Abstract
Autism Spectrum Disorder (ASD) is a group of neurodevelopmental disorders with complex genetic etiology. Recent studies have indicated that children with ASD may have altered folate or methionine metabolism, suggesting that the folate–methionine cycle may play a key role in the etiology of ASD. SLC19A1, also referred to as reduced folate carrier 1 (RFC1), is a member of the solute carrier group of transporters and is one of the key enzymes in the folate metabolism pathway. Findings from multiple genomic screens suggest the presence of an autism susceptibility locus on chromosome 21q22.3, which includes SLC19A1. Therefore, we performed a case-control study in a Japanese population. In this study, DNA samples obtained from 147 ASD patients at the Kanazawa University Hospital in Japan and 150 unrelated healthy Japanese volunteers were examined by the sequence-specific primer-polymerase chain reaction method pooled with fluorescence correlation spectroscopy. p < 0.05 was considered to represent a statistically significant outcome. Of 13 single nucleotide polymorphisms (SNPs) examined, a significant p-value was obtained for AA genotype of one SNP (rs1023159, OR = 0.39, 95% CI = 0.16–0.91, p = 0.0394; Fisher’s exact test). Despite some conflicting results, our findings supported a role for the polymorphism rs1023159 of the SLC19A1 gene, alone or in combination, as a risk factor for ASD. However, the findings were not consistent after multiple testing corrections. In conclusion, although our results supported a role of the SLC19A1 gene in the etiology of ASD, it was not a significant risk factor for the ASD samples analyzed in this study.
Keywords: autism spectrum disorder, reduced folate carrier, single nucleotide polymorphism
1. Introduction
Autism spectrum disorder (ASD) is a devastating neurodevelopmental disorder with a complex biological basis and is thought to involve multiple and variable gene–environment interactions. ASD is characterized by social impairments, communication problems, and restricted repetitive behaviors [1]. Most candidate genes currently implicated in ASD are involved in neurodevelopmental pathways, social-emotional behavior, or sex or neuropeptide hormonal signaling [2].
The SLC19A1 gene on human chromosome 21q22.3 [3] encodes one of the key enzymes in the folate metabolism pathway. SLC19A1, also referred to as reduced folate carrier 1 (RFC1), functions as a bidirectional anion exchanger, accepting folate cofactors and exporting various organic anions. SLC19A1 has five exons that contain the total open reading frame (ORF) [4,5,6]. The ORF of human SLC19A1 cDNA encodes a protein with 12 transmembrane domains and a single N-glycosylation site [3,7,8,9]. SLC19A1 mRNA is detectable in all human tissues [10].
Recent studies indicated that children with ASD may have changed folate or methionine metabolism, suggesting that the folate–methionine cycle may play an important role in the etiology of ASD [11]. Many important genes, including SLC19A1, are involved in the folate metabolism pathway and their roles in human diseases, such as gastric and esophageal cancers, have been studied in depth [12,13]. A marginal association with ASD was identified for a 19-bp deletion in the dihydrofolate reductase (DHFR) gene (odds ratio (OR): 2.69; 95% CI: 1.00–7.28; p < 0.05), which is involved in folate metabolism [14]. Common variants of the decreased folate carrier (RFC) and methylene tetrahydrofolate reductase (MTHFR) genes conferred increased susceptibility to ASD, suggesting a potential etiological role of impaired folate-dependent one-carbon metabolism in susceptibility to ASD [15].
However, the findings for genes involved in folate transport have been inconsistent between reports. Although the largest study to date found an important association between the SLC19A1 gene and ASD [15], a subsequent study failed to replicate this finding [16]. Other studies have not identified any mutations in genes included in folate transport in ASD populations [17,18,19].
Here, we hypothesized that genetic variants in SLC19A1 may play a role in the pathways that are altered in ASD and can therefore be considered candidate genes for testing in ASD patients. We performed a case-control study of 13 genetic variations to assess the involvement of SLC19A1 in ASD. The study was performed in a Japanese population, in which genetic variants of CD38 and BST-1/CD157 were reported to be associated with increased risk of ASD [20,21].
2. Results
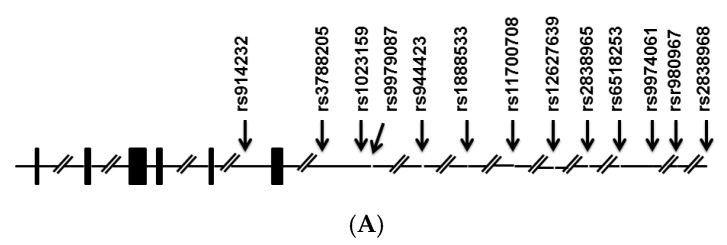
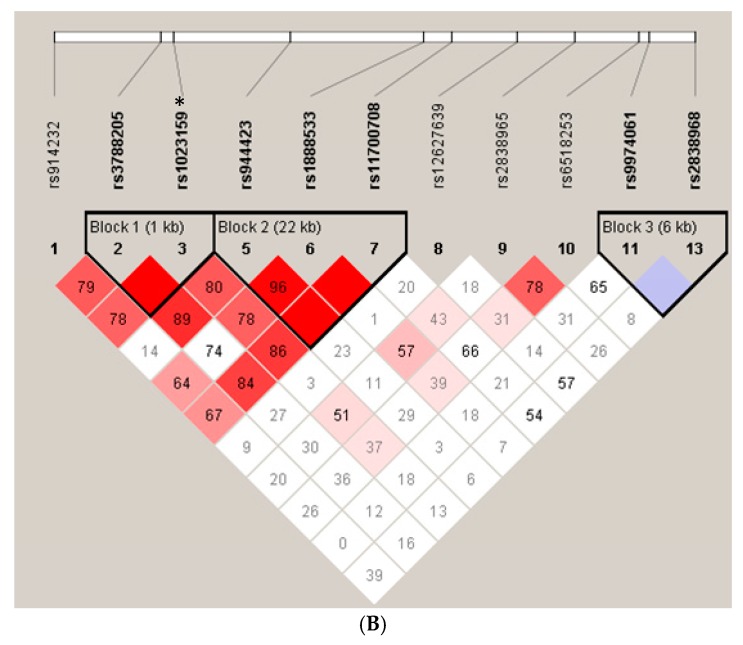
Thirteen SNPs were analyzed in this study, five of which (rs914232, rs3788205, rs1023159, rs944423, and rs9979087) were located in the SLC19A1 gene region; these were subjected to statistical analysis. The eight other SNPs (rs1888533, rs11700708, rs12627639, rs2838965, rs6518253, rs9974061, rs9980967, and rs2838968) were located in the adjacent region. Two SNPs (rs9980967, rs9979087) were excluded due to insufficient genotyping data. However, there were no significant associations between any of these SNPs and ASD, with the exception of rs1023159. As the results suggested a role (p = 0.0394; Table 1) of this polymorphism alone or in combination with others as a risk factor for ASD, this SNP was subjected to further analysis. No association was found after multiple testing corrections. Tests of Hardy–Weinberg equilibrium deviations were performed for each marker in two groups of case and control individuals, and polymorphisms showed evidence of deviation from Hardy–Weinberg equilibrium. The genotyping rate was above 95%. LD analysis of these SNPs identified three haplotype blocks, one of which (Block 1; Figure 1) consisted of two SNPs including one (rs1023159) with the lowest p-value (p = 0.0394; Table 1) among those analyzed.
Table 1.
Genotype and allele frequencies of rs1023159 at Kanazawa University Hospital for autism spectrum disorder (ASD).
| rs1023159 | Cases | Control | Odds Ratio (95% CI) | p |
|---|---|---|---|---|
| Genotype | (n = 144) | (n = 146) | ||
| G/G | 72 (50.0%) | 62 (42.5%) | Reference | |
| A/G | 63 (43.8%) | 64 (43.8%) | 0.85 (0.52–1.4) | 0.5368 |
| A/A | 9 (6.3%) | 20 (13.7%) | 0.39 (0.16–0.91) | 0.0394 |
| Allele | (n = 288) | (n = 292) | ||
| G | 207 (71.9%) | 188 (64.4%) | Reference | |
| A | 81 (28.1%) | 104 (35.6%) | 0.71 (0.50–1.0) | 0.0613 |
CI, confidence interval; p-values obtained by Fisher’s exact test are given; p < 0.05 is indicated in bold.
Figure 1.
The genomic structure of SLC19A1 (A). Bars, exons. Arrows, positions of single nucleotide polymorphisms (SNPs). Linkage disequilibrium plot of SNPs in the samples studied (B). Numbers in squares indicate D′ values. Reference Number (rs) with asterisk indicates the SNP with p < 0.05. The blocks are defined following the four-gamete rule [22]. Explanation of color scheme: If D′ < 1 and LOD (log of the likelihood odds ratio) <2, the cell color is white; if D′ = 1 and LOD < 2, the cell color is blue; if D′ < 1 and LOD ≥ 2, the cell color is shades of pink/red; if D′ = 1 and LOD ≥ 2, the cell color is bright red.
3. Discussion
In this population-based case-control study, we investigated the relationship between polymorphisms in the SLC19A1 gene and risk of ASD in a Japanese population. We identified no significant associations between SNPs of the SLC19A1 gene and ASD, with the exception of one SNP, although the results eventually did not support a role of the SLC19A1 gene in the etiology of ASD in our sample.
We have also calculated the genotype and allele frequencies of rs1023159 in Autism Genome Resources Exchange (AGRE) samples (Table 2). The frequency (15.5%) of the AA genotype in AGRE group of ASD cases, although with different ethnicity, was similar to the frequency (13.7%) observed in the group of Japanese controls.
Table 2.
Genotype and allele frequencies of rs1023159 in KU samples and AGRE samples for autism spectrum disorder (ASD).
| rs1023159 | KU | AGRE |
|---|---|---|
| Genotype | (n = 144) | (n = 191) |
| G/G | 72 (50.0%) | 63 (31.4%) |
| A/G | 63 (43.8%) | 104 (53.1%) |
| A/A | 9 (6.3%) | 30 (15.5%) |
| Allele | (n = 288) | (n = 394) |
| G | 207 (71.9%) | 230 (58.4%) |
| A | 81 (28.1%) | 164 (41.6%) |
KU, Kanazawa University; AGRE, Autism Genome Resources Exchange.
Recent genetic studies recognized the contribution of the SLC19A1 gene to neural tube defects (NTD) [23,24,25,26,27,28]. It was suggested that the maternal G allele may be a causative genetic risk factor for having a child with ASD independent of the child’s genotype [29]. In case-control analysis, a significant increase in the SLC19A1 G allele frequency was discovered among mothers of children with ASD, but not among affected children, and analysis of the SLC19A1 A80G genotype within family trios discovered that the maternal G allele was allied with a significant increase in risk of ASD, whereas the inherited genotype of the children was not [29].
Evidence indicates that expression of SLC19A1 in the intestine is subject to adaptive regulation in response to folate status [30]. Folic acid is the inactive, oxidized form of folate compounds that is important for many physiological systems of the body. Folate is the major one-carbon donor for de novo nucleotide synthesis for DNA replication and also for remethylation of homocysteine to methionine for essential methylation reactions [29]. The folate cycle interacts with the methionine cycle as well as the tetrahydrobiopterin construction and salvage pathways. Insufficiencies in folate can lead to anomalies in these pathways [31]. The methionine cycle is important for DNA methylation [15], a process that is important in regulating gene expression. Folate deficiency during various significant stages of fetal and infantile development upsets structural and functional alteration of the brain [32].
The reduced folate carrier is the principal mechanism by which folates and antifolates are delivered to mammalian cells and tissues [33]. As folate transport across cell membranes is mediated in part by the RFC, variants within this gene may affect the disease risk via an effect on folate and/or homocysteine levels [34]. Low levels of RFC could result in a number of pathophysiological states associated with folate deficiency, including cardiovascular disease, fetal anomalies, and neurological disorders [33]. Moretti et al. reported a 6-year-old girl with developmental delay, psychomotor regression, seizures, mental retardation, and autistic features associated with low cerebrospinal fluid (CSF) levels of 5-methyltetrahydrofolate, the biologically active form of folate in the CSF and blood [35]. Several studies reported considerably low CSF folate concentrations together with normal serum folate concentrations in children with autism [18,19,35,36,37].
SLC19A1 is situated on the CSF side of the choroid plexus, where it enables transport of concentrated folate into the CSF [11]. Taken together, variation in SLC19A1 expression may involve both neuronal structures and metabolism in the Central Nervous System (CNS). Defective transport of folate into the CNS is related to cerebral folate deficiency (CFD), a neurological disorder that is important in diagnosis of children with unexplained neurodevelopmental symptoms, which suggests the possible involvement of the folate-methionine pathway in ASD [31]. Further, early-onset low-functioning autism with neurological deficits has been suggested as a characteristic of children with both autism and CFD [18,19,35,38].
The major limitation of this study was the small sample size, resulting in deviation from Hardy–Weinberg equilibrium and limited power (68%) to reliably detect the role of SLC19A1 in ASD. We did not recognize any population stratification, admixture, and cryptic relation among the subjects in the present study, which may have contributed to the lack of association in this small sample. Another limitation was the lack of a replication cohort. Further studies with larger sample sizes and/or family-based association testing are needed to clarify the precise role of this gene in ASD. However, our findings were consistent with reports that SLC19A1 may not contribute to genetic susceptibility to ASD in some populations.
4. Materials and Methods
4.1. Study Population
The study population consisted of 147 ASD subjects (113 males, 34 females; 15.6 ± 0.6 years old, mean ± s.e.m.) from the Outpatient Psychiatry Department of Kanazawa University Hospital, as described previously [20,39]. All subjects satisfied the Diagnostic and Statistical Mannual of Mental Disorders-IV (DSM-IV) criteria for pervasive developmental ailment and Childhood Autism Rating Scale. Two experienced child psychiatrists established the diagnosis of ASD in all patients based on semi-structured behavioral observations and conversations with the subjects and their parents. The interview structure and clinical records were described previously [20]. One of the following methods was used as an aid to evaluate the autism-specific behaviors and symptoms during interviews with parents: the Asperger Syndrome Diagnostic Interview [40], Autism Diagnostic Interview-Revised (ADI-R) [41], Pervasive Developmental Disorders Autism Society Japan Rating Scale [42], Diagnostic Interview for Social and Communication Disorders [43], or Tokyo Autistic Behavior Scale [44]. A total of 150 individuals were recruited as controls (115 males, 35 females; 23.8 ± 0.3 years old). All patients and controls were Japanese with no non-Japanese parents or grandparents. These controls were part of a stock used frequently for single nucleotide polymorphism (SNP) analysis of ion channels related to arrhythmia at Kanazawa University Heart Center. This study was approved by the ethics committee of Kanazawa University School of Medicine (July 2015), and all participants and/or their caregivers provided informed consent. The study protocol was performed in accordance with the Declaration of Helsinki.
4.2. Genotyping
Genomic DNA was extracted as described previously [39] from venous blood samples using a commercial kit (Wizard Genomic DNA Purification kit; Promega, Madison, WI, USA) or from nails using an ISOHAIR DNA extraction kit (Nippon Gene, Tokyo, Japan). The genomic DNA samples were subjected to whole-genome amplification, and SNPs were determined by the sequence-specific primer-polymerase chain reaction (SSP-PCR) method followed by fluorescence correlation spectroscopy as described by Bannai et al. [45]. We selected a set of tagging SNPs that capture common variations and linkage disequilibrium (LD) structures across the SLC19A1 gene using the Tagger program incorporated with Haploview v4.2 software(Broad Institute of MIT and Harvard, Cambridge, MA, USA). The data source for tagging SNPs was the dbSNP database [46] and the HapMap genome browser, release 27 (operated by the National Institutes of Health (NIH), Bethesda, MD, USA) in the JPT (Japanese individuals from Tokyo, Japan), CHB (Han Chinese individuals from Beijing, China), ASW (African ancestry in Southwest USA), and CEU (Utah residents of northern and western European ancestry) populations. Selection of tagging SNPs was based on pairwise tagging only and the minor allele frequency was ≥5% in any one of the different ethnicities.
4.3. Statistical Analysis
Genotype and allele frequencies were examined using a contingency table and Fisher’s exact test (GraphPad Prism 6; GraphPad Software, San Diego, CA, USA), and p < 0.05 was taken to indicate statistical significance. We also used the method of Nyholt [47], which estimates an “effective number” of independent tests and then adjusts the smallest observed p-value using simulation based on this number of tests. In our samples, the estimated effective number for independent tests was 9 and the p-value was 0.005. The observed genotype frequency distributions were compared with those expected from the Hardy–Weinberg equilibrium and analyzed by the chi “χ” squared test.
Statistical power was calculated using the Genetic Power Calculator [48,49] assuming a population prevalence of 0.015 for ASD [50], and a D’ value of 1 between the marker and disease with a false positive rate of 5%.
5. Conclusions
This study showed no evidence supporting a role of the SLC19A1 gene in the etiology of ASD. The ethnic and cultural background may have influenced the results of our study. However, these findings warrant additional discussion and confirmation in subsequent studies. Further cellular and molecular studies are required to elucidate the precise role of this gene in ASD.
Acknowledgments
We acknowledge financial support from the Strategic Research Program for Brain Sciences from the Japan Agency for Medical Research and Development and Grants-in-Aid for Scientific Research (24590375 and 25461335) from the Japan Society for Promotion of Sciences. We thank AGRE for their cooperation.
Author Contributions
Haruhiro Higashida, Shigeru Yokoyama, Jian-Jun Huang and Li Liu conceived and designed the research. Shigeru Yokoyama and Naila Al Mahmuda performed experiments and analyzed data. Toshio Munesue, Kenshi Hayashi, Kunimasa Yagi, Masakazu Yamagishi and Hideo Nakatani contributed participant recruitment, clinical assessment and sample collections. Naila Al Mahmuda prepared the initial draft; and Shigeru Yokoyama, Naila Al Mahmuda, and Haruhiro Higashida revised the manuscript. All authors reviewed the final manuscript and approved its publication.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.American Psychiatric Association . Diagnostic and Statistical Mannual of Mental Disorders (DSM-V) 5th ed. American Psychiatric Pub.; Washington, DC, USA: 2013. [Google Scholar]
- 2.Chakrabarti B., Dudbridge F., Kent L., Wheelwright S., Hill-Cawthorne G., Allison C., Banerjee-Basu S., Baron-Cohen S. Genes related to sex steroids, neural growth, and social–emotional behavior are associated with autistic traits, empathy, and Asperger syndrome. Autism Res. 2009;2:157–177. doi: 10.1002/aur.80. [DOI] [PubMed] [Google Scholar]
- 3.Moscow J.A., Gong M., He R., Sgagias M.K., Dixon K.H., Anzick S.L., Meltzer P.S., Cowan K.H. Isolation of a gene encoding a human reduced folate carrier (RFC1) and analysis of its expression in transport-deficient, methotrexate-resistant human breast cancer cells. Cancer Res. 1995;55:3790–3794. [PubMed] [Google Scholar]
- 4.Tolner B., Roy K., Sirotnak F.M. Structural analysis of the human RFC-1 gene encoding a folate transporter reveals multiple promoters and alternatively spliced transcripts with 5′ end heterogeneity. Gene. 1998;211:331–341. doi: 10.1016/S0378-1119(98)00123-1. [DOI] [PubMed] [Google Scholar]
- 5.Williams F.M., Flintoff W.F. Structural organization of the human reduced folate carrier gene: Evidence for 5′ heterogeneity in lymphoblast mRNA. Somat. Cell Mol. Genet. 1998;24:143–156. doi: 10.1023/B:SCAM.0000007117.50428.63. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L., Wong S.C., Matherly L.H. Structure and organization of the human reduced folate carrier gene1. Biochim. Biophys. Acta. 1998;1442:389–393. doi: 10.1016/S0167-4781(98)00168-7. [DOI] [PubMed] [Google Scholar]
- 7.Prasad P.D., Ramamoorthy S., Leibach F.H., Ganapathy V. Molecular Cloning of the Human Placental Folate Transporter. Biochem. Biophys. Res. Commun. 1995;206:681–687. doi: 10.1006/bbrc.1995.1096. [DOI] [PubMed] [Google Scholar]
- 8.Williams F.M.R., Flintoff W.F. Isolation of a Human cDNA that Complements a Mutant Hamster Cell Defective in Methotrexate Uptake. J. Biol. Chem. 1995;270:2987–2992. doi: 10.1074/jbc.270.7.2987. [DOI] [PubMed] [Google Scholar]
- 9.Wong S.C., Proefke S.A., Bhushan A., Matherly L.H. Isolation of human cDNAs that restore methotrexate sensitivity and reduced folate carrier activity in methotrexate transport-defective Chinese Hamster ovary cells. J. Biol. Chem. 1995;270:17468–17475. doi: 10.1074/jbc.270.29.17468. [DOI] [PubMed] [Google Scholar]
- 10.Ganapathy V., Smith S., Prasad P. SLC19: The folate/thiamine transporter family. Pflugers Arch. 2004;447:641–646. doi: 10.1007/s00424-003-1068-1. [DOI] [PubMed] [Google Scholar]
- 11.Main P.A., Angley M.T., Thomas P., O’Doherty C.E., Fenech M. Folate and methionine metabolism in autism: A systematic review. Am. J. Clin. Nutr. 2010;91:1598–1620. doi: 10.3945/ajcn.2009.29002. [DOI] [PubMed] [Google Scholar]
- 12.Wang L., Chen W., Wang J., Tan Y., Zhou Y., Ding W., Hua Z., Shen J., Xu Y., Shen H. Reduced folate carrier gene G80A polymorphism is associated with an increased risk of gastroesophageal cancers in a Chinese population. Eur. J. Cancer. 2006;42:3206–3211. doi: 10.1016/j.ejca.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z., Xu Y., Zhou J., Wang X., Wang L., Hu X., Guo J., Wei Q., Shen H. Polymorphisms of thymidylate synthase in the 5′- and 3′-untranslated regions associated with risk of gastric cancer in South China: A case-control analysis. Carcinogenesis. 2005;26:1764–1769. doi: 10.1093/carcin/bgi143. [DOI] [PubMed] [Google Scholar]
- 14.Adams M., Lucock M., Stuart J., Fardell S., Baker K., Ng X. Preliminary evidence for involvement of the folate gene polymorphism 19 bp deletion-DHFR in occurrence of autism. Neurosci. Lett. 2007;422:24–29. doi: 10.1016/j.neulet.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 15.James S.J., Melnyk S., Jernigan S., Cleves M.A., Halsted C.H., Wong D.H., Cutler P., Bock K., Boris M., Bradstreet J.J., et al. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141B:947–956. doi: 10.1002/ajmg.b.30366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paşca S.P., Nemeş B., Vlase L., Gagyi C.E., Dronca E., Miu A.C., Dronca M. High levels of homocysteine and low serum paraoxonase 1 arylesterase activity in children with autism. Life Sci. 2006;78:2244–2248. doi: 10.1016/j.lfs.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 17.Gordon N. Cerebral folate deficiency. Dev. Med. Child Neurol. 2009;51:180–182. doi: 10.1111/j.1469-8749.2008.03185.x. [DOI] [PubMed] [Google Scholar]
- 18.Moretti P., Peters S.U., del Gaudio D., Sahoo T., Hyland K., Bottiglieri T., Hopkin R.J., Peach E., Min S.H., Goldman D., et al. Brief Report: Autistic Symptoms, Developmental Regression, Mental Retardation, Epilepsy, and Dyskinesias in CNS Folate Deficiency. J. Autism Dev. Disord. 2008;38:1170–1177. doi: 10.1007/s10803-007-0492-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramaekers V.T., Blau N., Sequeira J.M., Nassogne M.C., Quadros E.V. Folate receptor autoimmunity and cerebral folate deficiency in low-functioning autism with neurological deficits. Neuropediatrics. 2007;38:276–281. doi: 10.1055/s-2008-1065354. [DOI] [PubMed] [Google Scholar]
- 20.Munesue T., Yokoyama S., Nakamura K., Anitha A., Yamada K., Hayashi K., Asaka T., Liu H.-X., Jin D., Koizumi K., et al. Two genetic variants of CD38 in subjects with autism spectrum disorder and controls. Neurosci. Res. 2010;67:181–191. doi: 10.1016/j.neures.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama S., Al Mahmuda N., Munesue T., Hayashi K., Yagi K., Yamagishi M., Higashida H. Association Study between the CD157/BST1 Gene and Autism Spectrum Disorders in a Japanese Population. Brain Sci. 2015;5:188–200. doi: 10.3390/brainsci5020188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang N., Akey J.M., Zhang K., Chakraborty R., Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: The interplay of population history, recombination, and mutation. Am. J. Hum. Genet. 2002;71:1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Leary V.B., Pangilinan F., Cox C., Parle-McDermott A., Conley M., Molloy A.M., Kirke P.N., Mills J.L., Brody L.C., Scott J.M. Reduced folate carrier polymorphisms and neural tube defect risk. Mol. Genet. Metab. 2006;87:364–369. doi: 10.1016/j.ymgme.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 24.Pei L.J., Li Z.W., Zhang W., Ren A.G., Zhu H.P., Hao L., Zhu J.H., Li Z. Epidemiological study on reduced folate carrier gene (RFC1 A80G) polymorphism and other risk factors of neural tube defects. J. Peking Univ. Health Sci. 2005;37:341–345. [PubMed] [Google Scholar]
- 25.Pei L.J., Zhu H.P., Li Z.W., Zhang W., Ren A.G., Zhu J.H., Li Z. Interaction between maternal periconceptional supplementation of folic acid and reduced folate carrier gene polymorphism of neural tube defects. Chin. J. Med. Genet. 2005;22:284–287. [PubMed] [Google Scholar]
- 26.Zhang T., Lou J., Zhong R., Wu J., Zou L., Sun Y., Lu X., Liu L., Miao X., Xiong G. Genetic variants in the folate pathway and the risk of neural tube defects: A meta-analysis of the published literature. PLoS ONE. 2013;8:772. doi: 10.1371/journal.pone.0059570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang Y., Zhao H., Niu B., Li W.I., Zhou R., Zhang T., Xie J. Correlation of polymorphism of MTHFRs and RFC-1 genes with neural tube defects in China. Birth Defects Res. A Clin. Mol. Teratol. 2008;82:3–7. doi: 10.1002/bdra.20416. [DOI] [PubMed] [Google Scholar]
- 28.De Marco P., Calevo M.G., Moroni A., Merello E., Raso A., Finnell R.H., Zhu H., Andreussi L., Cama A., Capra V. Reduced folate carrier polymorphism (80A→G) and neural tube defects. Eur. J. Hum. Genet. 2003;11:245–252. doi: 10.1038/sj.ejhg.5200946. [DOI] [PubMed] [Google Scholar]
- 29.James S.J., Melnyk S., Jernigan S., Lehman S., Seidel L., Gaylor D.W., Cleves M.A. A functional polymorphism in the reduced folate carrier gene and DNA hypomethylation in mothers of children with autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B:1209–1220. doi: 10.1002/ajmg.b.31094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Said H.M., Chatterjee N., Haq R.U., Subramanian V.S., Ortiz A., Matherly L.H., Sirotnak F.M., Halsted C., Rubin S.A. Adaptive regulation of intestinal folate uptake: Effect of dietary folate deficiency. Am. J. Physiol. Cell Physiol. 2000;279:C1889–C1895. doi: 10.1152/ajpcell.2000.279.6.C1889. [DOI] [PubMed] [Google Scholar]
- 31.Frye R.E., Rossignol D.A. Cerebral Folate Deficiency in Autism Spectrum Disorders. [(accessed on 19 May 2016)];Autism Sci. Dig. J. Autsmone. 2011 Available online: http://www.autismone.org/content/cerebral-folate-deficiency-autism-spectrum-disorders-richard-frye-md-phd-and-daniel-rossigno. [Google Scholar]
- 32.Ramaekers V., Sequeira J.M., Quadros E.V. Clinical recognition and aspects of the cerebral folate deficiency syndromes. Clin. Chem. Lab. Med. 2013;51:497–511. doi: 10.1515/cclm-2012-0543. [DOI] [PubMed] [Google Scholar]
- 33.Matherly L., Hou Z., Deng Y. Human reduced folate carrier: Translation of basic biology to cancer etiology and therapy. Cancer Metastasis Rev. 2007;26:111–128. doi: 10.1007/s10555-007-9046-2. [DOI] [PubMed] [Google Scholar]
- 34.Stanislawska-Sachadyn A., Mitchell L.E., Woodside J.V., Buckley P.T., Kealey C., Young I.S., Scott J.M., Murray L., Boreham C.A., McNulty H., et al. The reduced folate carrier (SLC19A1) c.80G>A polymorphism is associated with red cell folate concentrations among women. Ann. Hum. Genet. 2009;73(Pt 5):484–491. doi: 10.1111/j.1469-1809.2009.00529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moretti P., Sahoo T., Hyland K., Bottiglieri T., Peters S., del Gaudio D., Roa B., Curry S., Zhu H., Finnell R.H., et al. Cerebral folate deficiency with developmental delay, autism, and response to folinic acid. Neurology. 2005;64:1088–1090. doi: 10.1212/01.WNL.0000154641.08211.B7. [DOI] [PubMed] [Google Scholar]
- 36.Lowe T.L., Cohen D.J., Miller S., Young J.G. Folic acid and B12 in autism and neuropsychiatric disturbances of childhood. J. Am. Acad. Child Psychiatry. 1981;20:104–111. doi: 10.1016/S0002-7138(09)60720-2. [DOI] [PubMed] [Google Scholar]
- 37.Ramaekers V.T., Rothenberg S.P., Sequeira J.M., Opladen T., Blau N., Quadros E.V., Selhub J. Autoantibodies to folate receptors in the cerebral folate deficiency syndrome. N. Eng. J. Med. 2005;352:1985–1991. doi: 10.1056/NEJMoa043160. [DOI] [PubMed] [Google Scholar]
- 38.Ramaekers V.T., Sequeira J.M., Blau N., Quadros E.V. A milk-free diet downregulates folate receptor autoimmunity in cerebral folate deficiency syndrome. Dev. Med. Child Neurol. 2008;50:346–352. doi: 10.1111/j.1469-8749.2008.02053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma W.-J., Hashii M., Munesue T., Hayashi K., Yagi K., Yamagishi M., Higashida H., Yokoyama S. Non-synonymous single-nucleotide variations of the human oxytocin receptor gene and autism spectrum disorders: A case-control study in a Japanese population and functional analysis. Mol. Autism. 2013;4:22. doi: 10.1186/2040-2392-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillberg C., Gillberg C., Råstam M., Wentz E. The Asperger Syndrome (and High-Functioning Autism) Diagnostic Interview (ASDI): A Preliminary Study of a New Structured Clinical Interview. Autism. 2001;5:57–66. doi: 10.1177/1362361301005001006. [DOI] [PubMed] [Google Scholar]
- 41.Lord C., Rutter M., le Couteur A. Autism Diagnostic Interview-Revised: A revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 42.Autism Society Japan . Pervasive Developmental Disorders Autism Society Japan Rating Scale (PARS) Spectrum Publishing Company; Tokyo, Japan: 2006. [Google Scholar]
- 43.Wing L., Leekam S.R., Libby S.J., Gould J., Larcombe M. The diagnostic interview for social and communication disorders: Background, inter-rater reliability and clinical use. J. Child Psychol. Psychiatry. 2002;43:307–325. doi: 10.1111/1469-7610.00023. [DOI] [PubMed] [Google Scholar]
- 44.Kurita H., Miyake Y. The Reliability and Validity of the Tokyo Autistic Behaviour Scale. Psychiatry Clin. Neurosci. 1990;44:25–32. doi: 10.1111/j.1440-1819.1990.tb00438.x. [DOI] [PubMed] [Google Scholar]
- 45.Nishida N., Tanabe T., Takasu M., Suyama A., Tokunaga K. Further development of multiplex single nucleotide polymorphism typing method, the DigiTag2 assay. Anal. Biochem. 2007;364:78–85. doi: 10.1016/j.ab.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 46.dbSNP: Short Genetic Variations. [(accessed on 24 June 2015)]; Available online: http://www.ncbi.nlm.nih.gov/SNP/
- 47.Nyholt D.R. A simple correction for multiple testing for SNPs in linkage disequilibrium with each other. Am. J. Hum. Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purcell S., Cherny S.S., Sham P.C. Genetic power calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics (Oxf. Engl.) 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 49.Genetic Power Calculator. [(accessed on 27 June 2015)]. Available online: http://pngu.mgh.harvard.edu/purcell/gpc/cc2.html.
- 50.Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators. Centers for Disease Control and Prevention (CDC) Prevalence of autism spectrum disorder among children aged 8 years—Autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill. Summ. 2014;63:1–21. [PubMed] [Google Scholar]