Abstract
The DNA damage response (DDR) is a molecular mechanism that cells have evolved to sense DNA damage (DD) to promote DNA repair, or to lead to apoptosis, or cellular senescence if the damage is too extensive. Recent evidence indicates that microRNAs (miRs) play a critical role in the regulation of DDR. Dietary bioactive compounds through miRs may affect activity of numerous genes. Among the most studied bioactive compounds modulating expression of miRs are epi-gallocatechin-3-gallate, curcumin, resveratrol and n3-polyunsaturated fatty acids. To compare the impact of these dietary compounds on DD/DDR network modulation, we performed a literature search and an in silico analysis by the DIANA-mirPathv3 software. The in silico analysis allowed us to identify pathways shared by different miRs involved in DD/DDR vis-à-vis the specific compounds. The results demonstrate that certain miRs (e.g., -146, -21) play a central role in the interplay among DD/DDR and the bioactive compounds. Furthermore, some specific pathways, such as “fatty acids biosynthesis/metabolism”, “extracellular matrix-receptor interaction” and “signaling regulating the pluripotency of stem cells”, appear to be targeted by most miRs affected by the studied compounds. Since DD/DDR and these pathways are strongly related to aging and carcinogenesis, the present in silico results of our study suggest that monitoring the induction of specific miRs may provide the means to assess the antiaging and chemopreventive properties of particular dietary compounds.
Keywords: food, bioactive compounds, n3-PUFA, resveratrol, curcumin, epi-gallocatechin-3gallate, chemoprevention
1. Introduction
The DNA in each of our cells accumulates thousands of lesions every day. Cells are continuously challenged by DNA damage stimuli from various exogenous environmental factors, such as ultraviolet (UV) radiation, ionizing radiation (IR) and numerous chemical agents, or endogenous sources mainly represented by products of cellular metabolism. DNA damage can interfere with essential cellular processes, such as transcription or replication, and can compromise the viability of the cell. Specific DNA lesions can also induce mutations that cause cancer or other diseases, as well as contribute to the aging process [1]. DNA damage at the telomeric chromosomal sections definitely has pro-aging consequences [2]. Fortunately, cells have developed elaborate and efficient response pathways to preserve genomic stability. The DNA damage response (DDR) [3] is an evolutionarily-conserved signaling cascade activated by DNA damage (DD), which directs cell fate toward DNA repair, senescence or apoptosis [4]. DDR is a signaling network initiated by lesion recognition and amplified by multiple mediator signaling proteins, which eventually activate downstream effectors to modulate cell fate. Multiple DNA repair pathways have evolved to resolve various DNA lesions, including: base excision repair that removes damaged bases; mismatch repair that recognizes base incorporation errors and base damage; nucleotide excision repair that removes bulky DNA adducts; and cross-link repair that removes inter-strand cross-links. In addition, breaks in the DNA backbone are repaired via double-strand break (DSB) repair pathways, including homologous recombination (HR) and nonhomologous end joining [5]. If DNA lesions are not properly repaired or amended during replication, they can be converted into permanent mutations. When this occurs at the sites of oncogenes or tumor suppressor genes, the risk of neoplastic cell transformation is significantly increased [1,6]. In higher organisms, successful DDR is thought to prevent neoplastic transformation in a cell-autonomous manner, by ensuring removal of cells with severely damaged DNA [7]. DDR signaling has been suggested as a key mechanism linking DNA damage accumulation, cell senescence and organismal aging. In fact, during the cell lifetime, the genomic DNA is continuously exposed to different exogenous or endogenous factors that destabilize its integrity and functionality. Therefore, genomic instability and decline of DNA repair efficiency is considered one of the main drivers of the aging process [8]. However, emerging data suggest that DDR signaling can also work through a paracrine/systemic mechanism, shaping the systemic environment through the regulation of tissue repair and immune responses [9]. Given the fundamental role of DDR in maintaining genome integrity, this complex signaling network requires accurate regulatory mechanisms to respond to different types of DNA lesions in different stages of the cell cycle.
Recently, microRNAs (miRs) have emerged as important players in regulatory networks affecting the DNA damage/repair process in a wide range of physiological and pathological conditions [5,10]. miRs are 18–25 nucleotide non-coding RNAs that post-transcriptionally regulate gene expression stalling the translation of the cognate mRNA or promoting its degradation [11]. miRs have been identified to influence physiological processes, such as development, growth and differentiation [12], and have also been implicated in a wide range of diseases [13]. Multiple miRs may target the same miR, and the majority of miRs contain multiple binding sites for miRs, generating a highly complex regulatory network system by which hundreds of genes involved in different signaling pathways can be regulated simultaneously [14].
Nutrients and their bioactive compounds can modulate the miRs’ expression involved in many physiological and pathological processes [15]. Nutrition is the process that offers different substances to an organism that can work as energy suppliers (carbohydrate and fat), as cell structure sources (proteins) and on metabolism control (vitamins and minerals), thereby maintaining its homeostasis. The importance of diet and nutrition in human health and disease is well established. Basic laboratory research, clinical trials and epidemiological studies demonstrated that nutrient-rich bioactive foods can induce epigenetic changes and alter genes’ expression by the alteration of the histone structure, DNA methylation and miRs’ modulation [16].
Epi-gallocatechin-3-gallate (EGCG), resveratrol (RSV), curcumin (CRC) and n3-PUFA (n3-polyunsaturated fatty acids) are among the most studied compounds shown to have beneficial effects on human health [17,18]. EGCG, RSV and CRC are polyphenols present in fruits and vegetables. n3-PUFAs, are polyunsaturated fatty acids found in plants, as linolenic acid (ALA), or in fish eicosapentaenoic acid (EPA); and docosahexaenoic acid (DHA). Each of these four dietary compounds, at a concentration that potentially may be achievable in the organism, has been shown to suppress cell proliferation and induce apoptosis in certain types of human cancer cells [18,19]. In addition to potential anticancer activity, they possess cardiovascular protective properties [20,21] and beneficial effects on degenerative diseases [22]. Furthermore, an anti-inflammatory and antioxidant activity has been frequently associated with these compounds [23]. Most importantly, all of them may exert these effects by modulating miRs’ expressions [24,25]. In order to investigate and compare the impact of these specific food-derived compounds on DDR processes, we performed a literature search to identify miRs involved in DD/DDR and modulated by these dietary compounds. An in silico analysis using the DIANA software web-server was applied to identify targets and pathways that play a major role in the DD/DDR modulation by these compounds [26]. The results of the analysis of the pathways allowed us to speculate how food intervention could modulate DD/DDR.
2. Results
2.1. miRs Involved in DD/DDR and Bioactive Compounds Modulated
The results of a literature search for miRs involved in DD/DDR processes are reported in Table 1. Table 2 shows the literature search results for miRs modulated by each of the four compounds: EGCG, CRC, RSV and n3-PUFA, including the tissue/cell type, dose/concentration and duration of exposure of cells/tissue to the compound used in the cited study. Most of the studies we found with our search criteria (see Materials and Methods) have been performed in vitro and most of them on different human cancer cells.
Table 1.
MicroRNAs involved in DD/DDR processes.
| miRs | Reference |
|---|---|
| miR-34a/b/c, miR-192, miR-215, miR-16-1, miR-143, miR-107, let-7, miR-200c, miR-16, miR-145, miR-134, miR-449a/b, miR-503, miR-21, miR-24, miR-421, miR-504, miR-125b, miR-106b, miR-21, miR-210, miR-373, miR-100, miR-195, miR-124a, miR-290 cluster (miR-291-3p, miR-294, miR-295) | [5,27] |
| miR-363, miR-25, miR-542 | [28] |
| miR-421, miR-24, miR-34a/b/c, miR-504, miR-125b, miR-302, miR-92, miR-192, miR-194, miR-215, miR-106a-92 cluster (miR-106a, miR-18b, miR-20b, miR-19b-2, miR-92a-2, miR-363), miR-106b/25 cluster (miR-106b, miR-25, miR-93), miR-210, miR-128, miR-20, miR-130b, miR-143, miR-145, miR-16-1, miR-16, miR-103, miR-26a, miR-206 | [27] |
| miR-15a, miR-29, miR-107, miR-605, miR-17-92 cluster (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, miR-92a-1), miR-21-605, miR-221, miR-222, miR-138, miR-223, miR-181a, miR-27a, miR-214, miR-101, miR-185, miR-100, miR-506, miR-545, miR-124, miR-9, miR-182, miR-146a | [10] |
Table 2.
MicroRNAs modulated by bioactive compounds; the effect on human normal or cancer cells.
| Compound | miRNA | Reference | Cells/Cancer | Dose/Duration |
|---|---|---|---|---|
| EGCG | miR-18, miR-16, let-7a, miR-221, miR-34b, miR-193b, miR-222, miR-342 | [29] | hepatic cancer | 100 μM, 24 h |
| miR-636, miR-3907 | [30] | normal dermal fibroblasts | 10 μM, 24 h | |
| miR-200c | [31] | colorectal cancer cells and colon cancer stem cells | 100 μM, 24 h | |
| miR-210, miR-98-5p | [32,33] | lung cancer | 40 μM, 9 h 10 μM, 24 h | |
| miR-1, miR-126 | [34,35] | osteosarcoma | 0.08 g/L, 48 h 0.2 g/L, 72 h | |
| miR-194 | [36] | hepatocarcinoma | 10 μg/mL, 48 h | |
| miR-7-1, miR-34a, miR-99a, miR-92, miR-93, miR-106b | [37,38] | neuroblastoma | 50 μM, 24 h | |
| miR-25, miR-92, miR-141, miR-200a | [39] | Hela cells and lymphoblasts | 1–5 μM, 24 h | |
| miR-33a, miR-122 | [40] | hepatocarcinoma | 50 μM, 1 h | |
| EGCG | miR-92, miR-93, miR-106b, miR-7-1, miR-34a, miR-99a | [41] | neuroblastoma | 50 μM, 24 h |
| miR-467bn, miR-487b, miR-197, miR-805, miR-374n, let-7f, miR-350, miR-24-1n, miR-137, miR-335-3p, let-7a, miR-222, miR-26b, miR-30c-1n, let-7d, miR-98, miR-30c, miR-30bn, miR-32, miR-674n, miR-532-5p, let-7g, miR-18a, miR-192, miR-302d, miR-30b, miR-802, let-7e, miR-322, miR-720, miR-146b, miR-340-3p, miR-185, miR-425, miR-10a, miR-126-5p, miR-101a, miR-30en, let-7c, miR-141, miR-33, miR-29an, miR-199b, miR-450a-5p, miR-21, miR-23a, miR-101b, miR-148a, miR-193, miR-23b, miR-107, miR-140, miR-551b, miR-466c-5p, miR-106a, miR-590-3p, miR-875-3p, miR-224, miR-292-5p, miR-678, miR-469, let-7bn, miR-463n, miR-574-3p, miR-201, miR-290-3p, miR-181a, miR-302a, miR-429, miR-133a, miR-190b, miR-710, miR-135b, miR-296-5p, miR-191n, miR-188-5p, miR-298, miR-181a-1n, miR-466g, miR-26bn, miR-466f-3p, miR-29bn, miR-1224, miR-291b-5p, miR-324-5p, miR-486, miR-128, miR-450b-3p, miR-135an, miR-294, miR-671-5p, miR-878-3p, miR-801, miR-370, miR-1, miR-494, miR-133b | [41] | hepatocarcinoma | 100 μM, 24 h | |
| CRC | miR-192-5p/215 | [42] | lung cancer | 15 μM, 48 h |
| miR-7 | [43] | pancreatic cancer | 3–6 μM, 72 h | |
| miR-22 | [44,45] | retinoblastoma | 20 μM, 48 h | |
| miR-27a | [46] | colon cancer | 2.5–10 μg/mL, 24 h | |
| miR-21 | [47] | |||
| let-7a, miR-21, miR-34a | [48] | esophageal cancer | 30 μM, 24 h | |
| miR-221 | [44,49] | pancreatic cancer | 500 nM of synthetic CRC analogue, 72 h | |
| CRC | miR-27a, miR-20a, miR-17-5p, miR-21 | [41] | colon carcinoma | 30 μM, 24 h |
| miR-203 | [41] | bladder carcinoma | 10 μM, 3 days | |
| miR-320, miR-26a, let-7i, miR-130a, miR-16, miR-125b, miR-23a, miR-27b, miR-155, miR-625, miR-576-3p, miR-186n, miR-9n, let-7i | [50] | lung adenocarcinoma | 15 μM, 48 h | |
| miR-15a, miR-16-1 | [41] | leukemic cells | 5–20 μM, 24-72 h | |
| miR15a, miR-16 | [51] | breast cancer | 10–60 μM, 24 h | |
| miR-103, miR-140, miR-146b, miR-148a, miR-15b, miR-181a, miR-181b, miR-181d, miR-195, miR-196a, miR-199an, miR-19a, miR-204, miR-20a, miR-21, miR-22, miR-23a, miR-23b, miR-24, miR-25, miR-26a, miR-27a, miR-34a, miR-374, miR-510, miR-7, miR-92, miR-93, miR-98 | [41] | pancreatic cancer | 10 μM, 72 h | |
| RSV | miR-663, miR-744m | [44,52] | breast cancer | 100 μM, 24 h |
| miR-21 | [44,53] | pancreatic cancer | 50 μM, 24 h | |
| miR-520h | [54] | lung cancer | 10–20 μM, 48 h | |
| miR-21, miR-181b, miR-663, miR-30c2 | [55] | peripheral blood mononuclear cells from hypertensive patients | RSV (8 mg) grape extract, one year daily intake ( in vivo study) | |
| miR-150, miR-296-5p | [56] | lymph node cancer prostate | 50 μM, 24 h | |
| miR-33a, miR-122 | [40] | hepatocarcinoma | 50 μM, 1 h | |
| miR-155 miR-663 | [57] | monocytic cells | 30–50 μM, 14 h | |
| RSV | miR-155, miR-34a | [58] | EBV-immortalized B cells | 25–50 μM, 24 h |
| miR-7, miR-17, miR-18b, miR-20a, miR-20b, miR-92b, miR-106a, miR106b, miR-17-5p, miR-20a, miR-106b, miR-17-92 cluster, miR-106ab clusters | [59] | prostate cancer | 50–100 μM, 24 h | |
| miR-622 | [41] | bronchial epithelial cells | 50 μM, 48 h | |
| miR-155, miR-633 | [41] | monocytes | 30 μM, 14 h | |
| let-7c, miR-106a, miR-106b, miR-1224-5p, miR-1228, miR-231, miR-1246, miR-1260, miR-1267, miR-1268, miR-129, miR-1290, miR-1308, miR-1469, miR-149, miR-150, miR-152, miR-15a, miR-17, miR-1825, miR-185, miR-18b, miR-1908, miR-1915, miR-197, miR-1972, miR-1973, miR-1974, miR-1975, miR-1977, miR-1979, miR-20a, miR-20b, miR-24, miR-296-5p, miR-483-5p, miR-513a-5p, miR-548q, miR-572, miR-575, miR-612, miR-638, miR-654-5p, miR-659, miR-671-5p, miR-7, miR-762, miR-764, miR-874, miR-92b, miR-939 | [41] | lymph node cancer prostate | 50 μM, 48 h | |
| miR-1, miR-100-1/2, miR-102, miR-103-1, miR-103-2, miR-146a, miR-146b-5p, miR-16-0, miR-17, miR-181a2, miR-194-2, miR-196a1, miR-205, miR-206, miR-21, miR-23a, miR-23b, miR-25, miR-26a, miR-29c, miR-30a-3p, miR-30c-1, miR-30d, miR-30e-5p, miR-323, miR-340, miR-363n-5p, miR-424, miR-494, miR-497, miR-560, miR-560, miR-565, miR-565, miR-572, miR-574, miR-594, miR-615, miR-622, miR-629, miR-631, miR-638, miR-639, miR-657, miR-659, miR-663, miR-801, miR-92a-2 | [41] | colorectal carcinoma | 50 μM, 14 h | |
| n3-PUFA | miR-192, miR-30c, miR-141-3p, miR-221-3p, miR-1283, let-7f, miR-181a-5p, miR-1, miR-30a | [60] | Caco-2 cells | 200 μM DHA in lipid micelles, 24 h |
| n3-PUFA | miR-26a, miR-26b | [61] | cholangiocarcinoma | 50 μM DHA, 12 h |
| miR-221 | [62] | endothelial progenitor cells | 25–125 μM EPA, 4 h | |
| miR-146, miR-181a | [63] | glioma | 25–50 μM DHA, 48 h | |
| miR-21 | [64] | breast cancer | 152 nM DHA, 24 h | |
| miR-30c, miR-20b, miR-16, miR-22, miR-145, miR-34, miR-25, miR-17, miR-26a, miR-29c, miR-200a, miR-206, miR-323, miR-16, miR-22, miR-20b, miR-30c, miR-183, miR-224, miR-145, miR-181a, miR-208, miR-143, miR-20a, miR-149, miR-125b | [19,65] | glioma | 50–100 μM DHA, 24 h |
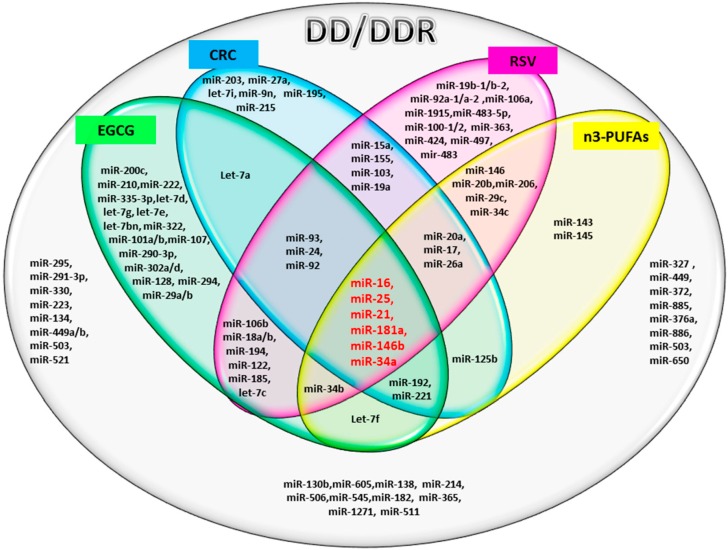
The Venn diagram in Figure 1, shows the common and distinct miRs modulated by bioactive compounds and DD/DDR processes. The literature analysis indicates that a large number of the DD-associated miRs can be modified by dietary bioactive compounds. Furthermore, this analysis also revealed that the expression of some miRs seems to be compound class specific, while others miRs seem to be modulated by more than one bioactive compound. Interestingly, we found six miRs that were common to all of the compounds (indicated in red in Figure 1).
Figure 1.
Venn diagram showing the microRNAs involved in DD/DDR (ellipse, grey) and identified as modulated by bioactive compounds: EGCG (epi-gallocatechin-3-gallate; green), CRC (curcumin; blue), RSV (resveratrol; pink) and n3-PUFAs (n3-polyunsaturated fatty acids, yellow). The common miRNAs, modulated by all four compounds, are indicated in red (miR-16, miR-25, miR-21, miR-181a, miR-146b, miR-34a).
2.2. In Silico Analysis of Pathways Shared by Different miRs Involved in DD/DDR and Modulated by Compounds
For the in silico analyses reported in Table 3, Table 4, Table 5, Table 6 and Table 7 and Figure 2, Figure 3, Figure 4, Figure 5 and Figure 6, common miRNAs between Table 1 and Table 2 and reported in Venn diagram (Figure 1) were used.
Table 3.
Enriched KEGG pathways for predicted targets of the DD/DDR microRNAs modulated by all four compounds.
| KEGG Pathway | p-Value | #of Genes | miRNAs |
|---|---|---|---|
| Fatty acid biosynthesis | 0 | 4 | miR-16 |
| Fatty acid metabolism | 2.23 × 10-5 | 7 | miR-16 |
| Thyroid hormone synthesis | 2.58 × 10-5 | 5 | miR-146b |
| Signaling pathways regulating pluripotency of stem cells | 0.0002346931 | 24 | miR-16 |
| Glioma | 0.000556152 | 15 | miR-16 miR-34a |
| Glycosphingolipid biosynthesis: lacto and neolacto series | 0.001440903 | 2 | miR-34a |
| Hippo signaling pathway | 0.00410767 | 24 | miR-16 miR-21 |
| Steroid hormone biosynthesis | 0.009497578 | 1 | miR-25 |
| Ovarian steroidogenesis | 0.01398717 | 1 | miR-25 |
| Melanoma | 0.01891852 | 14 | miR-16 |
| Prostate cancer | 0.02681897 | 16 | miR-16 |
| Cytokine-cytokine receptor interaction | 0.03260184 | 11 | miR-21 |
| mTOR signaling pathway | 0.03417768 | 13 | miR-16 miR-25 |
| Oocyte meiosis | 0.03995201 | 16 | miR-16 |
DIANA-miRPath v3.0 [66] was used to predict the targeted KEGG pathways by miRs involved in DD/DDR signaling and modulated by all considered compounds (EGCG, CRC, RSV and n3-PUFAs). The target prediction threshold was set at 0.85. p-value < 0.05. #: number.
Table 4.
Enriched KEGG pathways for predicted targets of the DD/DDR microRNAs modulated by EGCG.
| KEGG Pathway | p-Value | # of Genes | # of miRNAs |
|---|---|---|---|
| Fatty acid biosynthesis | 0 | 4 | 3 |
| ECM-receptor interaction | 0 | 42 | 13 |
| Signaling pathways regulating pluripotency of stem cells | 2.44 × 10−9 | 76 | 17 |
| Amebiasis | 3.53 × 10−6 | 24 | 6 |
| Proteoglycans in cancer | 1.17 × 10−4 | 117 | 12 |
| Mucin type O-glycan biosynthesis | 1.67 × 10−2 | 15 | 10 |
| Glioma | 1.43 × 10−1 | 39 | 10 |
| TGF-β signaling pathway | 1.85 × 100 | 40 | 8 |
| Fatty acid metabolism | 2.28 × 101 | 9 | 3 |
| Focal adhesion | 0.0002057474 | 110 | 7 |
| PI3K-Akt signaling pathway | 0.001829865 | 138 | 7 |
| Lysine degradation | 0.006232265 | 18 | 7 |
| ErbB signaling pathway | 0.02315742 | 45 | 7 |
| Protein digestion and absorption | 0.02773474 | 26 | 3 |
| Thyroid hormone signaling pathway | 0.03049808 | 57 | 8 |
| Glycosaminoglycan biosynthesis heparan sulfate/heparin | 0.05012798 | 9 | 5 |
DIANA-miRPath v3.0 was used to predict the targeted KEGG pathways by miRs involved in DDR signaling and that were EGCG modulated. The target prediction threshold was set at 0.85. p-value < 0.05. #: number.
Table 5.
Enriched KEGG pathways for predicted targets of the DDR microRNAs modulated by CRC.
| KEGG Pathway | p-Value | # of Genes | # of miRNAs |
|---|---|---|---|
| Fatty acid biosynthesis | 0 | 4 | 5 |
| ECM-receptor interaction | 0 | 19 | 7 |
| Fatty acid metabolism | 6.88 × 10−9 | 8 | 4 |
| Signaling pathways regulating pluripotency of stem cells | 2.16 × 10−5 | 55 | 9 |
| Glioma | 0.0002249224 | 20 | 5 |
| Proteoglycans in cancer | 0.0003169881 | 66 | 6 |
| TGF-β signaling pathway | 0.006985672 | 28 | 5 |
| Prostate cancer | 0.009151789 | 31 | 5 |
| Axon guidance | 0.01011865 | 45 | 3 |
| Melanoma | 0.01304415 | 20 | 4 |
| Prolactin signaling pathway | 0.01811186 | 28 | 5 |
| Pathways in cancer | 0.0206275 | 70 | 4 |
| Glycosaminoglycan biosynthesis-heparan sulfate/heparin | 0.02616968 | 3 | 5 |
DIANA-miRPath v3.0 was used to predict the targeted KEGG pathways by miRs involved in DDR signaling and that were CRC modulated. The target prediction threshold was set at 0.85. p-value < 0.05. #: number.
Table 6.
Enriched KEGG pathways for predicted targets of the DDR microRNAs modulated by RSV.
| KEGG Pathway | p-Value | # of Genes | # of miRNAs |
|---|---|---|---|
| Fatty acid biosynthesis | 0 | 5 | 7 |
| Fatty acid metabolism | 0 | 16 | 8 |
| Signaling pathways regulating pluripotency of stem cells | 1.07 × 10−6 | 64 | 13 |
| TGF-β signaling pathway | 1.39 × 10−4 | 34 | 9 |
| Proteoglycans in cancer | 8.1 × 10−1 | 70 | 10 |
| Axon guidance | 0.0009836851 | 55 | 6 |
| Hippo signaling pathway | 0.001005551 | 43 | 9 |
| Mucin type O-glycan biosynthesis | 0.002219981 | 12 | 8 |
| Glycosphingolipid biosynthesis: lacto and neolacto series | 0.006851962 | 6 | 7 |
| GABAergic synapse | 0.007317003 | 13 | 8 |
| Glioma | 0.03097475 | 21 | 7 |
DIANA-miRPath v3.0 was used to predict the targeted KEGG pathways by miRs involved in DDR signaling and that were RSV modulated. The target prediction threshold was set at 0.85. p-value < 0.05. #: number.
Table 7.
Enriched KEGG pathways for predicted DDR targets of the microRNAs modulated by n-3-PUFAs.
| KEGG Pathway | p-Value | # of Genes | # of miRNAs |
|---|---|---|---|
| ECM-receptor interaction | 0 | 27 | 4 |
| Fatty acid biosynthesis | 5.57 × 10-5 | 4 | 1 |
| Glycosphingolipid biosynthesis: lacto and neolacto series | 2.21 × 10-2 | 8 | 8 |
| Mucin type O-glycan biosynthesis | 0.0003249162 | 9 | 5 |
| Proteoglycans in cancer | 0.0004576113 | 76 | 8 |
| TGF-β signaling pathway | 0.001807704 | 27 | 5 |
| Thyroid hormone synthesis | 0.002952912 | 5 | 3 |
| Amebiasis | 0.00834241 | 24 | 2 |
| Signaling pathways regulating pluripotency of stem cells | 0.02548584 | 51 | 4 |
| Glioma | 0.02578235 | 31 | 6 |
DIANA-miRPath v3.0 was used to predict the targeted KEGG pathways by miRs involved in DDR signaling and that were n3-PUFAs modulated. The target prediction threshold was set at 0.85. p-value < 0.05. #: number.
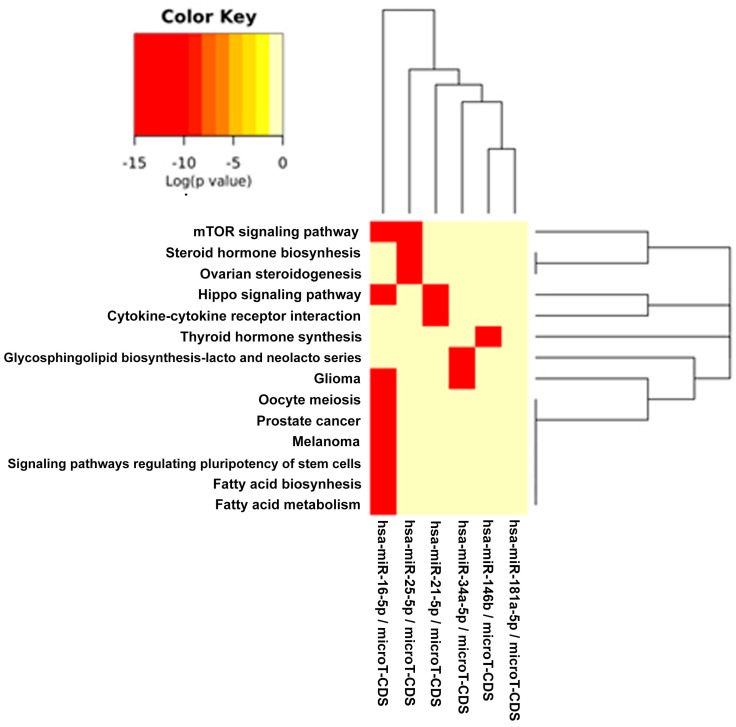
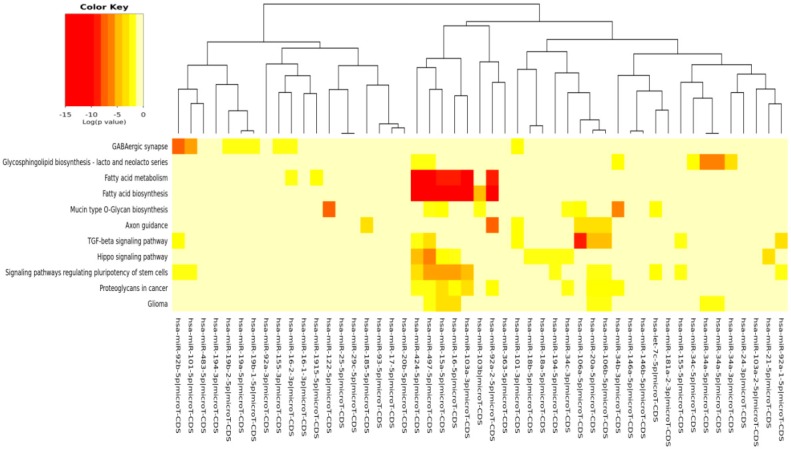
Figure 2.
Binary heat map of pathways related to the common microRNAs involved in DDR signaling and modulated by all of the compounds: EGCG, CRC, RSV, n3-PUFAs. In this plot, heat map calculation is based on binary p-values (0: not targeted, 1: targeted); all significantly targeted pathways are marked with deep red. The plot shows miRNAs targeting similar pathways and pathways being targeted by miRNAs.
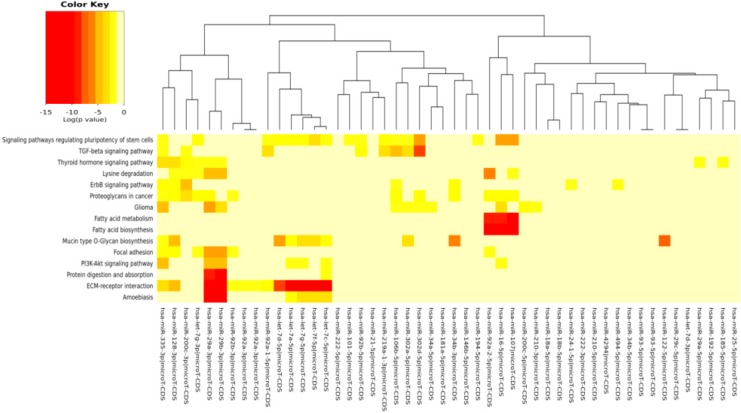
Figure 3.
MicroRNAs involved in DDR and modulated by EGCG versus the pathways’ heat map. In this plot, heat map calculation is based on absolute p-values. Darker colors represent lower p-values (higher significance). The plot shows miRNAs targeting similar pathway clusters and pathways being targeted by miRNA groups.
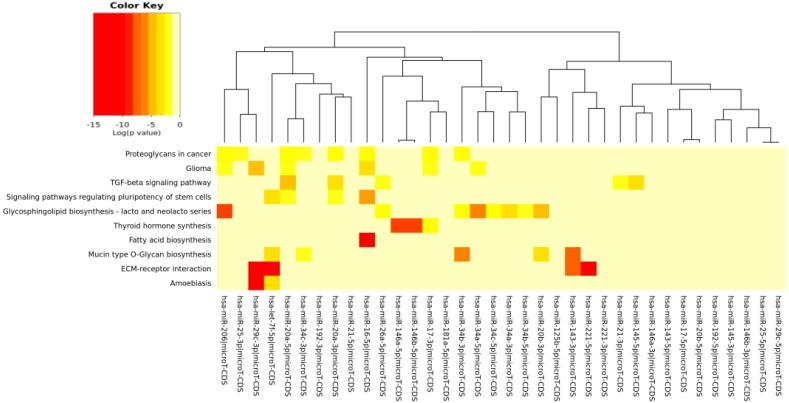
Figure 4.
MicroRNAs involved in DDR and modulated by CRC versus the pathways’ heat map. In this plot, heat map calculation is based on absolute p-values. Darker colors represent lower p-values (higher significance). The plot shows microRNAs targeting similar pathway clusters and pathways being targeted by miRNA groups.
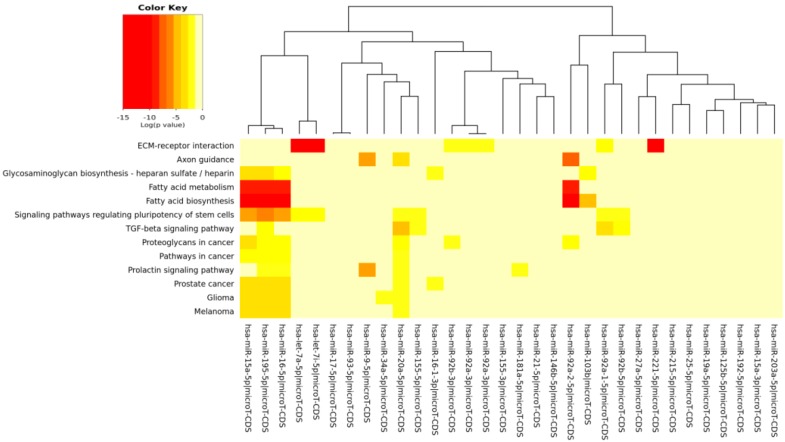
Figure 5.
MicroRNAs involved in DDR and modulated by RSV versus the pathways’ heat map. In this plot, heat map calculation is based on absolute p-values. Darker colors represent lower p-values (higher significance). The plot shows microRNAs targeting similar pathway clusters and pathways being targeted by miRNA groups.
Figure 6.
MicroRNAs involved in DDR and modulated by n3-PUFAs versus the pathways’ heat map. In this plot, heat map calculation is based on absolute p-values. Darker colors represent lower p-values (higher significance). The plot shows miRNAs targeting similar pathway clusters and pathways being targeted by miRNA groups.
Table 3 depicts the statistically-significant enriched KEGG pathways for miRs involved in DD/DDR and modulated by all compounds together (EGCG, CRC, RSV and n3-PUFA). The visual representation of the binary heat map showing the miRNAs/pathways interaction is reported in Figure 2. We found 14 KEGG pathways significantly related to genes targeted by miRs regulated by all four compounds. A consistent number of pathways were involved in energy metabolism, including the “mTOR signaling”, “fatty acid biosynthesis” and “fatty acid metabolism” pathways. Cancer-related pathways were also present, including glioma, melanoma and prostate cancer. In addition, we found significant pathways relevant in stem cell biology and tissue homeostasis, such as “signaling pathways regulating pluripotency of stem cells” and the “Hippo signaling pathway”.
The pathways significantly enriched (p < 0.05) for the target of miRs modulated by each specific compound (EGCG, CRC, RSV and n3-PUFA) are reported in Table 4, Table 5, Table 6 and Table 7, respectively. We found KEGG pathways, such as “fatty acid biosynthesis” and “signaling pathways regulating pluripotency of stem cells” significantly modulated by each compound. The visual representations of the heat maps showing the miR-pathway interaction, for each single compound, are reported in Figure 3, Figure 4, Figure 5 and Figure 6.
3. Discussion
The maintenance of genome integrity by an efficient DNA repair is of paramount importance in the prevention of cancer, attenuation of aging processes and age-related degenerative diseases. Effective DDR is the key mechanism providing genome stability. miRs play a critical role in the regulation of DDR. In this in silico study, therefore, we examined the influence of dietary bioactive compounds on the miRs involved in DDR, which potentially may offer clues on the role of these compounds as potential chemopreventive and antiaging (geroprotective) modalities.
The results of our analysis show a strong impact of the compounds considered on important miRs involved in DD/DDR pathways. In particular, the analysis suggests a synergic action of all compounds in the modulation of six miRs. Among these common miRs, we found miR-21 and miR-146. It has recently been highlighted that miR-146, miR-155 and miR-21 have a kay role in the interplay among DDR, cell senescence, inflammation and age-related diseases [9]. This trio of miRNAs, termed “inflamma-miRs”, has been primarily associated with chronic, low-grade inflammation known to characterize human aging and predisposing to age-related diseases [67]. We here find that two of these miRs, DD/DDR-associated (miR-146 and miR-21), are modulated by all four studied compounds and one of them, miR-155, by CRC and RSV [57]. miR-146 is one of the major miRs involved in orchestrating immune and inflammatory signaling via modulation of NF-kB activation [9]. Both miR-146a and miR-146b-5p have been found to target the DSB repair key protein BRCA1 [10].
miR-21 is a key modulator in many inflammatory pathways, and its aberrant expression in numerous cancers has led to its designation as an “onco-miR” [68]. miR-21 is induced by DD, negatively regulating G1/S transition. It also participates in the DNA damage-induced G2/M checkpoint [5]. It has been also shown that mR-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells [69]. In addition, miR-21 is upregulated during hepatitis C virus infection and negatively regulates IFN-α signaling through MyD88 and IRAK1; it may thus be a potential therapeutic target for antiviral intervention [70]. Recent observations suggest that miR-21 in cooperation with miR-145 (modulated by n3-PUFAs) play critical roles in the regulation of colon cancer stem cells [71]. miR-155 has been reported to regulate inflammation and immune responses [72]. miR-155 may have different functions in innate and adaptive immune responses, and the systemic diffusion of this DDR/-related miR may have either adverse or beneficial effects, depending on overall senescence/immunological host condition [9]. In DDR network, miR-155 modulates cell cycle after DD by targeting key genes involved in cell cycle control, such as WEE1 [73]. miR-155 has been shown to control the expression of TRF1, the protein that negatively regulates telomere length [74]. Indeed, the TRF1 gene is known to be a target of miR-155 [75]. Telomere length may also be regulated by miR-34a [76], which is in turn modulated by all four compounds, as demonstrated by our in silico analysis. miR-34b and miR-34c are also modulated by the three and two studied compounds, respectively. The miR-34 family, is a direct transcriptional target of p53, whose induction by DD and oncogenic stress pervades in diverse aspects of the DD response pathway [27]. Noteworthy, the compounds being evaluated here modulate members of the miR-17/92 cluster and miR-106a/b clusters (DDR modulated), having an important role in cancer and other numerous diseases [77]. In particular, our analysis reveals that RSV is able to modulate all of the miRs of these tree important clusters.
Our previous studies have demonstrated that RSV decreased the level of constitutive DD signaling by the reduced expression of γH2AX in proliferating A549, TK6 and WI-38 cells and in mitogenically-stimulated human lymphocytes [78]. The reduction of γ-H2AX was paralleled by a drop in the level of endogenous ROS and a decline in mTOR/S6K1 signaling [79]. H2AX is the initial sensor protein in the DSB response that can detect and mark DD by its phosphorylated form (γ-H2AX) [80,81]. H2AX is a target of miR-24 [80]. The present analysis suggests that RSV, as well as CRC and EGCG might regulate γH2AX expression by miR-24 mediation.
The DIANA program allowed us to investigate the common pathways associated with miRs. Among the most popular pathways involved in DD and DDR influenced by the compounds considered in this study, we found the fatty acids’ biosynthesis pathway and fatty acids’ metabolism. Alterations in fatty acid metabolism in cancer cells have received less attention, but are increasingly being recognized. Indeed, some studies have suggested that the DNA damage response is involved in the regulation of metabolic homeostasis. DNA damage could impair metabolic organ functions by causing cell death or senescence [82]. There is a strong relationship between DD and energy metabolism [83], and an implication for its role in tumorigenesis has been indicated. p53, the key factor in the DD/DDR network, has been reported to regulate fatty acid oxidation [84], and genes involved in fatty acid metabolism are regulated by p53 in different cell types and in response to both oncogenic stress and DD [85]. mTOR holds also an important role in lipid biosynthesis and metabolism [86,87] demonstrating a cross-talk among these pathways, and this fact has a significant meaning in cancerogenesis and chemoprevention. For example, by downregulating mTOR signaling and energy metabolism, it is possible to suppress the malignant phenotype of colorectal cancer cells [88,89]. Indeed, fatty acid synthase is a potential therapeutic target in cancer. There is strong evidence that constitutive mTOR signaling is the driving force of cellular and organismal aging and induction of senescence [90,91]. Extracellular matrix (ECM)-receptor interaction is also shown to be strongly involved in the DD/DDR and compounds interaction (EGCG, n3-PUFA, CRC). This is of interest in light of the evidence that ECM proteins are involved in cancer progression and outcome. The initiation of cell transformation is generally associated with genetic alterations in normal cells that lead to the loss of intercellular- and/or extracellular-matrix (ECM)-mediated cell adhesion [92]. There is a connection between ECM proteins, such integrin signaling and DNA repair [93]. There is also strong evidence that hyaluronic acid (HA; hyaluronate), the key component of ECM, by neutralizing the ROS, is reducing the induction of DNA damage by endogenous and exogenous oxidants [94,95,96]. Particularly interesting is the role of HA, which is a large constituent of stem cells’ niche, in protecting the integrity of genome of stem cells against reactive oxidants [95]. Our in silico results evidence that signaling regulating the pluripotency of stem cells pathway is clearly targeted by many miRs selected in this study, as evidenced by the heat maps of specific compounds. miRs represent an important layer of regulation for stem cell self-renewal and differentiation [97,98]. Among DDR/compound-modulated miRs, in our analysis, let-7, miR-302 and miR-17-92 have been found in previous studies as regulators of cellular pluripotency [99]. Signaling regulating the pluripotency of stem cells converges towards the activation of the transcriptional network and of many different pathways, which often show cross-talk in the determination of stem cell function. Indeed, FOXO, e.g., transcription factors, is required for DNA damage-induced growth arrest checkpoints [100], and as a further example, the mTOR pathway has been shown to be a major regulator of both ROS levels and autophagy in human stem cells [101]. The life-long persistence of stem cells in the body makes them particularly susceptible to the accumulation of cellular damage, which ultimately can lead to cell death, senescence or loss of regenerative function. Indeed, stem cells in many tissues have been found to undergo profound changes with age, exhibiting blunted responsiveness to tissue injury, dysregulation of proliferative activities and declining functional capacities. These changes translate into reduced effectiveness of cell replacement and tissue regeneration in aged organisms [102]. Noteworthy, miR-146a/b and miR-195, which we found modulated by all compounds and by CRC, respectively, have been reported as age-induced miRNAs involved in stem cell senescence [103]. These recent findings have demonstrated that silencing miR-195 reverses the senescence clock in aged stem cells by telomerase reactivation. The regulation of telomere length performed by the above-mentioned miR-155 may also have an important role in the cancer stem cell fate and cancer tissue homeostasis. miR-155 is upregulated in many cancers and can promote cancer stem cell phenotypes in liver [104]. RSV has been demonstrated to be able to decrease its levels, supporting our prediction analysis [57].
The availability of nutritional agents that can target specific miRs regulating stem cells’ function can improve regenerative potential in the body and, finally, counteract aging and prevent diseases, such as cancer.
It must be considered that most of the literature here analyzed refers to experimental research carried out on in vitro models. The results indicated that exposure time and concentrations of compounds can differently affect miRNA expression in different human cells. In addition another important issue evident in the literature [41] is the poor bioavailability of phytochemicals due to their metabolism, absorption and, consequently, the fact that most phytochemical compounds reach the targeted sites at sub-therapeutic concentrations [105]. These observations pave the way for in vivo dietary intervention studies and clinical trials focused on setting the appropriate dosage of the bioactive compounds that can effectively affect the miRs’ expression in humans.
The comparison of pathways identified, with our in silico analysis, to those identified experimentally in the study examples utilized here interestingly demonstrated that new pathways were evidenced from our prediction. Even more interesting, however, is that some of our predicted pathways were indeed found as directly implicated in some cancer studies and modulated by the compounds here considered.
The fact that experimental literature results have found that pathways, such as, for example, “fatty acid biosynthesis” and “fatty acid metabolism”, were actually affected by the compounds taken into account in this paper validated our prediction. This positive inter-comparison proves that the pathways computationally found by us were, indeed, the same found experimentally involved in some cancer models and in some senescence studies, confirming the validity of our prediction study. For examples, in breast and lung cancer cells, EGCG affected fatty acid metabolism [106,107]. Resveratrol was found to suppress cancer cell proliferation by inhibiting the fatty acid synthase signaling pathway [108]. Curcumin, as well, induces apoptosis, inhibiting intracellular fatty acid synthase in human breast cancer [109]. Clinical studies corroborate these findings [110], and trials were also carried out [111]. Resveratrol and EGCG are also found to improve the functional activity of the membrane lipids in aged liver models by influencing lipid composition and metabolism [112,113].
Presently, miRs have been already found to be modulated by the compounds here considered. On the other hand, other studies found that some of the pathways here evidenced are modulated by our compounds, but we provide here the link between miRs’ expression and the pathways’ modulation by the respective compounds. Thus, the importance of our results consists, indeed, of the possibility of detecting experimentally the link between miRs’ expression and pathways’ modulation.
Some authors suggested an interplay among miRNA, DD/DDR, specific signaling pathways and cancer promotion and invasion [114,115], and bioactive compounds play an important role in this interplay.
Taken together, the clinical relevance of such observations could be related to the bioactive compounds’ chemopreventive value in some type of tumors, as for example in breast cancers or lung cancers, where enough evidence corroborates this hypothesis.
In conclusion, since DDR and these pathways are strongly related to aging and carcinogenesis, the in silico results of our study suggest that monitoring the induction of specific miRs may provide the means to assess the antiaging and chemopreventive properties of particular bioactive compounds. Furthermore, the results of this in silico study indicate the most important pathways potentially modulated by dietary compounds and provide a framework for the generation of new strategies to design experimental studies and, finally, new anti-cancer and anti-aging therapies.
4. Materials and Methods
4.1. Search Strategy and Selection Criteria
A literature search was performed using PubMed databases to identify miRNAs with experimental evidence of involvement in DD and DDR (last accessed on 15 January 2016). The following keywords were used: “microRNA” and “DNA damage”. Due to the vast amount of literature, we selected in this paper the most significant and extensive reviews. We subsequently replaced “DNA damage” with “DNA damage response” or with “DNA damage repair” (see Table 1 in the Results).
Regarding miRs and the specific compounds considered in this paper, we did additional searches using “microRNA” and “epigallocatechin gallate” as key terms, and we substituted “epigallocatechin gallate” with “curcumin”, “resveratrol” or “n3-PUFA”. The literature search was limited to reports in English and in humans (see Table 2 in the Results).
The miRNAs involved in the DD/DDR network (from Table 1) and modulated by bioactive compounds (from Table 2) were selected to build the Venn diagram (Figure 1).
4.2. In Silico Analysis
All miRs used in the in silico analysis were selected as common between Table 1 and Table 2 and reported in the Venn diagram (Figure 1).
In Table 3 and Figure 2, the miRs found modulated by all four compounds were used (red in Figure 1). In Table 4, Table 5, Table 6 and Table 7 and in Figure 3, Figure 4, Figure 5 and Figure 6, the miRs modulated by a single compound were used.
To identify molecular pathways potentially altered by the expression of specific miRNAs selected from the literature search, the online software DIANA-mirPathv3 [26] was used. The software has the capacity to analyze the combinatorial effect of different miRNAs on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. DIANA-mirPathv3 combines the gene targets of the selected miRs into a superset (union) performing the enrichment analysis and calculating the significance levels (p-values) between each miRNA and every pathway. The p-value is a measure of the association between a selected gene from the list of pathways. In pathway analysis, generally, p-values higher than 0.05 indicate that the association is not statistically significant, and the pathway could be rejected. In our analyses of pathways prediction, miR-gene interactions are derived from the in silico miRNA target prediction algorithms: DIANA-microT-CDS. The settings utilized were: p-value threshold of 0.05; DIANA-microT-CDS threshold of 0.85; merging method: pathways union. Fisher’s method was used to combine the results of more than one independent test bearing on the same hypothesis. All of the results were corrected in the combined enrichment analysis for multiple hypotheses testing, by applying Benjamini and Hochberg’s algorithm [116].
The results of the miR-pathway interaction are presented as “heat maps” (graphical representations of data where values in a matrix are represented as colors). They enable the visualization of a very large number of variables. The software DIANA-mirPathv3 utilizes the hierarchical clustering results on both axes (pathways and miRNAs), in order to construct the heat map visualization.
Acknowledgments
Laura Teodori funded by Fondazione San Raffaele, Ceglie Messapica Italy Research Project MERIT-RBNE08HM7T, Italian Ministry of Health. The authors would like to thank . Mauro Sansò, Chief Operating Officer at Fondazione San Raffaele, for his excellent management of the MERIT #RBNE08HM7T funds. Zbigniew Darzynkiewicz is supported by the Robert A. Welke Cancer Research Foundation. Dario Coletti is supported by ANR (# 13-BSV1-0005), NIH (# 5R01CA180057-02), AFM (# 2012-0773), UPMC Emergence 2011 and IBPS 2015.
Abbreviations
| EGCG | Epi-gallocatechin-3-gallate |
| RSV | Resveratrol |
| CRC | Curcumin |
| n3-PUFAs | n3-polyunsaturated fatty acids |
| miR | micro-RNA |
| DD | DNA damage |
| DDR | DNA damage response |
Author Contributions
Laura Teodori and Felicia Carotenuto conceived of and designed the in silico study. Maria C. Albertini, Dario Coletti and Luigi Campanella analyzed the data. Alessandra Vilmercati contributed analysis tools. Laura Teodori, Felicia Carotenuto and Zbigniew Darzynkiewicz wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hoeijmakers J.H. DNA damage, aging, and cancer. N. Engl. J. Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 2.Rossiello F., Herbig U., Longhese M.P., Fumagalli M., d’Adda di Fagagna F. Irreparable telomeric DNA damage and persistent DDR signalling as a shared causative mechanism of cellular senescence and ageing. Curr. Opin. Genet. Dev. 2014;26:89–95. doi: 10.1016/j.gde.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirbu B.M., Cortez D. DNA damage response: Three levels of DNA repair regulation. Cold Spring Harb. Perspect. Biol. 2013;5:a012724. doi: 10.1101/cshperspect.a012724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malaquin N., Carrier-Leclerc A., Dessureault M., Rodier F. DDR-mediated crosstalk between DNA-damaged cells and their microenvironment. Front. Genet. 2015;6 doi: 10.3389/fgene.2015.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu H., Gatti R.A. MicroRNAs: New players in the DNA damage response. J. Mol. Cell Biol. 2011;3:151–158. doi: 10.1093/jmcb/mjq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campisi J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodier F., Campisi J. Four faces of cellular senescence. J. Cell Biol. 2011;192:547–556. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moskalev A.A., Shaposhnikov M.V., Plyusnina E.N., Zhavoronkov A., Budovsky A., Yanai H., Fraifeld V.E. The role of DNA damage and repair in aging through the prism of Koch-like criteria. Ageing Res. Rev. 2013;12:661–684. doi: 10.1016/j.arr.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Olivieri F., Albertini M.C., Orciani M., Ceka A., Cricca M., Procopio A.D., Bonafe M. DNA damage response (DDR) and senescence: Shuttled inflamma-miRNAs on the stage of inflamm-aging. Oncotarget. 2015;6:35509–35521. doi: 10.18632/oncotarget.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang C., Peng G. Non-coding RNAs: An emerging player in DNA damage response. Mutat. Res. Rev. Mutat. Res. 2015;763:202–211. doi: 10.1016/j.mrrev.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Yan X., Chao T., Tu K., Zhang Y., Xie L., Gong Y., Yuan J., Qiang B., Peng X. Improving the prediction of human microRNA target genes by using ensemble algorithm. FEBS Lett. 2007;581:1587–1593. doi: 10.1016/j.febslet.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 12.Luo W., Nie Q., Zhang X. MicroRNAs involved in skeletal muscle differentiation. J. Genet. Genom. 2013;40:107–116. doi: 10.1016/j.jgg.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., Li M., Wang G., Liu Y. MiR2 Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peter M.E. Targeting of mrnas by multiple miRNAs: The next step. Oncogene. 2010;29:2161–2164. doi: 10.1038/onc.2010.59. [DOI] [PubMed] [Google Scholar]
- 15.Bishop K.S., Ferguson L.R. The interaction between epigenetics, nutrition and the development of cancer. Nutrients. 2015;7:922–947. doi: 10.3390/nu7020922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi S.W., Friso S. Epigenetics: A new bridge between nutrition and health. Adv. Nutr. 2010;1:8–16. doi: 10.3945/an.110.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chedraui P., Perez-Lopez F.R. Nutrition and health during mid-life: Searching for solutions and meeting challenges for the aging population. Climacteric. 2013;16:85–95. doi: 10.3109/13697137.2013.802884. [DOI] [PubMed] [Google Scholar]
- 18.Hosseini A., Ghorbani A. Cancer therapy with phytochemicals: Evidence from clinical studies. Avicenna J. Phytomed. 2015;5:84–97. [PMC free article] [PubMed] [Google Scholar]
- 19.Farago N., Feher L.Z., Kitajka K., Das U.N., Puskas L.G. MicroRNA profile of polyunsaturated fatty acid treated glioma cells reveal apoptosis-specific expression changes. Lipids Health Dis. 2011;10 doi: 10.1186/1476-511X-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez-Leyva D., Dupasquier C.M., McCullough R., Pierce G.N. The cardiovascular effects of flaxseed and its omega-3 fatty acid, α-linolenic acid. Can. J. Cardiol. 2010;26:489–496. doi: 10.1016/S0828-282X(10)70455-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carotenuto F., Minieri M., Monego G., Fiaccavento R., Bertoni A., Sinigaglia F., Vecchini A., Carosella L., di Nardo P. A diet supplemented with Ala-rich flaxseed prevents cardiomyocyte apoptosis by regulating caveolin-3 expression. Cardiovasc. Res. 2013;100:422–431. doi: 10.1093/cvr/cvt211. [DOI] [PubMed] [Google Scholar]
- 22.Carotenuto F., Costa A., Albertini M.C., Rocchi M.B.L., Rudov A., Coletti D., Minieri M., di Nardo P., Teodori L. Dietary flaxseed mitigates impaired skeletal muscle regeneration: In vivo, in vitro and in silico studies. Int. J. Med. Sci. 2016;13:206–219. doi: 10.7150/ijms.13268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruijters E.J., Haenen G.R., Willemsen M., Weseler A.R., Bast A. Food-derived bioactives can protect the anti-inflammatory activity of cortisol with antioxidant-dependent and -independent mechanisms. Int. J. Mol. Sci. 2016;17:239. doi: 10.3390/ijms17020239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer J.D., Soule B.P., Simone B.A., Zaorsky N.G., Jin L., Simone N.L. MicroRNA expression altered by diet: Can food be medicinal? Ageing Res. Rev. 2014;17:16–24. doi: 10.1016/j.arr.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Miceli M., Bontempo P., Nebbioso A., Altucci L. Natural compounds in epigenetics: A current view. Food Chem. Toxicol. 2014;73:71–83. doi: 10.1016/j.fct.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-mirpath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan G., Mathur R., Hu X., Zhang X., Lu X. miRNA response to DNA damage. Trends Biochem. Sci. 2011;36:478–484. doi: 10.1016/j.tibs.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vrijens K., Bollati V., Nawrot T.S. MicroRNAs as potential signatures of environmental exposure or effect: A systematic review. Environ. Health Perspect. 2015;123:399–411. doi: 10.1289/ehp.1408459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsang W.P., Kwok T.T. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J. Nutr. Biochem. 2010;21:140–146. doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 30.An I.S., An S., Park S., Lee S.N., Bae S. Involvement of MicroRNAs in epigallocatechin gallate-mediated UVB protection in human dermal fibroblasts. Oncol. Rep. 2013;29:253–259. doi: 10.3892/or.2012.2083. [DOI] [PubMed] [Google Scholar]
- 31.Toden S., Tran H.M., Tovar-Camargo O.A., Okugawa Y., Goel A. Epigallocatechin-3-gallate targets cancer stem-like cells and enhances 5-fluorouracil chemosensitivity in colorectal cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., Bian S., Yang C.S. Green tea polyphenol egcg suppresses lung cancer cell growth through upregulating miR-210 expression caused by stabilizing HIF-1α. Carcinogenesis. 2011;32:1881–1889. doi: 10.1093/carcin/bgr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou D.H., Wang X., Feng Q. EGCG enhances the efficacy of cisplatin by downregulating HSA-miR-98-5p in NSCLC A549 cells. Nutr. Cancer. 2014;66:636–644. doi: 10.1080/01635581.2014.894101. [DOI] [PubMed] [Google Scholar]
- 34.Zhu K., Wang W. Green tea polyphenol EGCG suppresses osteosarcoma cell growth through upregulating miR-1. Tumour Biol. 2016;37:4373–4382. doi: 10.1007/s13277-015-4187-3. [DOI] [PubMed] [Google Scholar]
- 35.Jiang L., Tao C., He A., He X. Overexpression of miR-126 sensitizes osteosarcoma cells to apoptosis induced by epigallocatechin-3-gallate. World J. Surg. Oncol. 2014;12 doi: 10.1186/1477-7819-12-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mekky R.Y., El-Ekiaby N.M., Hamza M.T., Elemam N.M., El-Sayed M., Esmat G., Abdelaziz A.I. miR-194 is a hepatocyte gate keeper hindering HCV entry through targeting CD81 receptor. J. Infect. 2015;70:78–87. doi: 10.1016/j.jinf.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Chakrabarti M., Khandkar M., Banik N.L., Ray S.K. Alterations in expression of specific MicroRNAs by combination of 4-HPR and EGCG inhibited growth of human malignant neuroblastoma cells. Brain Res. 2012;1454:1–13. doi: 10.1016/j.brainres.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakrabarti M., Ai W., Banik N.L., Ray S.K. Overexpression of miR-7-1 increases efficacy of green tea polyphenols for induction of apoptosis in human malignant neuroblastoma SH-SY5Y and SK-N-DZ cells. Neurochem. Res. 2013;38:420–432. doi: 10.1007/s11064-012-0936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon M.W., Yan F., Zhong X., Mazumder P.B., Xu-Monette Z.Y., Zou D., Young K.H., Ramos K.S., Li Y. Regulation of p53-targeting MicroRNAs by polycyclic aromatic hydrocarbons: Implications in the etiology of multiple myeloma. Mol. Carcinog. 2015;54:1060–1069. doi: 10.1002/mc.22175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baselga-Escudero L., Blade C., Ribas-Latre A., Casanova E., Suarez M., Torres J.L., Salvado M.J., Arola L., Arola-Arnal A. Resveratrol and EGCG bind directly and distinctively to miR-33a and miR-122 and modulate divergently their levels in hepatic cells. Nucleic Acids Res. 2014;42:882–892. doi: 10.1093/nar/gkt1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milenkovic D., Jude B., Morand C. MiRNA as molecular target of polyphenols underlying their biological effects. Free Radic. Biol. Med. 2013;64:40–51. doi: 10.1016/j.freeradbiomed.2013.05.046. [DOI] [PubMed] [Google Scholar]
- 42.Ye M., Zhang J., Zhang J., Miao Q., Yao L., Zhang J. Curcumin promotes apoptosis by activating the p53-miR-192–5p/215-XIAP pathway in non-small cell lung cancer. Cancer Lett. 2015;357:196–205. doi: 10.1016/j.canlet.2014.11.028. [DOI] [PubMed] [Google Scholar]
- 43.Ma J., Fang B., Zeng F., Pang H., Zhang J., Shi Y., Wu X., Cheng L., Ma C., Xia J., et al. Curcumin inhibits cell growth and invasion through up-regulation of miR-7 in pancreatic cancer cells. Toxicol. Lett. 2014;231:82–91. doi: 10.1016/j.toxlet.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 44.Phuah N.H., Nagoor N.H. Regulation of microRNAs by natural agents: New strategies in cancer therapies. BioMed Res. Int. 2014;2014:804510. doi: 10.1155/2014/804510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sreenivasan S., Thirumalai K., Danda R., Krishnakumar S. Effect of curcumin on miRNA expression in human Y79 retinoblastoma cells. Curr. Eye Res. 2012;37:421–428. doi: 10.3109/02713683.2011.647224. [DOI] [PubMed] [Google Scholar]
- 46.Noratto G.D., Jutooru I., Safe S., Angel-Morales G., Mertens-Talcott S.U. The drug resistance suppression induced by curcuminoids in colon cancer SW-480 cells is mediated by reactive oxygen species-induced disruption of the microRNA-27A-ZBTB10-SP axis. Mol. Nutr. Food Res. 2013;57:1638–1648. doi: 10.1002/mnfr.201200609. [DOI] [PubMed] [Google Scholar]
- 47.Chen J., Xu T., Chen C. The critical roles of miR-21 in anti-cancer effects of curcumin. Ann. Transl. Med. 2015;3 doi: 10.3978/j.issn.2305-5839.2015.09.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramaniam D., Ponnurangam S., Ramamoorthy P., Standing D., Battafarano R.J., Anant S., Sharma P. Curcumin induces cell death in esophageal cancer cells through modulating notch signaling. PLoS ONE. 2012;7:752. doi: 10.1371/journal.pone.0030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarkar S., Dubaybo H., Ali S., Goncalves P., Kollepara S.L., Sethi S., Philip P.A., Li Y. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(KIP1), p57(KIP2), and PUMA. Am. J. Cancer Res. 2013;3:465–477. [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J., Du Y., Wu C., Ren X., Ti X., Shi J., Zhao F., Yin H. Curcumin promotes apoptosis in human lung adenocarcinoma cells through miR-186* signaling pathway. Oncol. Rep. 2010;24:1217–1223. doi: 10.3892/or_00000975. [DOI] [PubMed] [Google Scholar]
- 51.Yang J., Cao Y., Sun J., Zhang Y. Curcumin reduces the expression of BCL-2 by upregulating miR-15a and miR-16 in MCF-7 cells. Med. Oncol. 2010;27:1114–1118. doi: 10.1007/s12032-009-9344-3. [DOI] [PubMed] [Google Scholar]
- 52.Vislovukh A., Kratassiouk G., Porto E., Gralievska N., Beldiman C., Pinna G., El’skaya A., Harel-Bellan A., Negrutskii B., Groisman I. Proto-oncogenic isoform A2 of eukaryotic translation elongation factor EEF1 is a target of miR-663 and miR-744. Br. J. Cancer. 2013;108:2304–2311. doi: 10.1038/bjc.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu P., Liang H., Xia Q., Li P., Kong H., Lei P., Wang S., Tu Z. Resveratrol induces apoptosis of pancreatic cancers cells by inhibiting miR-21 regulation of BCL-2 expression. Clin. Transl. Oncol. 2013;15:741–746. doi: 10.1007/s12094-012-0999-4. [DOI] [PubMed] [Google Scholar]
- 54.Yu Y.H., Chen H.A., Chen P.S., Cheng Y.J., Hsu W.H., Chang Y.W., Chen Y.H., Jan Y., Hsiao M., Chang T.Y., et al. miR-520h-mediated FOXC2 regulation is critical for inhibition of lung cancer progression by resveratrol. Oncogene. 2013;32:431–443. doi: 10.1038/onc.2012.74. [DOI] [PubMed] [Google Scholar]
- 55.Tome-Carneiro J., Larrosa M., Yanez-Gascon M.J., Davalos A., Gil-Zamorano J., Gonzalvez M., Garcia-Almagro F.J., Ruiz Ros J.A., Tomas-Barberan F.A., Espin J.C., et al. One-year supplementation with a grape extract containing resveratrol modulates inflammatory-related microRNAs and cytokines expression in peripheral blood mononuclear cells of type 2 diabetes and hypertensive patients with coronary artery disease. Pharmacol. Res. 2013;72:69–82. doi: 10.1016/j.phrs.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 56.Dhar S., Hicks C., Levenson A.S. Resveratrol and prostate cancer: Promising role for microRNAs. Mol. Nutr. Food Res. 2011;55:1219–1229. doi: 10.1002/mnfr.201100141. [DOI] [PubMed] [Google Scholar]
- 57.Tili E., Michaille J.J., Adair B., Alder H., Limagne E., Taccioli C., Ferracin M., Delmas D., Latruffe N., Croce C.M. Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting junb and jund. Carcinogenesis. 2010;31:1561–1566. doi: 10.1093/carcin/bgq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Espinoza J.L., Takami A., Trung L.Q., Kato S., Nakao S. Resveratrol prevents EBV transformation and inhibits the outgrowth of EBV-immortalized human b cells. PLoS ONE. 2012;7:752. doi: 10.1371/journal.pone.0051306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dhar S., Kumar A., Rimando A.M., Zhang X., Levenson A.S. Resveratrol and pterostilbene epigenetically restore PTEN expression by targeting oncomiRs of the miR-17 family in prostate cancer. Oncotarget. 2015;6:27214–27226. doi: 10.18632/oncotarget.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gil-Zamorano J., Martin R., Daimiel L., Richardson K., Giordano E., Nicod N., Garcia-Carrasco B., Soares S.M., Iglesias-Gutierrez E., Lasuncion M.A., et al. Docosahexaenoic acid modulates the enterocyte CACO-2 cell expression of microRNAs involved in lipid metabolism. J. Nutr. 2014;144:575–585. doi: 10.3945/jn.113.189050. [DOI] [PubMed] [Google Scholar]
- 61.Yao L., Han C., Song K., Zhang J., Lim K., Wu T. Omega-3 polyunsaturated fatty acids upregulate 15-PGDH expression in cholangiocarcinoma cells by inhibiting miR-26A/B expression. Cancer Res. 2015;75:1388–1398. doi: 10.1158/0008-5472.CAN-14-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiu S.C., Chiang E.P., Tsai S.Y., Wang F.Y., Pai M.H., Syu J.N., Cheng C.C., Rodriguez R.L., Tang F.Y. Eicosapentaenoic acid induces neovasculogenesis in human endothelial progenitor cells by modulating C-KIT protein and PI3-K/AKT/ENOS signaling pathways. J. Nutr. Biochem. 2014;25:934–945. doi: 10.1016/j.jnutbio.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 63.Antal O., Hackler L., Jr., Shen J., Man I., Hideghety K., Kitajka K., Puskas L.G. Combination of unsaturated fatty acids and ionizing radiation on human glioma cells: Cellular, biochemical and gene expression analysis. Lipids Health Dis. 2014;13 doi: 10.1186/1476-511X-13-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mandal C.C., Ghosh-Choudhury T., Dey N., Choudhury G.G., Ghosh-Choudhury N. miR-21 is targeted by omega-3 polyunsaturated fatty acid to regulate breast tumor CSF-1 expression. Carcinogenesis. 2012;33:1897–1908. doi: 10.1093/carcin/bgs198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Visioli F., Giordano E., Nicod N.M., Davalos A. Molecular targets of omega 3 and conjugated linoleic fatty acids—“Micromanaging” cellular response. Front. Physiol. 2012;3 doi: 10.3389/fphys.2012.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DIANA-miRPath v3.0. [(accessed on 1 March 2016)]. Available online: http://snf-515788.vm.okeanos.grnet.gr/dianauniverse/index.php?r=mirpath.
- 67.Quinn S.R., O’Neill L.A. A trio of microRNAs that control toll-like receptor signalling. Int. Immunol. 2011;23:421–425. doi: 10.1093/intimm/dxr034. [DOI] [PubMed] [Google Scholar]
- 68.Kumarswamy R., Volkmann I., Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8:706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang P., Zou F., Zhang X., Li H., Dulak A., Tomko R.J., Jr., Lazo J.S., Wang Z., Zhang L., Yu J. MicroRNA-21 negatively regulates CDC25A and cell cycle progression in colon cancer cells. Cancer Res. 2009;69:8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu Y.R., Liu X.J., Li X.J., Shen Z.Z., Yang B., Wu C.C., Li J.F., Miao L.F., Ye H.Q., Qiao G.H., et al. MicroRNA miR-21 attenuates human cytomegalovirus replication in neural cells by targeting CDC25A. J. Virol. 2015;89:1070–1082. doi: 10.1128/JVI.01740-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu Y., Nangia-Makker P., Farhana L., Rajendra S.G., Levi E., Majumdar A.P. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol. Cancer. 2015;14 doi: 10.1186/s12943-015-0372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marques-Rocha J.L., Samblas M., Milagro F.I., Bressan J., Martinez J.A., Marti A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015;29:3595–3611. doi: 10.1096/fj.14-260323. [DOI] [PubMed] [Google Scholar]
- 73.Tili E., Michaille J.J., Wernicke D., Alder H., Costinean S., Volinia S., Croce C.M. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc. Natl. Acad. Sci. USA. 2011;108:4908–4913. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dinami R., Ercolani C., Petti E., Piazza S., Ciani Y., Sestito R., Sacconi A., Biagioni F., le Sage C., Agami R., et al. miR-155 drives telomere fragility in human breast cancer by targeting TRF1. Cancer Res. 2014;74:4145–4156. doi: 10.1158/0008-5472.CAN-13-2038. [DOI] [PubMed] [Google Scholar]
- 75.Smogorzewska A., de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 76.Xu X., Chen W., Miao R., Zhou Y., Wang Z., Zhang L., Wan Y., Dong Y., Qu K., Liu C. miR-34a induces cellular senescence via modulation of telomerase activity in human hepatocellular carcinoma by targeting FOXM1/C-myc pathway. Oncotarget. 2015;6:3988–4004. doi: 10.18632/oncotarget.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mogilyansky E., Rigoutsos I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Halicka H.D., Zhao H., Li J., Lee Y.S., Hsieh T.C., Wu J.M., Darzynkiewicz Z. Potential anti-aging agents suppress the level of constitutive MTOR- and DNA damage-signaling. Aging. 2012;4:952–965. doi: 10.18632/aging.100521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Darzynkiewicz Z., Zhao H., Halicka H.D., Li J., Lee Y.S., Hsieh T.C., Wu J.M. In search of antiaging modalities: Evaluation of MTOR- and ROS/DNA damage-signaling by cytometry. Cytometry A. 2014;85:386–399. doi: 10.1002/cyto.a.22452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lal A., Pan Y., Navarro F., Dykxhoorn D.M., Moreau L., Meire E., Bentwich Z., Lieberman J., Chowdhury D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanaka T., Halicka H.D., Huang X., Traganos F., Darzynkiewicz Z. Constitutive histone H2AX phosphorylation and atm activation, the reporters of DNA damage by endogenous oxidants. Cell Cycle. 2006;5:1940–1945. doi: 10.4161/cc.5.17.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shimizu I., Yoshida Y., Suda M., Minamino T. DNA damage response and metabolic disease. Cell Metab. 2014;20:967–977. doi: 10.1016/j.cmet.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 83.Vose S., Mitchell J. Relationship between DNA Damage and Energy Metabolism: Evidence from DNA Repair Deficiency Syndromes. INTECH Open Access Publisher; Rijeka, Croatia: 2011. [Google Scholar]
- 84.Assaily W., Rubinger D.A., Wheaton K., Lin Y., Ma W., Xuan W., Brown-Endres L., Tsuchihara K., Mak T.W., Benchimol S. ROS-mediated p53 induction of LPIN1 regulates fatty acid oxidation in response to nutritional stress. Mol. Cell. 2011;44:491–501. doi: 10.1016/j.molcel.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 85.Jiang D., LaGory E.L., Kenzelmann Broz D., Bieging K.T., Brady C.A., Link N., Abrams J.M., Giaccia A.J., Attardi L.D. Analysis of p53 transactivation domain mutants reveals ACAD11 as a metabolic target important for p53 pro-survival function. Cell Rep. 2015;10:1096–1109. doi: 10.1016/j.celrep.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Laplante M., Sabatini D.M. An emerging role of MTOR in lipid biosynthesis. Curr. Biol. 2009;19:R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soliman G.A. The integral role of mtor in lipid metabolism. Cell Cycle. 2011;10:861–862. doi: 10.4161/cc.10.6.14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang L., Wu P., Senthilkumar R., Tian X., Liu H., Shen X., Tao Z., Huang P. Loss of fatty acid synthase suppresses the malignant phenotype of colorectal cancer cells by down-regulating energy metabolism and MTOR signaling pathway. J. Cancer Res. Clin. Oncol. 2016;142:59–72. doi: 10.1007/s00432-015-2000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Flavin R., Peluso S., Nguyen P.L., Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6:551–562. doi: 10.2217/fon.10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leontieva O.V., Demidenko Z.N., Blagosklonny M.V. Dual MTORC1/C2 inhibitors suppress cellular geroconversion (a senescence program) Oncotarget. 2015;6:23238–23248. doi: 10.18632/oncotarget.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blagosklonny M.V. Rapamycin extends life and health span because it slows aging. Aging. 2013;5:592–598. doi: 10.18632/aging.100591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jinka R., Kapoor R., Sistla P.G., Raj T.A., Pande G. Alterations in cell-extracellular matrix interactions during progression of cancers. Int. J. Cell Biol. 2012;2012:219196. doi: 10.1155/2012/219196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dickreuter E., Cordes N. Cell-ECM interactions control DDR. Oncoscience. 2015;2:679–680. doi: 10.18632/oncoscience.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao H., Tanaka T., Mitlitski V., Heeter J., Balazs E.A., Darzynkiewicz Z. Protective effect of hyaluronate on oxidative DNA damage in WI-38 and A549 cells. Int. J. Oncol. 2008;32:1159–1167. doi: 10.3892/ijo_32_6_1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Darzynkiewicz Z., Balazs E.A. Genome integrity, stem cells and hyaluronan. Aging. 2012;4:78–88. doi: 10.18632/aging.100438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Halicka H.D., Mitlitski V., Heeter J., Balazs E.A., Darzynkiewicz Z. Attenuation of the oxidative burst-induced DNA damage in human leukocytes by hyaluronan. Int. J. Mol. Med. 2009;23:695–699. doi: 10.3892/ijmm_00000182. [DOI] [PubMed] [Google Scholar]
- 97.Gangaraju V.K., Lin H. MicroRNAs: Key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilson K.D., Venkatasubrahmanyam S., Jia F., Sun N., Butte A.J., Wu J.C. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sharma A., Wu J.C. MicroRNA expression profiling of human-induced pluripotent and embryonic stem cells. Methods Mol. Biol. 2013;936:247–256. doi: 10.1007/978-1-62703-083-0_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lei H., Quelle F.W. Foxo transcription factors enforce cell cycle checkpoints and promote survival of hematopoietic cells after DNA damage. Mol. Cancer Res. 2009;7:1294–1303. doi: 10.1158/1541-7786.MCR-08-0531. [DOI] [PubMed] [Google Scholar]
- 101.Laplante M., Sabatini D.M. MTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oh J., Lee Y.D., Wagers A.J. Stem cell aging: Mechanisms, regulators and therapeutic opportunities. Nat. Med. 2014;20:870–880. doi: 10.1038/nm.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Okada M., Kim H.W., Matsu-Ura K., Wang Y.G., Xu M., Ashraf M. Abrogation of age-induced microRNA-195 rejuvenates the senescent mesenchymal stem cells by reactivating telomerase. Stem Cells. 2016;34:148–159. doi: 10.1002/stem.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu F., Kong X., Lv L., Gao J. TGF-β1 acts through miR-155 to down-regulate TP53INP1 in promoting epithelial-mesenchymal transition and cancer stem cell phenotypes. Cancer Lett. 2015;359:288–298. doi: 10.1016/j.canlet.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 105.Aqil F., Munagala R., Jeyabalan J., Vadhanam M.V. Bioavailability of phytochemicals and its enhancement by drug delivery systems. Cancer Lett. 2013;334:133–141. doi: 10.1016/j.canlet.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Puig T., Vazquez-Martin A., Relat J., Petriz J., Menendez J.A., Porta R., Casals G., Marrero P.F., Haro D., Brunet J., et al. Fatty acid metabolism in breast cancer cells: Differential inhibitory effects of epigallocatechin gallate (EGCG) and C75. Breast Cancer Res. Treat. 2008;109:471–479. doi: 10.1007/s10549-007-9678-5. [DOI] [PubMed] [Google Scholar]
- 107.Relat J., Blancafort A., Oliveras G., Cufi S., Haro D., Marrero P.F., Puig T. Different fatty acid metabolism effects of (−)-epigallocatechin-3-gallate and C75 in adenocarcinoma lung cancer. BMC Cancer. 2012;12 doi: 10.1186/1471-2407-12-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khan A., Aljarbou A.N., Aldebasi Y.H., Faisal S.M., Khan M.A. Resveratrol suppresses the proliferation of breast cancer cells by inhibiting fatty acid synthase signaling pathway. Cancer Epidemiol. 2014;38:765–772. doi: 10.1016/j.canep.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 109.Fan H., Liang Y., Jiang B., Li X., Xun H., Sun J., He W., Lau H.T., Ma X. Curcumin inhibits intracellular fatty acid synthase and induces apoptosis in human breast cancer MDA-MB-231 cells. Oncol. Rep. 2016;35:2651–2656. doi: 10.3892/or.2016.4682. [DOI] [PubMed] [Google Scholar]
- 110.Crew K.D., Ho K.A., Brown P., Greenlee H., Bevers T.B., Arun B., Sneige N., Hudis C., McArthur H.L., Chang J., et al. Effects of a green tea extract, polyphenon e, on systemic biomarkers of growth factor signalling in women with hormone receptor-negative breast cancer. J. Hum. Nutr. Diet. 2015;28:272–282. doi: 10.1111/jhn.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carroll R.E., Benya R.V., Turgeon D.K., Vareed S., Neuman M., Rodriguez L., Kakarala M., Carpenter P.M., McLaren C., Meyskens F.L., Jr., et al. Phase IIA clinical trial of curcumin for the prevention of colorectal neoplasia. Cancer Prev. Res. 2011;4:354–364. doi: 10.1158/1940-6207.CAPR-10-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Momchilova A., Petkova D., Staneva G., Markovska T., Pankov R., Skrobanska R., Nikolova-Karakashian M., Koumanov K. Resveratrol alters the lipid composition, metabolism and peroxide level in senescent rat hepatocytes. Chem. Biol. Interact. 2014;207:74–80. doi: 10.1016/j.cbi.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 113.Liu H.W., Chan Y.C., Wang M.F., Wei C.C., Chang S.J. Dietary (−)-epigallocatechin-3-gallate supplementation counteracts aging-associated skeletal muscle insulin resistance and fatty liver in senescence-accelerated mouse. J. Agric. Food Chem. 2015;63:8407–8417. doi: 10.1021/acs.jafc.5b02501. [DOI] [PubMed] [Google Scholar]
- 114.Niu J., Shi Y., Tan G., Yang C.H., Fan M., Pfeffer L.M., Wu Z.H. DNA damage induces NF-κB-dependent microRNA-21 up-regulation and promotes breast cancer cell invasion. J. Biol. Chem. 2012;287:21783–21795. doi: 10.1074/jbc.M112.355495. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Zhang X., Wan G., Mlotshwa S., Vance V., Berger F.G., Chen H., Lu X. Oncogenic WIP1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70:7176–7186. doi: 10.1158/0008-5472.CAN-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]