Abstract
In the last several years, nanoscale vesicles that originate from tumor cells and which can be found circulating in the blood (i.e. exosomes and microvesicles) have been discovered to contain a wealth of proteomic and genetic information to monitor cancer progression, metastasis, and drug efficacy. However, the use of exosomes and microvesicles as biomarkers to improve patient care has been limited by their small size (30 nm–1 μm) and the extensive sample preparation required for their isolation and measurement. In this Critical Review, we explore the emerging use of micro and nano-technology to isolate and detect exosomes and microvesicles in clinical samples and the application of this technology to the monitoring and diagnosis of cancer.
Introduction
Cancer is often localized in difficult to access parts of the body, such as in the brain, ovaries, or pancreas, making measurements of molecular biomarkers on tumor cells impractical for routine clinical monitoring or for disease diagnostics.1 In the last several years, nanoscale exosomes (30 nm–100 nm) and microvesicles (100 nm to 1 μm), which originate from tumor cells and can be found circulating in the blood, have been discovered to contain a wealth of proteomic and genetic information for disease diagnostics as well as the monitoring of cancer progression, metastasis, and drug efficacy.2 Unfortunately, establishing the clinical utility of exosomes and microvesicles as biomarkers to improve patient care has been limited by fundamental technical challenges that stem from their small size and the extensive sample preparation required prior to measurement. The scarcity of tumor-derived exosomes and microvesicles relative to those originating from healthy cells, their overlap in size with other nanoscale objects (e.g. protein aggregates, cell debris) present in clinical samples, and the short half-life of protein surface-markers once removed from the body further complicate these measurements.3
Platforms that use microscale structures (e.g. microfluidics), where dimensions are designed to match those of cells, have been used with great success to selectively and sensitively sort4–8 and detect9–12 cells. It is challenging, however, to translate these approaches to the nanoscale sizes optimal for the sorting and detection of microvesicles and exosomes, due to the expense of nanolithography, the inherently low throughput and susceptibility to clogging of nanoscale fluid channels, and the unfavorably strong scaling of many of the forces used to sort objects in microfluidics as they become nanoscale.
In this manuscript, we will review the current state of the art in using micro and nano-devices to isolate and detect exosomes and microvesicles and we will share our vision for the future translational and clinical relevance of this rapidly growing field.
What are exosomes? What are microvesicles? What’s the difference?
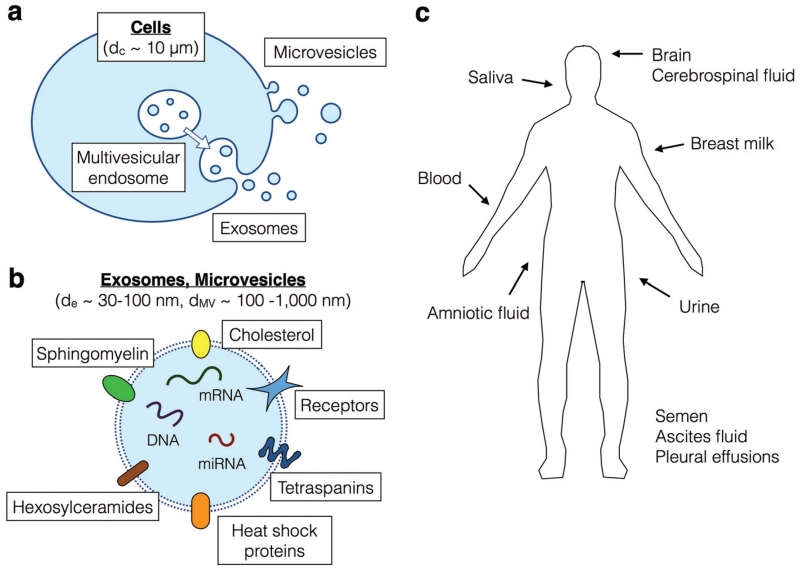
Exosomes are 30–100 nm vesicles released during fusion of the multivesicular endosomes (MVEs) with the plasma membrane.13(Fig. 1a) These nanoscale membrane bound vesicles carry specific surface markers that are representative of their cells of origin, including surface proteins and nucleic acids, such as micro RNA (miRNA), messenger RNA (mRNA), and DNA.14,15 Due to their endosomal origins, exosomes tend to be enriched for proteins related to transport and fusion (e.g. flotillin, caveolin-1), tetraspanins (e.g. CD63, CD9, CD81), heat shock proteins (e.g. Hsp90), and lipid-related proteins.16,17(Fig. 1b) To date, 4500 proteins have been measured in exosomes, many of which have been found to serve a role in intercellular communication.16 In addition to proteins, exosomes have been found to contain a greater amount of cholesterol, sphingomyelin, and hexosylceramides and a lesser amount of phosphatidylethanolamine and phosphatidylcholine than found in the cellular membrane.18
Fig. 1.
a. The biogenesis of exosomes and microvesicles from cells. b. A schematic showing the contents of exosomes and microvesicles. c. A cartoon showing where circulating exosomes and microvesicles can be found in non-invasively obtainable clinical samples.
In addition to proteins and lipids, exosomes have also been found to contain nucleic acids such as miRNA and mRNA,14 as well as double stranded DNA.15 Most of the RNAs found within exosomes are 20–200 base-pairs in length, including full-length miRNA and tRNA, and fragments of mRNA and rRNA.19 Different RNA isolation methods have resulted in inconsistent reports as to both the total quantity of exosomal RNA and the relative quantity of RNA types, and as such a detailed accounting of the nucleic acid cargo remains an open question.18
In contrast to exosomes, which originate from endosomes, microvesicles are fragments of plasma membrane which range in size from 100 nm to 1 μm.(Fig. 1a) The origins of microvesicles are diverse and include budding of microvesicles directly from the plasma membrane and blebbing of apoptotic bodies during cell death.20 In the literature, there has been some confusion over the definition of exosomes versus microvesicles, and they have sometimes been used interchangeably.18 As it stands, there are not satisfactory detection or isolation techniques that discriminate microvesicles versus exosomes. Therefore, it is currently not possible to accurately describe their differences in cargo. Professor Suresh Mathivanan from the La Trobe Institute for Molecular Science in Australia has created a valuable online resource that catalogs the cargo of exosomes and microvesicles from various origins, with the database including lipids, proteins, and RNAs (http://www.exocarta.org).
Circulating exosomes and microvesicles as diagnostic biomarkers
The sparse molecular markers that are shed from tumor cells into peripheral circulation have great potential for routine clinical monitoring of the molecular state of difficult to access tumor sites.21 These markers, including soluble proteins, cell-free DNA, circulating tumor cells (CTCs), and circulating microvesicles and exosomes (Table 1) have been shown to contain valuable information on the molecular state of a cancer.22,23
Table 1.
Comparison of soluble proteins, cell-free DNA, circulating tumor cells (CTCs), and circulating exosomes and microvesicles for non-invasive cancer monitoring
| Markers | Size | Occurence (7.5 mL blood) |
Cargo |
|---|---|---|---|
| Soluble proteins | ~5 nm | 100 fM (1011 molecules) |
NA |
| Cell free DNA | 200 base pairs |
100 copies | NA |
| Circulating tumor cells |
5–30 μm | 0–1000+ cells | Proteins nucleic acid |
| Exosomes/ microvesicles |
30 nm– 1 μm |
106 exosomes or microvesicles |
Proteins and nucleic acid from mother cell |
Engineers have devised many ingenious strategies to isolate and measure both rare CTCs4,5,8,9 and sparse soluble proteins in blood.24–27 However, fundamental limitations of these detection modalities impede their clinical application. CTCs are limited by their extremely sparse concentrations in blood (0–1000 + CTC in 7.5 mL of blood), resulting in long run-times, excessive consumption of valuable clinical samples, and Poisson counting error.28 The detection of soluble protein based biomarkers, such as prostate specific antigen (PSA) in prostate cancer, has been limited by issues of specificity. Because the biogenesis of soluble protein found in blood cannot be determined, diagnostics based on protein detection suffer from high false-positive rates.29,30
Isolation of intact CTCs offers the promise of combining phenotypic characterization with molecular analysis of extracted nucleic acids. However, CTCs can be difficult to detect using existing platforms, especially in the setting of minimal residual disease or early-stage cancer.31,32 The FDA-approved CellSearch device has had limited sensitivity in diseases such as melanoma, pancreatic, and ovarian cancer,31,32 even in the context of advanced or metastatic disease. In contrast, exosomes have been readily detectable in early- and late-stage pancreatic cancer,33,34 melanoma,35 glioblastoma,36 and other cancers for which CTC isolation is currently difficult or impossible.
Cell-free DNA (cfDNA), which consists of fragments of DNA emitted into the bloodstream by dying cancer cells, has shown great promise in recent years as an alternative biomarker to soluble proteins or CTCs.37 There are somatic mutations only present in tumor cell DNA, which provide a highly specific biomarker that can be specifically detected using quantitative PCR or next-generation sequencing. It was recently demonstrated by Luis Diaz’s group that these DNA fragments can be used for diagnostics and drug-resistance screening.38 In contrast with CTCs, cfDNA is relatively easy to isolate and sequence, and can be readily detected in patients with many different types of cancer.38 However, there is evidence that cfDNA may better reflect the nucleic acid complement of dying or apoptotic cells39 rather than viable solid tumor cells or CTCs.
Early studies on exosomes and microvesicles show them to have great potential as an accessible biomarker for cancer detection and monitoring.2,3,36,40–43 Microvesicles and exosomes are remarkably stable in circulation and have been found in blood, urine, saliva, breast milk, semen, ascites fluid, amniotic fluid, and cerebrospinal fluid.16 (Fig. 1c) Exosomes offer many of the benefits of CTCs: they are discrete units, containing multiple molecular markers that indicate their cellular origin and provide a rich proteomic and genetic profile of the cell. However, unlike CTCs, exosomes are present in the circulation in quantities more similar to soluble proteins (~105 mL−1 compared to ~0–100 mL−1 for CTCs).44,45
The clinical value of exosomes and microvesicles
The clinical value of tumor derived exosomes and microvesicles can be considered either by themselves or in combination with other “liquid biopsy” assays, including CTC, cfDNA, and soluble proteins. In making this consideration, it is worth noting that the relative importance of these liquid biopsies in the clinic is a matter of ongoing debate.21,46
Exosomal contents, including nucleic acids and proteins, are thought to be representative of the cell of origin, and thus can be interrogated for the purposes of targeting therapy and monitoring response to treatment. An important aspect of personalized medicine is the detection of genetic variants that can be treated with targeted therapeutic agents, either as mono-therapy or together with chemotherapeutic agents. For instance, driver mutations, such as BRAF V600E in melanoma, EGFR L858R in lung cancer, and HER2 amplification in breast cancer can all be treated with targeted pharmaceutical agents. Recently, Thakur and colleagues reported the specific detection of the BRAF V600E mutation in melanoma exosomes, and the EGFR L858R and T790M mutations in lung cancer exosomes, while another group was able to detect gene amplification in exosomal DNA.47 Taken together, these experiments suggest the potential for the development of exosome-based genotyping to guide therapy selection when CTCs or cfDNA are undetectable.
The abundance of detectable exosomes, and their nucleic acid payloads, may also be relevant for the early detection of molecular resistance to targeted therapy. CTCs and cfDNA are often undetectable for patients who have undergone multiple rounds of therapy, and repeat surgical access to tumor is not typically warranted nor well-tolerated by the patient.48 Therefore, the expression of mutations such as EGFR T790M in non-small cell lung cancer49 and ALK F1174L in neuroblastoma,50 which confer resistance to their respective targeted therapies, may go unnoticed. Importantly, such clinically relevant exosome-based DNA analysis will also likely be possible using biosamples besides whole blood. Tumor-derived exosomes have been isolated from lung pleural effusions,51 ascites,52 and bladder cancer urine,53 suggesting additional non-invasively accessible sources of tumor exosomes to guide therapeutic decision making.
Perhaps the most promising area for the clinical use of exosomes is for detection and monitoring of early stage disease. It is thought that if cancers such as pancreatic and ovarian could be detected before the advanced stage at which most patients are initially diagnosed, that significant improvements in the dismal survival rates of these diseases might be achievable.54–56 While current approaches for CTC isolation and variant detection in cfDNA are fairly sensitive in patients with advanced or metastatic disease, it is an open question in the field as to whether current assays can achieve the sensitivity necessary for early detection of or even screening for cancer. In contrast, multiple investigators have recently reported not only readily detectable tumor exosomes in early stage pancreatic and other cancers, but also the presence of protein markers associated with disease progression. In one study of Stage I pancreatic ductal adenoma cancer (PDAC) patients, higher levels of exosome-resident macrophage migration inhibitory factor (MIF) were found to predict an increased risk of the eventual development of liver metastasis.34 Intriguingly, this group also showed in a PDAC mouse model, that MIF blockade could prevent liver metastasis, suggesting that tumor exosome contents may prove to be clinically relevant for both prognosis as well as the development of novel targeted therapies. Using mass spectrometry to assess the protein cargo of early stage pancreatic cancer exosomes, Melo and colleagues recently identified glypican-1 (GPC1) as a cell surface marker highly expressed on tumor-derived exosomes.33 By evaluating exosomes from the serum of cancer-bearing mice and humans, they were able to show that numbers of GPC1+ exosomes correlated with tumor burden and survival. These results suggest that, just as CTC counts and the allelic fraction of variants detected in cell-free DNA have been shown to correlate with tumor volume, exosomes can also function as a tumor volume surrogate for real-time monitoring of response to therapy. Finally, and perhaps the most exciting result reported by this team, was the finding that measurement of GPC1+ exosomes could distinguish between patients with a precursor form of pancreatic cancer, intraductal papillary mucinous neoplasm (IPMN) and healthy donors.33 Further development of these assays could, therefore, lead to highly sensitive screening tests to identify one of the earliest, and most treatable forms, of one of the most lethal cancers.
Isolation of exosomes and microvesicles
In this section, we discuss conventional isolation of exosomes and microvesicles and new approaches that use micro- and nano-based devices to improve performance. The advantages and disadvantages of the isolation techniques that we highlight are summarized in Table 2.
Table 2.
Comparison of isolation techniques for exosomes and microvesicles
| Technology | Recovery rate | Purity | Sample volume | Integrated detection | Dynamic size selection | Time |
|---|---|---|---|---|---|---|
| Ultracentrifugation65 | 5–25% | Low | Low | − | − | 4–5 h |
| Density-gradient60 | 36–65% | High | Low | − | − | 4–6 h |
| Ciliated nanowire-on-microvillar63 | 60% | Not given | ~30 μl | − | − | 10 min |
| Acoustic-based64 | >80% | Not given | 0.4–0.7 μl min−1 | − | + | <30 min |
| Inertial lift force-based68 | Not given | >99% | 70 μl min−1 | + | − | >4 h |
| Surface-modified57,65–67 | 42–94% | >85% | 4–16 μl min−1 | − | − | <1 h |
| Nanoshearing67 | Not given | Not given | Not given | − | − | <3 h |
Background: conventional isolation techniques
Ultracentrifugation
Conventional exosome isolation techniques separate exosomes and microvesicles based on their size and buoyant density. In the most common separation technique, differential centrifugation, exosomes and microvesicles are isolated using a sequence of centrifugation steps. In the first step, 1000g centrifugation is used to remove all objects larger than exosomes and microvesicles, including dead cells and cell debris with a diameter larger than 1 μm. Next, ultracentrifugation at > 100 000g is used to pellet exosomes from the remaining supernatant.57 Centrifugation is time consuming (>4 h), results in co-purification with non-exosomal or microvesicle debris, has a low recovery yield and low specificity, and requires expensive laboratory equipment and highly trained technicians.58,59
Density-gradient separation
Density-gradient separation offers an improvement in purity and recovery rate over differential centrifugation. In density-gradient separation, a sample is spun in a tube that contains a density gradient of a viscous material, such that objects separate based on their isopycnic point.57 While this technique can achieve higher purity and recovery rate than conventional differential centrifugation,60 it cannot separate exosomes from viruses or microvesicles due to their similar buoyant density.57 The run times for density gradient separation are similar to conventional ultracentrifugation, and requires the same ultracentrifugation equipment, making it impractical for many clinical applications.3,57
Isolating exosomes using micro- and nano-systems
The micrometer-scale dimensions of microfluidics, and the control over microscale objects it affords,61 have great potential to improve the isolation of exosomes and microvesicles.16 In the last few years, new approaches have emerged to scale microfluidic techniques to the nanoscale for their application to microvesicles and exosomes. There are inherent challenges to this scaling, such as the expense of nanoscale lithography, the susceptibility of nanoscale channels to clogging, and the unfavorably strong scaling of many forces on objects in microfludics as they become nanoscale.62 These challenges have generated many new creative approaches.
Nanowire-on-micropillar
One particularly creative example of work in this field is by the Liu group at University of Texas. This group has developed a ciliated nanowire-on-micropillar structure to isolate exosomes, to achieve the nanoscale features necessary to trap exosomes using only conventional microfabrication techniques.63 Using silicon microfabrication, electroplating, and electroless metal-assisted nanowire etching techniques, they fabricated micropillars and etched porous silicon nanowire into the sidewalls of these micropillars to physically trap exosomes. They also demonstrated that the trapped exosomes can be released by dissolving the nanowires using PBS buffer. Because of the device’s microscale fluid channel, a small sample volume (~100 μl) can be efficiently processed. In addition, the high number of micropillars and nanowire decreases the chance of clogging, as the clogging of any particular part of the device will not dramatically affect overall device performance.
Acoustic sorting
Another promising method to sort exosomes and microvesicles in microfluidic devices is to use applied fields (e.g. acoustic) to exert forces on the exosomes and microvesicles, rather than nanofabricated physical barriers. The advantage of these methods is that they do not require nanolithography and are less susceptible to clogging, because the force fields are reconfigurable. Recently, the Lee and Weissleder group showed label-free purification of exosomes using an acoustic-based microfluidic device.64 Their acoustic nanofilter separates exosomes from other biological components based on their size. Particles in the applied acoustic field move toward the pressure nodes due to radiation forces, with a force proportional to the particle’s volume. Larger particles migrate faster across the microfluidic channel than smaller particles, enabling differently sized objects to be separated into different laminar flows. This device can achieve >80% recovery of exosomes and >90% for larger microvesicles from cell cultured media and the diameter of particles that it isolates can be adjusted electronically by adjusting the acoustic field or flow velocity.
Immunoaffinity-based isolation
In contrast to strategies that isolate exosomes or microvesicles based on their size, immunoaffinity-based approaches sort them instead by the expression of specific proteins on their surface. This approach has the advantage, compared to size-based purification, of reduced co-purification with cell-debris and protein aggregates as well as the ability to isolate specific subpopulations of exosomes or microvesicles based on the expression of a specific surface marker.57,65–67 One non-microfluidic example of immunoaffinity-based sorting of is the use of conventional magnetic activated cell sorting (MACS) columns.43 These columns are designed to separate cells directly from biological samples by taking advantage of strong magnetic forces on magnetic nanoparticle (MNP) labeled cells and the lack of magnetism of biological samples. The Taylor group repurposed MACS to isolate exosomes from serum samples from normal controls, patients with benign disease, and early stage ovarian cancer.43 In this technique, epithelial cell adhesion molecule (EpCAM) expressing exosomes were incubated with anti-EpCAM magnetic microbeads and trapped using a conventional MACS Separator. After the unlabeled material was passed through and discarded, the exosomes were released and collected. Using the enriched exosomes, they profiled exosomal miRNAs to see which ones were elevated in exosomes compared to cells. The levels of tumor-derived exosomes were found to increase with progression of ovarian cancer.
By performing immunoaffinity-based isolation of exosomes on a microfluidic chip, recovery rate can be improved and smaller sample volumes can be processed.61 The Toner group presented an immunoaffinity-based microfluidic device that rapidly and specifically isolates exosomes from cell cultured media or serum samples. The surface of the microfludic channel was modified to coat the surface with biotinylated anti-CD63, a pan-exosome marker. Exosomes were captured from serum by binding to the antibody. Since the isolation takes place in a handheld microfluidic device, the technique has potential for use as a point-of-care tool if coupled with a downstream analysis technique.
The Trau group also developed a device to isolate exosomes using immunoaffinity, with improved specificity using a novel method.67 Coined nanoshearing, on this chip an electro-hydrodynamic lateral fluid flow is generated within a few nanometers of an electrode surface to remove non-specifically bound exosomes from an immunocapture site. This nanoshearing technique resulted in a 3 fold improvement in specificity compared to conventional hydrodynamic flow and it showed limit of detection of >2760 exosomes per μl. On this chip, the captured exosomes were incubated with anti-fluorescein HRP antibody and tetramethylbenzidine (TMB) to induce colorimetric read-out. Therefore, the presence of the targeted exosomes can be detected using only the naked eye.
Another strategy that has been used to isolate exosomes and microvesicles is to make use of the forces on particles in microfluidic channels that naturally arise due to inertia. The DiCarlo group demonstrated a device that uses the inertial lift force to isolate exosomes from other biological particles.68 The DiCarlo group fabricated a high aspect ratio micro-channel, which creates an inertial lift force on traveling particles that forces them to the channel centerline. In this device, exosomes were captured onto 20 μm streptavidin microbeads using immunocapture. The inertia forces the 20 μm beads to travel to the centerline while other debris, smaller than 20 μm, are unaffected, enabling the beads to be separated. Downstream of this sorting module, each bead is detected by an integrated flow cytometer to quantify the captured exosomes. The combination of isolation and quantification gives this technique potential for practical clinical use.
Detection of exosomes and microvesicles
In this section, we discuss conventional detection of exosomes and microvesicles and new approaches that use micro- and nano-based devices to improve performance. The advantages and disadvantages of the detection techniques that we highlight are summarized in Table 3.
Table 3.
Summary of exosome and microvesicle detection technologies
| Technology | Size range | Specificity | Run time |
|---|---|---|---|
| Nanoparticle tracking analysis | 10 nm to 2 μm | Size and immunoaffinity | <1 hour |
| Dynamic light scattering45,73 | 0.3 nm to 10 μma | Size | <1 hour |
| Flow cytometry75 | 300 nm and above | Size and immunoaffinity | >1 hour |
| Electron microscopy45 | 0.1 nm and above | Size | >1 hour |
| Holographic imaging77 | 40 nm and above | Size | <1 hour |
| Nanopore81,85 | 10 nm and aboveb | Size | <1 hour |
| Magnetic resonance36 | NA | Immunoaffinity | <1 hour |
| Plasmonic93–95 | NA | Immunoaffinty | <1 hour |
Size measurement accuracy is sensitive to particle size homogeneity.
Dynamic range is a function of nanopore size.
Background: conventional detection techniques
Nanoparticle tracking analysis
Nanoparticle tracking allows exosomes and microvesicles to be sized and counted by combining light microscopy and software that analyzes the particles’ Brownian motion. The software tracks the motion of exosomes and microvesicles while they diffuse through the field-of-view, and calculates the diameter of the vesicle based on its rate of Brownian motion. Although the exosomes and microvesicles are smaller than the diffraction-limited resolution of the microscope, the particle’s size can be resolved by analyzing its motion. The Brownian motion can be related to particle size using the Stokes–Einstein relationship, which requires knowledge of only the temperature and the viscosity of the suspending fluid.69 Both light scattering and fluorescence modes have been demonstrated,70 with scattering used to measure size and fluorescence used to profile a labeled molecular marker. Nanoparticle tracking analysis has been used extensively to measure exosomes and microvesicles, and has quickly become a gold standard in the field.70–72 Malvern produces a commercial nanoparticle tracking product, branded the Nanosight, specified to measure particles in the size range of 10 nm–2 μm and concentrations within the range of 106 to 109 particles per mL (Malvern).
Dynamic light scattering (DLS)
In DLS, the diameter of microvesicles and exosomes is determined by measuring the dynamic changes in fluctuations from the scattering of coherent light (i.e. from a laser) through a suspension of exosomes and microvesicles.45,73 These changes arise from Brownian motion, which causes the distance between the light scattering vesicles to fluctuate. By quantifying the time-scale of the decay of this auto-correlation, the rate of diffusion of the particles and thereby the size of the particles, can be calculated.73
DLS is a commonly used laboratory tool, which in addition to microvesicles and exosomes, is used to characterize the size and concentration of a variety of particles including nanoparticles, polymers, and proteins.74 For exosomes and microvesicles, the DLS is specified to measure particles in the size range of 0.3 nm to 10 μm (Malvern). The DLS’s accuracy can be distorted by the presence of only a small number of large particles (e.g. platelets), and as such DLS measurements on exosomes require careful sample preparation.45,74 DLS is not amenable to molecular labeling, and as such cannot be used to profile molecular specific information about the exosomes or microvesicles.
Flow cytometry
In flow cytometry, the workhorse technique for high-throughput analysis of cells, each vesicle or exosome passes individually through a laser spot and its emitted scattered and fluorescent light is measured.75 Due to the 100× smaller size of exosomes and microvesicles relative to cells, the challenge in applying flow cytometry to exosomes and microvesicles is the difficulty of recovering such weak signals. As such, the disadvantage is that only particles larger than 300 nm can be resolved.45 The advantage of flow cytometry is that it allows individual exosomes to be resolved and it allows multiple surface markers to be measured per exosome.76 Flow cytometry machines now entering the market are pushing the detection limit to 100 nm and beyond, (e.g. A50-Micro-PLUS, Apogee) potentially expanding the utility of flow cytometers to analyze exosomes.
Transmission electron microscope (TEM)
Due to the diameter of exosomes being less than optical wavelengths, they cannot be resolved using conventional light microscope. Using the short wavelength of electrons, TEM can resolve individual exosomes.45 Complementary to the techniques described above, which can provide size, concentration, and analysis of surface markers, TEM imaging allows the morphology and heterogeneity of exosomes to be resolved. Extensive sample processing is required including dehydration, fixation, and metallization.
Micro and nano-based detection techniques
On-chip nano holographic imaging
The Ozcan group at UCLA recently demonstrated a handheld platform that can detect nanoscale objects in a mobile format that can be used at the point of medical care.77 Holographic imaging has been used previously for low-cost, cell-phone based high resolution imaging of cells.78 In holographic imaging, no lenses, lasers or other bulky optical components are required. On these platforms, instead of a coherent light source (i.e. laser) as is used in conventional holographic imaging, much less expensive and bulky light sources with short coherent times (i.e. LED) are used. The sample scatters and refracts the light from the LED, creating interference patterns that generate a hologram of each cell. This interference pattern expands to a size much larger than the cell and can be readout with a CCD or CMOS camera chip mounted below the sample. Based on the captured hologram, an image of the cell can be reconstructed. Super-resolution techniques of hologram imaging have been demonstrated, achieving resolution <1 μm.79
Recently, the Ozcan group has demonstrated an updated version of their holographic imaging system that achieves nanoscale resolution appropriate for exosome and microvesicle imaging.77 This system achieves nanoscale resolution by combining on-chip holographic microscopy with the addition of self-assembled nanolenses. An array of self-assembled nanolenses are constructed, which increase the scattering from the particle and direct the scattered light toward the image sensor. These lenses boost the signal of the particle’s holographic image, enabling the detection of nanoparticles as small as 40 nm. On this chip, Ozcan’s group has demonstrated imaging of synthetic nanoparticles and viruses. Compared to Nanoparticle Tracking Analysis (NTA), holographic imaging has several key advantages. The most relevant advantage is that holographic imaging can achieve similar results as NTA (resolution of particles as small as 40 nm), but in a handheld format that can be used in practical clinical settings. Additionally, because holographic imaging does not depend on the Brownian motion of the particles, it can measure objects larger than possible using NTA, up to 1 mm that diffuse too slowly for NTA to observe.
Nanopore ion occlusion based sensing
In resistive pulse sensing (RPS), single nanoparticles are measured as they are driven, one-be-one, through a nanopore.80 As the particle flows through the pore, it creates a transient change in the ionic current flow. The change in current is proportional to the size of the particle, enabling quantitative particle sizing with appropriate calibration. Tunable Resistive Pulse Sensing (tRPS) is an adaptation of resistive pulse-sensing, in which the size of the pore can be elastically stretched to the size of the particle to improve sensitivity.81
By measuring the duration, magnitude, and frequency of nanopore occlusions, exosome and microvesicle diameter and concentration can be determined for exosomes and microvesicles as small as 90 nm.80 However, there is a tradeoff between sensitivity to small exosomes and microvesicles and robustness against clogging.80 There are a variety of strategies that have been demonstrated in RPS that can enhance dynamic range in exosome and microvesicle measurement. For instance, the effective sample aperture can be reduced, without risking clogging, by using a low conductivity sheath flow.82,83 Additionally, by using two symmetric RPS channels on a microfluidic chip connected to a differential amplifier, nanopore occlusions as small as 1% could be resolved, enabling larger RPS channels to measure smaller exosomes, and thus extending dynamic range.83,84
Nanopore ion occlusion sensing has several key advantages compared to conventional techniques. Compared to techniques, such as DLS that measure the ensemble average of many exosomes and microvesicles, much smaller sample volumes (~100×) can be interrogated. Additionally, far lower concentrations of exosomes and microvesicles can be resolved, with the ultimate limit of detection being a single exosome or microvesicle. Compared to electron microscopy, which can also resolve individual exosomes and microvesicles, nanopore based measurements are much faster, less expensive, and higher throughput.
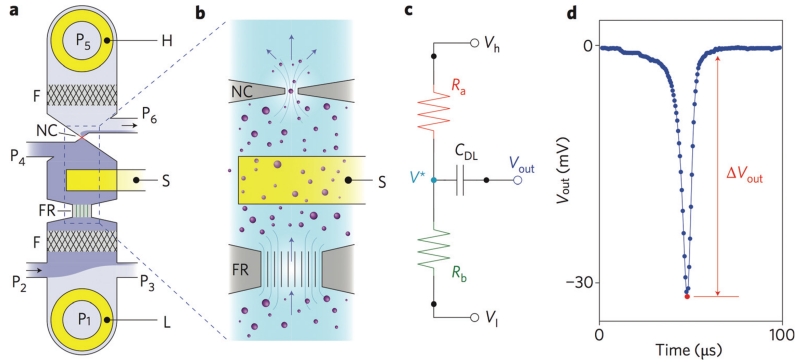
In one recent work, conventional RPS was improved upon by integrating a nanopore-occlusion based device into a microfabricated chip-based format. (Fig. 2a) The Cleland group at University of California at Santa Barbara developed a micro-machine-based ion occlusion device, in which a fluidic voltage divider (Fig. 2b and c) is used to optimize bandwidth and sensitivity to achieve a throughput of 500 000 particles per second (>10× improvement over conventional devices).80,85 Additionally, the microfluidic design enables volumes as small as 1 μL to be analyzed. On this chip, bacteriophage and nanoparticles were detected, (Fig. 2d) demonstrating great potential for application to exosomes and microvesicles. Compared to conventional RPS or tRPS techniques, the Cleland lab’s chip, due to its use of conventional fabrication techniques, has potential to be economically fabricated for use as a clinical diagnostic.80,85
Fig. 2.
Nanopore Cleland Group. a. Layout of the nanopore device. External voltage bias electrodes (H, L) and sensing electrode (S); embedded nanometre-scale filters (F); fluid resistor (FR); nanoconstriction (NC); pressure-regulated fluidic ports (P1–P6). b. Zoomed in view of the nanoparticle sensing components. Nanoparticles flow in the direction of the arrows, and change the electrical potential of the fluid adjacent to the nanoconstriction, which are detected by the sensing electrode S. c. Circuit model of the nanopore device. d. Vout as a function of time as a single nanoparticle traverses the nanopore.85
Diagnostic magnetic resonance for exosome detection
The Lee and Weissleder group at Massachusetts General Hospital have developed a miniaturized nuclear magnetic resonance based platform (μNMR) to sensitively measure exosomes.86 μNMR has previously been used with great success for the ultrasensitive measurements of rare cells,87 bacteria,88 soluble proteins,89 small molecules,89 and nucleic acid.90 The adoption of μNMR to the detection of exosomes was made challenging due to the much smaller size of exosomes relative to cells, resulting in far fewer surface markers that can be labeled. To overcome this drop in signal, the Weissleder/Lee group developed a microfluidic platform that was integrated with the μNMR system to concentrate and purify exosomes prior to the measurement, and thus boost the signal.36 On this chip, exosomes were first labeled with MNPs targeted to microvesicle protein markers. These exosomes were then captured onto a membrane filter where they were trapped based on their size, and subsequently washed. On this filter, μNMR detection of the surrounding H2 molecules was performed, and the change in the NMR dephasing time (T2 relaxation) due to the presence of MNPs in the detection region, was used to profile the exosomes.
The Weissleder and Lee technique had much higher sensitivity than conventional detection (>100× nanoparticle tracking analysis). To demonstrate its clinical utility, they used their device to differentiate glioblastoma multiforme (GBM) exosomes from healthy host cell derived exosomes. On their chip, they profiled the protein expression of these GBM derived exosomes and showed that they could detect tumor mutations, which were clinically relevant to glioblastoma treatment options.36 Following up on this work, the Weissleder and Lee group demonstrated in a next-generation chip the monitoring of drug-resistance in glioblastoma by analyzing the exosomal RNA.91
Plasmonic exosome detection
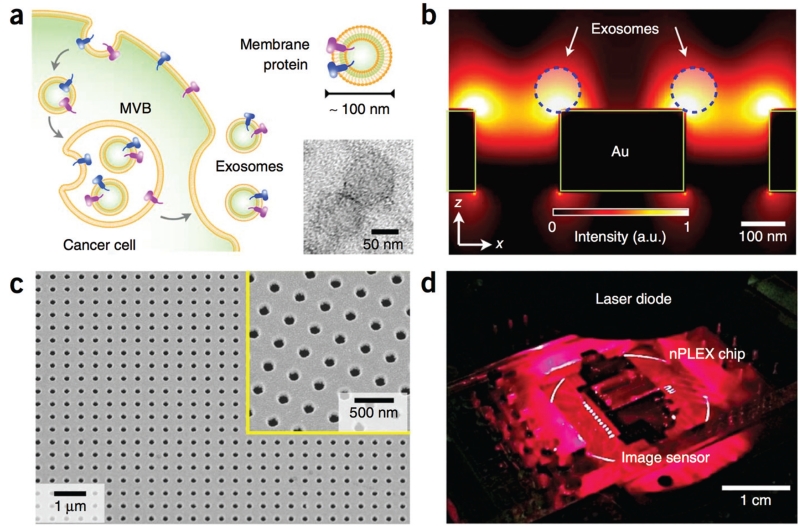
Surface Plasmon resonance (SPR)-based nanosensors have gained a great deal of attention in recent years for their ability to sensitively detect the binding of exceedingly small numbers of molecules, down to as few as 1 molecule.92 Because the region of sensitivity of these nanohole arrays extends only a small distance above the SPR surface ~200 nm, these sensors are extremely well suited for exosome and microvesicle detection.93–95 In a recent publication by the Weissleder/Lee group, they developed an SPR based exosome sensor, which they called the nano-plasmonic exosome (nPLEX).(Fig. 3a) The nPLEX consists of a series of nanohole arrays, each nanohole array coated with affinity ligands to capture a specific type of exosome.(Fig. 3d) Each nanohole had a diameter of 200 nm and a periodicity of 450 nm. When an exosome would bind to one of the nanopore arrays, it causes a spectral shift in the nanopore optical transmittance. (Fig. 3c) By combining these nanopore arrays with a miniaturized imaging setup (Fig. 3b), the Weisslder/Lee group demonstrated that their chip can be scaled for massively parallel measurements (105 independent nanopore arrays), with each nanopore array functionalized with a different affinity ligand. To demonstrate the utility of nPLEX, samples from ovarian cancer patients were measured and could be readily differentiated from healthy controls. Each measurement required a sample volume of only 0.3 μl.
Fig. 3.
Nanoplasmonic exosome detection. a. A scanning electron microscope (SEM) image of the periodic nanoholes in the nanoplasmonic sensor. Each nano-pore has a diameter of 200 nm with a periodicity of 450 nm, and consists of 200 nm thick gold. The inset shows a zoomed-in image. b. A photograph of the nanoplasmonic exosome sensor prototype. c. A schematic of changes in transmission spectra, illustrating nanoplasmonic exosome detection. The nanopore surface is functionalized by a layer of polyethylene glycol (PEG). Antibody conjugation and specific exosome binding were monitored by transmission spectral shifts as shown. (a.u.: arbitrary unit) d. SEM image shows exosome capture by functionalized nanoplasmonic device.93
Conclusions
Noninvasive diagnostics and monitoring of cancer continue to make inroads into the clinic, resulting in increased personalization of therapy, higher sensitive disease monitoring, and less patient discomfort from invasive procedures. Tumor-derived exosomes offer new clinical opportunities for minimally invasive diagnostics and disease monitoring, which complement existing circulating biomarkers. To further advance the development of exosome-based clinical assays, validation studies to establish the sensitivity, reproducibility, and specificity of such tests need to be conducted in large cohorts of patients and for multiple cancers of interest. Moreover, it will be critical to the field to establish the relative sensitivity and clinical utility of exosomes versus CTCs and cell-free DNA, and use further testing of patient samples to match the best type of test or tests to the clinical question being posed. Finally, our understanding of the biology of tumor-derived exosomes has only scratched the surface. As discoveries continue to be made as to the role of exosomes in promoting tumor growth and metastatic seeding, this will undoubtedly drive the development of new therapeutic strategies and more robust and sensitive tests to aid clinical decision making.
While the recent work highlighted in this review shows the promise of exosomes as a biomarker for cancer, the extensive processing that is required to isolate and detect them have limited their clinical use.3 The use of micro- and nano-technologies designed to overcome these technical challenges are ongoing. By enabling rapid sample preparation and molecular analyses from small sample volumes, these platforms can help fully harness the clinical potential of these circulating biomarkers in cancer and beyond.
Biographies

Jina is a graduate student of Bioengineering at the University of Pennsylvania. Her research focuses on the early detection of lethal diseases, such as pancreatic cancer and traumatic brain injury, using rare circulating cells and exosomes. She is interested in developing point-of-care platforms that are low cost, small, fast, and highly accessible. Her PhD work is done in strong collaboration with UPenn’s medical school, which is making it possible for her to rapidly translate her research into clinical testing and use.

Erica L. Carpenter, MBA, PhD, is the director of the Circulating Tumor Material Laboratory and Research Assistant Professor in the Department of Medicine, Division of Hematology/Oncology at the University of Pennsylvania. She completed her doctoral studies in the Immunology program of the Biomedical Graduate Studies program at the University of Pennsylvania School of Medicine and her postdoctoral training in the field of cancer genetics at the Oncology Division of the Children’s Hospital of Philadelphia. Prior to completing her studies in medical science, Dr Carpenter had obtained a Masters in Business Administration at the Tuck School at Dartmouth College, and worked as a business executive for a Fortune 50 manufacturer. Over the course of her business career, she had responsibility for divisions with revenues up to $2 billion, and participated in the management of both domestic and overseas business units. Dr Carpenter’s research programs focus on the identification, capture, and analysis of circulating tumor cells and cell-free DNA from cancer patients.

David is an Assistant Professor of Bioengineering and Electrical and Systems Engineering at the University of Pennsylvania. His research focuses on the integration of microelectronics, microfluidics, nanomaterials and molecular targeting, and their application to medicine. This multidisciplinary approach enables Issadore’s lab to explore new technologies to bring medical diagnostics from expensive, centralized facilities, directly to clinical and resource-limited settings for applications including early detection of pancreatic cancer, Tuberculosis diagnosis in patients co-infected with HIV, and prognosis of traumatic brain injury. His academic background in electrical engineering and applied physics (PhD, Harvard 2009) and his research experience in a hospital research laboratory (MGH) have prepared him to work and collaborate effectively on these inherently cross-disciplinary problems.
References
- 1.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. Nat. Rev. Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 2.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Biochim. Biophys. Acta, Gen. Subj. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Simpson RJ, Lim JWE, Moritz RL, Mathivanan S. Expert Review of Proteomics. 2009;6:267. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 4.Nagrath S, et al. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozkumur E, et al. Sci. Transl. Med. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muluneh M, Shang W, Issadore D. Adv. Healthcare Mater. 2014;3:1078–1085. doi: 10.1002/adhm.201300502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Issadore D, Westervelt RM. Point-of-care Diagnostics on a Chip. Springer; 2013. [Google Scholar]
- 8.Carpenter EL, Rader J, Ruden J, Rappaport EF, Hunter KN, Hallberg PL, Krytska K, O’Dwyer PJ, Mosse YP. Front. Oncol. 2014;4:201. doi: 10.3389/fonc.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Issadore D, Chung J, Shao H, Liong M, Ghazani AA, Castro CM, Weissleder R, Lee H. Sci. Transl. Med. 2012;4:141ra92. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee H, Sun E, Ham D, Weissleder R. Nat. Med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huh D, Gu W, Kamotani Y, Grotberg JB, Takayama S. Physiol. Meas. 2005;26:R73. doi: 10.1088/0967-3334/26/3/R02. [DOI] [PubMed] [Google Scholar]
- 12.Gawad S, Schild L, Renaud P. Lab Chip. 2001;1:76–82. doi: 10.1039/b103933b. [DOI] [PubMed] [Google Scholar]
- 13.Johnstone RM, Adam M, Pan BT. Can. J. Biochem. Cell Biol. 1984;62:1246–1254. doi: 10.1139/o84-159. [DOI] [PubMed] [Google Scholar]
- 14.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 15.Thakur BK, et al. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeringer E, Barta T, Li M, Vlassov AV. Cold Spring Harbor Protoc. 2015;2015 doi: 10.1101/pdb.top074476. pdb–top074476. [DOI] [PubMed] [Google Scholar]
- 17.Logozzi M, et al. PloS One. 2009;4(4):e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raposo G, Stoorvogel W. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeringer E, Li M, Barta T, Schageman J, Pedersen KW, Neurauter A, Magdaleno S, Setterquist R, Vlassov AV. World J. Methodol. 2013;3:11. doi: 10.5662/wjm.v3.i1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Nedawi K, Meehan B, Rak J. Cell Cycle. 2009;8:2014–2018. doi: 10.4161/cc.8.13.8988. [DOI] [PubMed] [Google Scholar]
- 21.Haber DA, Velculescu VE. Cancer Discovery. 2014;4:650–661. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pantel K, Alix-Panabieres C. Cancer Res. 2013;73:6384–6388. doi: 10.1158/0008-5472.CAN-13-2030. [DOI] [PubMed] [Google Scholar]
- 23.Schwarzenbach H, Hoon DSB, Pantel K. Nat. Rev. Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 24.Gaster RS, Xu L, Han S-J, Wilson RJ, Hall DA, Osterfeld SJ, Yu H, Wang SX. Nat. Nanotechnol. 2011;6:314–320. doi: 10.1038/nnano.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pekin D, et al. Lab Chip. 2011;11:2156–2166. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- 26.Sardesai NP, Barron JC, Rusling JF. Anal. Chem. 2011;83:6698–6703. doi: 10.1021/ac201292q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chin CD, et al. Nat. Med. 2011;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 28.Lang JM, Casavant BP, Beebe DJ. Sci. Transl. Med. 2012;4:141ps13. doi: 10.1126/scitranslmed.3004261. [DOI] [PubMed] [Google Scholar]
- 29.U.S. Preventive Services Task Force Ann. Intern. Med. 2008;149:185. [Google Scholar]
- 30.Kosaka N, Iguchi H, Ochiya T. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allard B, Sandra P, Smyth MJ, Stagg J. Clin. Cancer Res. 2013;19:5626–5635. doi: 10.1158/1078-0432.CCR-13-0545. [DOI] [PubMed] [Google Scholar]
- 32.Khoja L, Lorigan P, Zhou C, Lancashire M, Booth J, Cummings J, Califano R, Clack G, Hughes A, Dive C. J. Invest. Dermatol. 2013;133:1582–1590. doi: 10.1038/jid.2012.468. [DOI] [PubMed] [Google Scholar]
- 33.Melo SA, et al. Nature. 2015;523:177. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa-Silva B, et al. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peinado H, et al. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Nat. Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Pancreas. 1998;17:89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 38.Bettegowda C, et al. Sci. Transl. Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. Clin. Chim. Acta. 2001;313:139–142. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 40.Clayton A, Court J, Navabi H, Adams M, Mason MD, Hobot JA, Newman GR, Jasani B. J. Immunol. Methods. 2001;247:163–174. doi: 10.1016/s0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- 41.Kahlert C, et al. J. Biol. Chem. 2014;289:3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, Widmark A. Br. J. Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor DD, Gercel-Taylor C. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 44.Grant R, Ansa-Addo E, Stratton D, Antwi-Baffour S, Jorfi S, Kholia S, Krige L, Lange S, Inal J. J. Immunol. Methods. 2011;371:143–151. doi: 10.1016/j.jim.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 45.Van DP, Hoekstra EAG, Sturk A, Otto C, Van L, Nieuwland TGR. J. Thromb. Haemostasis. 2010;8:2596–2607. doi: 10.1111/j.1538-7836.2010.04074.x. [DOI] [PubMed] [Google Scholar]
- 46.Dawson S-J, et al. N. Engl. J. Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 47.Balaj L, Lessard R, Dai L, Cho Y-J, Pomeroy SL, Breakefield XO, Skog J. Nat. Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ignatiadis M, et al. PloS One. 2011;6:e15624. doi: 10.1371/journal.pone.0015624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gazdar AF. Oncogene. 2009;28:S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carpenter EL, Mosse YP. Nat. Rev. Clin. Oncol. 2012;9:391–399. doi: 10.1038/nrclinonc.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park JO, et al. Proteomics. 2013;13:2125–2134. doi: 10.1002/pmic.201200323. [DOI] [PubMed] [Google Scholar]
- 52.Runz S, Keller S, Rupp C, Stoeck A, Issa Y, Koensgen D, Mustea A, Sehouli J, Kristiansen G, Altevogt P. Gynecol. Oncol. 2007;107:563–571. doi: 10.1016/j.ygyno.2007.08.064. [DOI] [PubMed] [Google Scholar]
- 53.Beckham CJ, Olsen J, Yin P-N, Wu C-H, Ting H-J, Hagen FK, Scosyrev E, Messing EM, Lee Y-F. J. Urol. 2014;192:583–592. doi: 10.1016/j.juro.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 54.Klapman J, Malafa MP. Cancer Control. 2008;15:280–287. doi: 10.1177/107327480801500402. [DOI] [PubMed] [Google Scholar]
- 55.Rhim AD, et al. Gastroenterology. 2014;146:647–651. doi: 10.1053/j.gastro.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sopik V, Rosen B, Giannakeas V, Narod SA. Gynecol. Oncol. 2015;138:757–761. doi: 10.1016/j.ygyno.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 57.Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 58.Thery C, Amigorena S, Raposo G, Clayton A. Curr. Protoc. Cell Biol. 2006:3–22. doi: 10.1002/0471143030.cb0322s30. DOI: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 59.Alvarez ML, Khosroheidari M, Ravi RK, DiStefano JK. Kidney Int. 2012;82:1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Z, Wang C, Li T, Liu Z, Li L. Oncol. Lett. 2014;8:1701–1706. doi: 10.3892/ol.2014.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen C, Skog J, Hsu C-H, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M, Irimia D. Lab Chip. 2010;10:505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Napoli M, Eijkel JCT, Pennathur S. Lab Chip. 2010;10:957–985. doi: 10.1039/b917759k. [DOI] [PubMed] [Google Scholar]
- 63.Wang Z, Wu H.-j., Fine D, Schmulen J, Hu Y, Godin B, Zhang JXJ, Liu X. Lab Chip. 2013;13:2879–2882. doi: 10.1039/c3lc41343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee K, Shao H, Weissleder R, Lee H. ACS Nano. 2015;9:2321–2327. doi: 10.1021/nn506538f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bobrie A, Colombo M, Krumeich S, Raposo G, Thery C. J. Extracell. Vesicles. 2012;1:18397. doi: 10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathivanan S, Lim JWE, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Mol. Cell. Proteom. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vaidyanathan R, Naghibosadat M, Rauf S, Korbie D, Carrascosa LG, Shiddiky MJA, Trau M. Anal. Chem. 2014;86:11125–11132. doi: 10.1021/ac502082b. [DOI] [PubMed] [Google Scholar]
- 68.Dudani JS, Gossett DR, Henry TK, Lamm RJ, Kulkarni RP, Di Carlo D. Biomicrofluidics. 2015;9:014112. doi: 10.1063/1.4907807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mason TG, Ganesan K, van Zanten JH, Wirtz D, Kuo SC. Phys. Rev. Lett. 1997;79:3282. [Google Scholar]
- 70.Dragovic RA, et al. Nanomed.: Nanotechnol., Biol. Med. 2011;7:780–788. [Google Scholar]
- 71.Soo CY, Song Y, Zheng Y, Campbell EC, Riches AC, Gunn-Moore F, Powis SJ. Immunology. 2012;136:192–197. doi: 10.1111/j.1365-2567.2012.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gercel-Taylor C, Atay S, Tullis RH, Kesimer M, Taylor DD. Anal. Biochem. 2012;428:44–53. doi: 10.1016/j.ab.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 73.Berne BJ, Pecora R. Dynamic light scattering with applications to chemistry, biology, and physics. Courier Corporation; 2000. [Google Scholar]
- 74.Filella M, Zhang J, Newman ME, Buffle J. Colloids Surf., A. 1997;120:27–46. [Google Scholar]
- 75.Lyons AB, Parish CR. J. Immunol. Methods. 1994;171:131–137. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 76.Orozco AF, Lewis DE. Cytometry, Part A. 2010;77:502–514. doi: 10.1002/cyto.a.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McLeod E, Dincer TU, Veli M, Ertas YN, Nguyen C, Luo W, Greenbaum A, Feizi A, Ozcan A. ACS Nano. 2015;9:3265–3273. doi: 10.1021/acsnano.5b00388. [DOI] [PubMed] [Google Scholar]
- 78.Tseng D, Mudanyali O, Oztoprak C, Isikman SO, Sencan I, Yaglidere O, Ozcan A. Lab Chip. 2010;10:1787–1792. doi: 10.1039/c003477k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bishara W, Sikora U, Mudanyali O, Su T-W, Yaglidere O, Luckhart S, Ozcan A. Lab Chip. 2011;11:1276–1279. doi: 10.1039/c0lc00684j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maas SLN, De Jeroen V, Broekman MLD. J. Visualized Exp. 2014;92:51623. [Google Scholar]
- 81.Platt M, Willmott GR, Lee GU. Small. 2012;8:2436–2444. doi: 10.1002/smll.201200058. [DOI] [PubMed] [Google Scholar]
- 82.Nieuwenhuis JH, Kohl F, Bastemeijer J, Sarro PM, Vellekoop MJ. Sens. Actuators, B. 2004;102:44–50. [Google Scholar]
- 83.Pol E, Coumans F, Varga Z, Krumrey M, Nieuwland R. J. Thromb. Haemostasis. 2013;11:36–45. doi: 10.1111/jth.12254. [DOI] [PubMed] [Google Scholar]
- 84.Wu X, Kang Y, Wang Y-N, Xu D, Li D, Li D. Electrophoresis. 2008;29:2754–2759. doi: 10.1002/elps.200700912. [DOI] [PubMed] [Google Scholar]
- 85.Fraikin J-L, Teesalu T, McKenney CM, Ruoslahti E, Cleland AN. Nat. Nanotechnol. 2011;6:308–313. doi: 10.1038/nnano.2011.24. [DOI] [PubMed] [Google Scholar]
- 86.Issadore D, Min C, Liong M, Chung J, Weissleder R, Lee H. Lab Chip. 2011;11:2282–2287. doi: 10.1039/c1lc20177h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Castro CM, Ghazani AA, Chung J, Shao H, Issadore D, Yoon T-J, Weissleder R, Lee H. Lab Chip. 2014;14:14–23. doi: 10.1039/c3lc50621e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liong M, Fernandez-Suarez M, Issadore D, Min C, Tassa C, Reiner T, Fortune SM, Toner M, Lee H, Weissleder R. Bioconjugate Chem. 2011;22:2390–2394. doi: 10.1021/bc200490r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perez JM, Josephson L, O’Loughlin T, Hogemann D, Weissleder R. Nat. Biotechnol. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- 90.Liong M, Im H, Majmudar MD, Aguirre AD, Sebas M, Lee H, Weissleder R. Adv. Healthcare Mater. 2014;3:1015–1019. doi: 10.1002/adhm.201300672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R. Nat. Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kravets VG, et al. Nat. Mater. 2013;12:304–309. doi: 10.1038/nmat3537. [DOI] [PubMed] [Google Scholar]
- 93.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H. Nat. Biotechnol. 2014;32:490–495. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu L, et al. Anal. Chem. 2014;86:8857–8864. doi: 10.1021/ac5023056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rupert DLM, Lässer C, Eldh M, Block S, Zhdanov VP, Lotvall JO, Bally M, Höök F. Anal. Chem. 2014;86:5929–5936. doi: 10.1021/ac500931f. [DOI] [PubMed] [Google Scholar]