dimer is colored in green and cyan, respectively, with the catalytic triad (DDE) of the inner subunit shown in red. The other proximal IN dimer is colored similarly but in more pale colors. Arrows indicate the two proximal IN dimers. A distal IN dimer is colored in slate and orange. DNA is omitted in (a). (b) shows the same view as Fig. 3c. c, A close-up view around the active site of the inner IN subunit in the RSV intasome. The DNA strands are colored as in Fig. 1, and the catalytic triad residues (DDE) are shown in red sticks. d,e, Protein arrangement in the tetrameric PFV intasome 2,3. The color scheme follows that used for the proximal IN dimers of RSV IN in (a, b). Arrows indicate the two IN dimers. DNA is omitted in (d). (e) shows the same view as Fig. 3f. f, A close-up view around the active site of the inner IN subunit in the PFV intasome (PDB ID: 3OS0 3). The color scheme follows that in (c).
Extended Data Figure 7. Composite omit maps.
Simulated annealing composite omit 2mFo−DFc density contoured at 1.0 σ, shown for area within 3.5 Å from any protein or DNA atom in the final model. In a and b, electron densities around protein and DNA are colored differently (blue and green, respectively).
Extended Data Figure 8. DNA conformations in the RSV and PFV intasomes.
a,b, DNA structure in the RSV (a) or PFV (b) intasome, alternatively referred to as the strand-transfer complex (STC). The PFV intasome model is PDB ID: 3OS0 3. c, A comparison of DNA structures between the RSV and PFV intasomes (STCs). The integration product DNAs (RSV in cyan, PFV in red) superimposed at a viral DNA terminus are shown in three different view angles. Note the significant deviation in overall trajectory of the target DNA, and difference in the orientation of the second viral DNA molecule. The region spanning the two integration sites on opposing target DNA strands is 6 bp for RSV and 4 bp for PFV.
Extended Data Figure 9. Electron density for the central 6 bp of the target DNA.
The sigma A-weighted 2mFo−DFc map contoured at 1.5 σ (a) or 2.5 σ (b), overlaid with the final model for the central 6 bp region between the two integration sites.
Extended Data Table 1.
Data collection and refinement statistics
| RSV intasome | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 124.9, 157.8, 126.6 |
| α, β, γ (°) | 90.0, 110.9, 90.0 |
| Resolution (Å) | 49.3-3.80(3.90–3.80) |
| Rmerge (%) | 14.7 (247.6) |
| I/σI | 6.0 (0.6) |
| CC1/2 | 0.994 (0.287) |
| Completeness (%) | 99.2 (99.3) |
| Redundancy | 5.5(5.5) |
| Refinement | |
| Resolution (Å) | 49.3 - 3.80 |
| No. reflections | 44627 |
| Reflections for Rfree | 2183 |
| Rwork/Rfree | 25.4/29.4 |
| No. atoms | |
| Protein | 19309 |
| Ligand/ion | 44 |
| Water | 0 |
| B-factors | |
| Protein | 206.9 |
| Ligand/ion | 205.0 |
| Water | N/A |
| R.m.s deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.72 |
Statistics for the highest resolution shell are shown in parentheses.
Reference:
CC1/2: P.A. Karplus and K. Diederichs, Linking crystallographic model and data quality. (2012) Science 336, 1030–1033 (Ref. 41).
Supplementary Material
A video showing the overall structure of the RSV intasome and positioning of the three structural domains of IN within the intasome.
Acknowledgments
We thank K. Kurahashi for generating many mutant IN expression plasmids, J. Kankanala and Z. Wang for synthesizing Pt-modified oligonucleotides, and J. Nix for help with X-ray data collection. X-ray data were collected at the Advanced Photon Source (APS) NE-CAT beamlines, which are supported by the NIGMS (P41 GM103403). APS is a US Department of Energy Office of Science User Facility operated by Argonne National Laboratory under Contract DE-AC02-06CH11357. This research was supported by the National Institutes of Health grants GM109770, AI087098 to H.A. and AI100682 to D.P.G.
Footnotes
Author Contributions
Z.Y. designed strategy and developed protocols for producing and crystallizing the RSV intasome, and prepared all crystals. Z.Y., K.S., and S. Banerjee collected X-ray diffraction data. K.S. analyzed the data and determined the crystal structure. K.P. and S. Bera examined behaviors of the RSV intasome in solution and analyzed mutant IN activities. D.P.G. and H.A. conceived the project and supervised the research. H.A. wrote the manuscript with important input from all authors.
Atomic coordinates and structure factors have been deposited with the Protein Data Bank under accession code 5EJK.
The authors declare no competing financial interests.
References
- 1.Craigie R, Bushman FD. HIV DNA integration. Cold Spring Harbor perspectives in medicine. 2012;2:a006890. doi: 10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maskell DP, et al. Structural basis for retroviral integration into nucleosomes. Nature. 2015 doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta K, et al. Solution conformations of prototype foamy virus integrase and its stable synaptic complex with U5 viral DNA. Structure. 2012;20:1918–1928. doi: 10.1016/j.str.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi K, et al. A possible role for the asymmetric C-terminal domain dimer of Rous sarcoma virus integrase in viral DNA binding. PloS one. 2013;8:e56892. doi: 10.1371/journal.pone.0056892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin Z, Lapkouski M, Yang W, Craigie R. Assembly of prototype foamy virus strand transfer complexes on product DNA bypassing catalysis of integration. Protein science : a publication of the Protein Society. 2012;21:1849–1857. doi: 10.1002/pro.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang ZN, Mueser TC, Bushman FD, Hyde CC. Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. Journal of molecular biology. 2000;296:535–548. doi: 10.1006/jmbi.1999.3463. [DOI] [PubMed] [Google Scholar]
- 9.Vora A, Bera S, Grandgenett D. Structural organization of avian retrovirus integrase in assembled intasomes mediating full-site integration. The Journal of biological chemistry. 2004;279:18670–18678. doi: 10.1074/jbc.M314270200. [DOI] [PubMed] [Google Scholar]
- 10.Gao K, Butler SL, Bushman F. Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes. The EMBO journal. 2001;20:3565–3576. doi: 10.1093/emboj/20.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peletskaya E, et al. Localization of ASV integrase-DNA contacts by site-directed crosslinking and their structural analysis. PloS one. 2011;6:e27751. doi: 10.1371/journal.pone.0027751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutzke RA, Plasterk RH. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. Journal of virology. 1998;72:4841–4848. doi: 10.1128/jvi.72.6.4841-4848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu R, Grandgenett DP. Molecular and genetic determinants of rous sarcoma virus integrase for concerted DNA integration. Journal of virology. 2003;77:6482–6492. doi: 10.1128/JVI.77.11.6482-6492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Wei SQ, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. The Journal of biological chemistry. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Craigie R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. The Journal of biological chemistry. 2005;280:29334–29339. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bojja RS, et al. Architecture of a full-length retroviral integrase monomer and dimer, revealed by small angle X-ray scattering and chemical cross-linking. The Journal of biological chemistry. 2011;286:17047–17059. doi: 10.1074/jbc.M110.212571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lubkowski J, et al. Atomic Resolution Structures of the Core Domain of Avian Sarcoma Virus Integrase and Its D64N Mutant. Biochemistry. 1999;38:15060. doi: 10.1021/bi995092n. [DOI] [PubMed] [Google Scholar]
- 18.HIV Drug Resistance Database: Integrase Inhibitor (INI) Resistance Notes. 2014 [Google Scholar]
- 19.Quashie PK, et al. Characterization of the R263K mutation in HIV-1 integrase that confers low-level resistance to the second-generation integrase strand transfer inhibitor dolutegravir. Journal of virology. 2012;86:2696–2705. doi: 10.1128/JVI.06591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montano SP, Pigli YZ, Rice PA. The mu transpososome structure sheds light on DDE recombinase evolution. Nature. 2012;491:413–417. doi: 10.1038/nature11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harper AL, Sudol M, Katzman M. An amino acid in the central catalytic domain of three retroviral integrases that affects target site selection in nonviral DNA. Journal of virology. 2003;77:3838–3845. doi: 10.1128/JVI.77.6.3838-3845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 23.Serrao E, Ballandras-Colas A, Cherepanov P, Maertens GN, Engelman AN. Key determinants of target DNA recognition by retroviral intasomes. Retrovirology. 2015;12:39. doi: 10.1186/s12977-015-0167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. Journal of virology. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aiyer S, et al. Structural and sequencing analysis of local target DNA recognition by MLV integrase. Nucleic acids research. 2015;43:5647–5663. doi: 10.1093/nar/gkv410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballandras-Colas A. Cryo-EM reveals a novel octameric integrase structure for beta-retrovirus intasome function. Nature. 2016;530:358–361. doi: 10.1038/nature16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Temin HM. The participation of DNA in Rous sarcoma virus production. Virology. 1964;23:486–494. doi: 10.1016/0042-6822(64)90232-6. [DOI] [PubMed] [Google Scholar]
- 28.Grandgenett DP, Vora AC, Schiff RD. A 32,000-dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology. 1978;89:119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 29.Donehower LA, Varmus HE. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panganiban AT, Temin HM. The retrovirus pol gene encodes a product required for DNA integration: identification of a retrovirus int locus. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartzberg P, Colicelli J, Goff SP. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984;37:1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- 32.Vora AC, et al. Avian retrovirus U3 and U5 DNA inverted repeats. Role of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases. The Journal of biological chemistry. 1997;272:23938–23945. doi: 10.1074/jbc.272.38.23938. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski ZaMW. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Vol. 276. Academic Press; 1997. p. 307. [DOI] [PubMed] [Google Scholar]
- 34.Kabsch W. Xds. Acta crystallographica. Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy AJ, et al. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teplyakov AVaA. MOLREP: an Automated Program for Molecular Replacement. J Appl Cryst. 1997;30:4. doi: 10.1107/S0021889897006766. [DOI] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of molecular biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 40.Wang JY, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. The EMBO journal. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic acids research. 2014;42:W320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey KK, et al. Rous sarcoma virus synaptic complex capable of concerted integration is kinetically trapped by human immunodeficiency virus integrase strand transfer inhibitors. The Journal of biological chemistry. 2014;289:19648–19658. doi: 10.1074/jbc.M114.573311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A video showing the overall structure of the RSV intasome and positioning of the three structural domains of IN within the intasome.