Abstract
Alzheimer's disease (AD) is a multifactorial, fatal neurodegenerative disorder characterized by the abnormal accumulation of Aβ and Tau deposits in the brain. There is no cure for AD, and failure at different clinical trials emphasizes the need for new treatments. In recent years, significant progress has been made toward the development of miRNA-based therapeutics for human disorders. This study was designed to evaluate the efficiency and potential safety of miRNA replacement therapy in AD, using miR-15/107 paralogues as candidate drug targets. We identified miR-16 as a potent inhibitor of amyloid precursor protein (APP) and BACE1 expression, Aβ peptide production, and Tau phosphorylation in cells. Brain delivery of miR-16 mimics in mice resulted in a reduction of AD-related genes APP, BACE1, and Tau in a region-dependent manner. We further identified Nicastrin, a γ-secretase component involved in Aβ generation, as a target of miR-16. Proteomics analysis identified a number of additional putative miR-16 targets in vivo, including α-Synuclein and Transferrin receptor 1. Top-ranking biological networks associated with miR-16 delivery included AD and oxidative stress. Collectively, our data suggest that miR-16 is a good candidate for future drug development by targeting simultaneously endogenous regulators of AD biomarkers (i.e., Aβ and Tau), inflammation, and oxidative stress.
Keywords: Alzheimer's disease, brain delivery, miR-16, microRNA, therapy
Introduction
Alzheimer's disease (AD) is the most common form of dementia characterized by the progressive decline of cognitive and behavioral abilities.1 Most (>95%) AD cases are of sporadic origin with unknown cause.2,3 The two major hallmarks of the disease are senile plaques and neurofibrillary tangles in the brain. Plaques are composed of depositions of β-amyloid (Aβ) peptides, generated by the cleavage of the amyloid precursor protein (APP) by BACE1/β-secretase and γ-secretase (a multiprotein complex composed of Presenilin, Nicastrin, PEN2, and Aph-1).4 Neurofibrillary tangles are composed of intraneuronal inclusions of hyperphosphorylated and aggregated Tau protein.5
Up to now, most therapeutic efforts in AD have been concentrated toward the development of drugs against Aβ (e.g., APP, BACE1, and γ-secretase) or Tau independently. AD pathology is complex and likely multifactorial, where it has been suggested that the conventional “one protein, one drug, one disease” theory for AD would not be effective.6 Failure of recent clinical trials is in line with this hypothesis.6,7,8 Strategies that target simultaneously multiple disease components and/or pathways could therefore address this issue.9,10
MiRNAs comprise the largest group of small (~22 nt) endogenous noncoding RNAs driving gene silencing in cells,11 predicted to regulate more than 60% of protein-coding genes.12,13 Once incorporated into the RNA-induced silencing complex, miRNAs function to repress translation and/or promote RNA degradation through imperfect base-pairing with specific mRNA sequences, generally located in the 3' untranslated region (3'UTR).14 The aberrant expression of miRNAs in many human diseases and involvement in key biological pathways has made them attractive drug targets.15,16,17 This is well recognized in the cancer field, where miRNAs can function as oncogenes or tumor suppressors.16,18 The interest for miRNA replacement therapy is rapidly growing,19,20,21 which involves the “reintroduction” of a missing miRNA into cells to compensate for a loss-of-function. miRNA mimics used in replacement therapy have the same sequence and structure as the depleted, endogenously expressed miRNA. Thus, off-target effects are less likely to occur as the mimics behave like their natural counterparts by fine-tuning the expression of targets through conserved miRNA:mRNA interactions.19 In contrast to conventional gene therapy that involves relatively large DNA plasmids or viral vectors, miRNA mimics are substantially smaller in size, and they merely need to enter the cytoplasm of target cells to be active. Another strong rational to use miRNAs in replacement therapy is based on the fact that a single miRNA can regulate multiple genes simultaneously, therefore acting on “disease pathways”.22
The miR-15/107 superfamily controls a number of fundamental processes including metabolism, cell cycle regulation, inflammation, and the stress response.23,24 Interestingly, several members of this family have been documented to be misregulated in AD brain, including miR-16,25,26 miR-15a/b,27,28,29,30,31 miR-195,32,33 and miR-103/107.34,35,36 Studies in vitro have implicated miR-16, miR-15a, miR-195 in the regulation of BACE1 and APP expression, Aβ production, and Tau phosphorylation.31,33,37 More recent studies in vivo have implicated miR-195 in memory formation.33 Collectively, these studies point to the potential therapeutic use of miRNAs in AD by targeting genes involved in both Aβ production and Tau metabolism. To date, however, a detailed comparative analysis of miR-15/107 superfamily members has not yet been conducted.
A defined protocol has been proposed before entering miRNAs into the clinic, including the optimization of suitable candidates.38 However, these procedures have been developed mainly for peripheral disorders (e.g., inhibition of liver-specific miR-122),39 leaving neurodegenerative diseases largely unexploited. Several critical questions remain to be addressed: Is widespread delivery to the brain possible? Are miRNAs functional in the brain, and particularly in neurons? What are potential side effects? In attempt to address these issues, we sought to evaluate the therapeutic applicability of miRNAs in AD, using miR-15/107 family members as candidate drug targets. Specifically, we wanted to determine the efficiency and potential safety of miRNA mimics toward the regulation of AD-related genes in vivo with a focus on endogenous APP, BACE1, and Tau.
Results
Comparative analysis of miR-15/107 family members in vitro and in cells
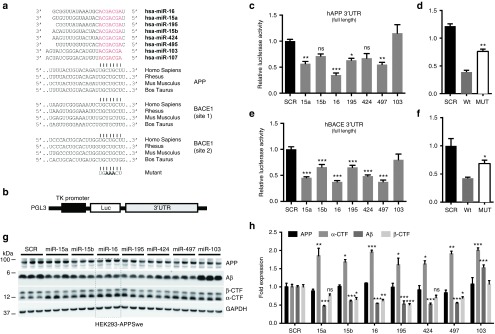
Our experimental strategy is presented in see Supplementary Figure S1. We first evaluated the effects of miR-15a, -15b, -16, -195, -424, -497, and -103 mimics on human APP and BACE1 expression in luciferase-based assays. In contrast to previous studies,33,40,41 we used the full-length 3'UTR of tested genes to better mimic physiological conditions. As shown in Figure 1a, the predicted miRNA binding sites in APP and BACE1 are highly conserved. We cotransfected the wild-type 3'UTR reporter constructs (Figure 1b) with candidate miRNA mimics into native HEK293 cells. Compared to a scrambled control, most miRNAs significantly reduced luciferase signal (expression) of both APP and BACE1 (Figure 1c,e). Among tested miRNAs, miR-16 showed the strongest negative effects on both APP and BACE1. To validate the specificity of these results, we generated mutant APP (CTG546-548AAA) and BACE1 (CTG269-271AAA and CTG1798-1800AAA) reporter constructs. As expected, disruption of miR-16 binding sites partly rescued the effects on luciferase activity (Figure 1d,f).
Figure 1.
Comparative analysis of miR-15/107 family members in vitro. (a) Mature miRNA sequences are shown. Seed sequences are shown in red. The corresponding binding sites within the amyloid precursor protein (APP) and BACE1 3'UTRs are shown in gray. (b) Schematic representation (not to scale) of the luciferase reporter construct. Luc, luciferase gene; TK, thymidine kinase promoter. (c) APP 3'UTR regulation by selected miR-15/107 family members. HEK293 cells were transfected with 50 nmol/l final concentration of candidate mimics. Twenty-four hours post-transfection luciferase signal was measured. Signals were normalized for transfection efficiency, and graph represents the relative luciferase signals compared to the scrambled control (SCR). Statistical significance was assessed by one-way analysis of variance (ANOVA) with Bonferroni multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001. (d) Luciferase assays were performed using a mutant 3'UTR construct for APP. Here, cells were treated with and 12.5 nmol/l of miR-16 mimics. A significant difference was observed between wild-type and mutant constructs (**P < 0.01). (e) BACE1 3'UTR regulation by selected miR15/107 family members. HEK293 cells were transfected with 50 nmol/l final concentration of candidate miRNA mimics. Twenty-four hours post-transfection luciferase signal was measured. Graph represents the relative luciferase signals compared to the SCR. Statistical significance was assessed by one-way ANOVA with Bonferroni multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001. (f) Luciferase assay using a 3'UTR BACE1 double mutant construct. Cells were treated with 25 nmol/l of miR-16 oligos. Statistical significance was calculated by one-way ANOVA with Bonferroni multiple comparison test. (g,h) HEK293-APPSwe cells were treated with candidate mimics at a final concentration of 50 nmol/l. A strong effect on soluble Aβ levels (measured in cell medium) was observed after 24 hours treatment. This is in agreement with the downregulation of APP β-CTFs (the direct BACE1 substrates) and concomitant increase in APP α-CTFs. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
We next performed functional studies in HEK293 cells overexpressing the APP Swedish (KM670/671NL) mutation (hereinafter referred to as HEK293-APPSwe), with a purpose to measure human Aβ levels.28 As before, we introduced equal concentrations of miR-15a, -15b, -16, -195, -424, -497, and -103 mimics in this cell line. These experiments showed that miR-16, -15, and -195 mimics similarly suppressed Aβ production (Figure 1g). This effect was unrelated to overall miRNA levels in the cells (see Supplementary Figure S2). APP β-CTF levels, the direct products of BACE1, were significantly downregulated in these conditions. Inversely, APP α-CTF levels were increased. Endogenous BACE1 protein was below detection levels in this cell line. Mutant APP levels remained unchanged following mimic overexpression (Figure 1h), which was expected since expressed approximately fivefold over endogenous APP (ref. 42 and data not shown) and it does not contain a 3'UTR. On the other hand, all members of this family could downregulate endogenous APP in native HEK293 cells (see Supplementary Figure S3).
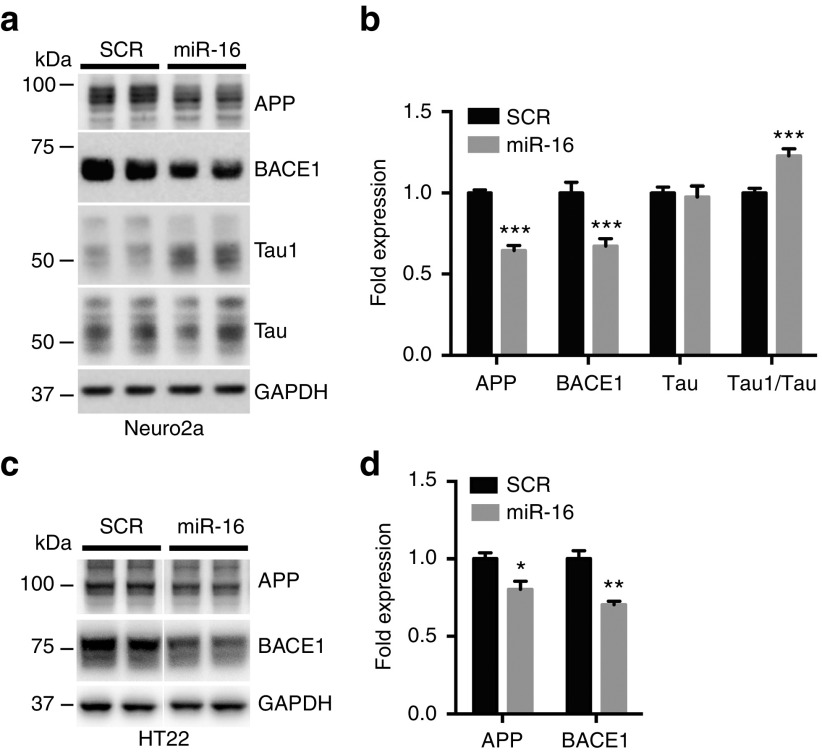
Given its strong regulatory effects on APP, BACE1, APP β-CTFs, and Aβ (both direct substrates of BACE1), we focused our studies on miR-16. We investigated the effects of miR-16 mimics on endogenous APP and BACE1 in neuronal cells. We observed a concomitant reduction of APP and BACE1 protein levels following miR-16 overexpression in native Neuro2a cells (Figure 2a,b). Notably, the introduction of miR-16 induced also a significant decrease in total Tau phosphorylation (as measured using the Tau1 epitope, which specifically labels nonphosphorylated Tau43) (Figure 2a,b). We also validated the effects of miR-16 on human Aβ in Neuro2a cells expressing APPSwe (hereinafter referred to as Neuro2a-APPSwe) (see Supplementary Figure S2). Finally, we observed lower levels of endogenous APP and BACE1 in miR-16-expressing native HT22 cells, an independent neuronal cell line (Figure 2c,d). Unfortunately, Tau protein was below detection levels in this cell type. Taken together, these results identified miR-16 as an endogenous regulator of both Aβ production and Tau phosphorylation.
Figure 2.
Effects of miR-16 overexpression on amyloid precursor protein (APP), BACE1, and Tau in neuronal cells. (a,b) Representative western blot analysis of Neuro2a cells treated with miR-16 mimics (50 nmol/l final concentration). Results are shown 24 hours post-transfection. Shown here is a combined regulatory effect of miR-16 on APP, BACE1, and Tau. (c,d) Western analysis of BACE1 following miR-16 overexpression in HT22 cells. Here, results are shown 48 hours post-transfection. A scrambled oligonucleotide sequence (SCR) was used as negative control in all experiments. Error bars represent standard errors derived from three or more independent experiments performed in triplicate. Statistical significance between SCR- and miRNA-treated cells was determined using unpaired t-test with Bonferroni multiple comparison test as a post-test. Data are shown as mean ± standard error of the mean. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Loss of the DLEU2/miR-15a/16-1 in sporadic AD
The above-mentioned results prompted us to re-evaluate the expression levels of miR-16 in AD. The miR-16 and miR-15a cluster is encoded within the DLEU2 noncoding gene on chromosome 13.44 To exclude any bias toward one of the two miRNAs, we measured the DLEU2 transcript in human brain tissues. By real-time quantitative RT-PCR (qRT-PCR), we observed a significant downregulation of DLEU2 mRNA in AD patients when compared to nondemented controls in both the temporal and frontal cortex (see Supplementary Figure S4). These results strengthen the notion that miR-16 (and miR-15a) is lower in sporadic AD, and further validate the use of miR-16 in therapeutic applications.
Effective delivery of miR-16 mimics into the mammalian brain
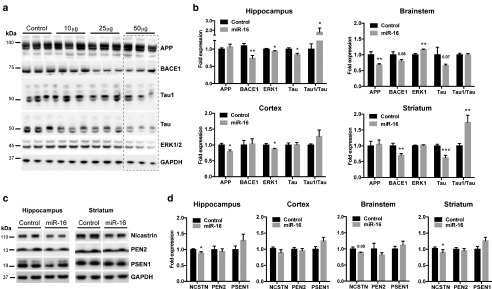
Oligonucleotide delivery using osmotic pumps is a recognized technique with potential therapeutic applications in mammals including humans.45,46,47,48 We therefore used this strategy to deliver miR-16 mimics into the mouse brain. We chose wild-type mice—instead of AD mice—for these in vivo preclinical studies since harboring all physiological regulatory elements (e.g., 3'UTR) of genes of interest. We first treated mice with increasing doses of miR-16 mimics for 7 days (n = 3/group). As control we used vehicle alone (saline 0.9%). Following delivery, the mice were sacrificed and the hippocampi were isolated for functional analyses. In these conditions, we observed a dose-dependent decrease in endogenous BACE1 and Tau (Figure 3a). Tau1 epitope was significantly increased (mirroring lower Tau phosphorylation), consistent with our cell-based studies. We used the previously recognized miR-16 target ERK1 as internal control.31 These effects were specific, as they were not reproduced using a chemically-modified nonfunctional miR-16 mimic (see Supplementary Figure S5).
Figure 3.
In vivo regulation of AD genes by miR-16 mimics. (a) Dose-dependent effects of miR-16 mimics on BACE1, Tau, and ERK1/2 in the hippocampus. As control we used vehicle alone (saline 0.9%). Amyloid precursor protein remained unaffected in this region. (b) Region-dependent effects of miR-16 mimics on AD-related genes. Data are shown as mean ± standard error of the mean. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as a normalizing control. (c,d) Representative western analysis and quantification of γ-secretase complex members. Overall changes in protein levels were calculated by parametric unpaired t test with Welch's correction, where *P < 0.05, **P < 0.01, ***P < 0.001.
Based on the aforementioned observations, we chose a dose of 50 μg/day to pursue our in vivo studies. An independent group of mice received miR-16 mimics for 7 days (n = 10/group). We first evaluated the levels of miR-16 mimics in the treated mice. By qRT-PCR, we observed a strong increase of miR-16 in the hippocampus (106-fold), cortex (34-fold), striatum (27-fold), and brainstem (27-fold) (see Supplementary Figure S6). Such increases were independent of miR-16 baseline levels (see Supplementary Figure S6). We also performed RIP-Chip (i.e., anti-Ago2) assays to determine the enrichment of miR-16 directly in the RNA-induced silencing complex following its overexpression. These experiments showed a 3.63- and 2.66-fold enrichment of miR-16 in the cortex and brainstem, respectively (see Supplementary Figure S6). Thus, the absolute increase of (functional) miR-16 is physiologically relevant.
Western blot analysis on APP, BACE1, Tau, and ERK1 was next performed on four different brain regions, including hippocampus, cortex, striatum, and brainstem (Figure 3b). Interestingly, the effects of miR-16 mimics on BACE1, APP, Tau, and ERK1 were region-dependent. For instance, APP protein levels were downregulated in the cortex, brainstem, and striatum but not in hippocampus. BACE1 protein levels were reduced in the hippocampus, brainstem, and striatum. We also marked a significant downregulation of total Tau in the hippocampus, brainstem, and striatum followed by a modest increase in unphosphorylated Tau in the hippocampus and striatum. We confirmed an overall decrease of phosphorylated Tau (PHF13 epitope) in the hippocampus of treated mice (see Supplementary Figure S7). ERK1 was downregulated mainly in the hippocampus and cortex. We also investigated mRNA levels for APP, BACE1, and Tau in these regions. These experiments showed that only APP and Tau mRNA expression was significantly lower in the cortex and brainstem respectively, but not in other regions (see Supplementary Figure S7).
In the course of these studies, we also observed a significant downregulation of Nicastrin in the treated mice (Figure 3c,d). These effects were not observed on other members of the γ-secretase complex, including Presenilin-1 and PEN2. Using the miRWalk algorithm,49 we identified one putative miR-16 binding site located in 3'UTR of Nicastrin (see Supplementary Figure S8). We therefore repeated the luciferase-based experiments using the full-length Nicastrin 3'UTR as before.50 As hypothesized, miR-16 could significantly downregulate luciferase activity (Nicastrin expression) in these conditions (see Supplementary Figure S8).
Assessment of potential side effects related to miR-16 mimic brain delivery
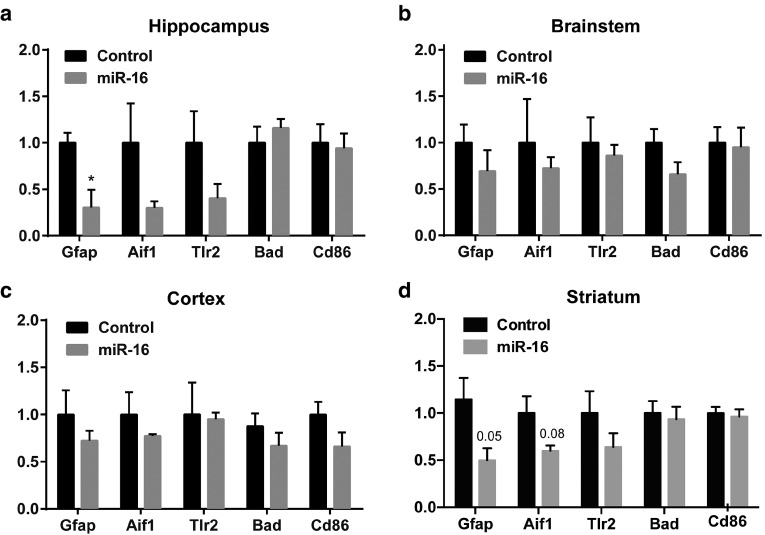
Previous studies have associated miR-16 with the inflammation response.51,52 We therefore wanted to determine whether miR-16 overexpression was associated with an increase (or decrease) in inflammation markers. Compared to controls, treated mice displayed a significant downregulation of glial fibrillary acidic protein (Gfap) in the hippocampus and striatum (Figure 4). A tendency for reduced Allograft inflammatory factor 1(Aif1) was also observed in these regions. Other inflammation markers including Toll-like receptor 2 (Tlr2), Bcl-2-associated death promoter (Bad), and T-lymphocyte activation antigen (CD86) remained unchanged in these conditions, with an overall nonstatistical trend for lower levels. These results strongly suggest that miR-16 delivery to the brain per se is not associated with overt inflammation.
Figure 4.
Analysis of inflammation markers following miR-16 mimic treatment. Seven days after delivery, mRNA levels of inflammatory markers were measured by qRT-PCR in (a) hippocampus, (b) brainstem, (c) cortex, and (d) striatum. Overall changes were calculated by parametric unpaired t-test with Welch's correction, where P < 0.05 is considered as statistical significant. Data are shown as mean ± standard error of the mean. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as normalization control.
Given no or little effects of miR-16 mimics on mRNA levels of candidate genes (see Supplementary Figure S7), we next thought to perform proteomics studies. The purpose here was twofold: (i) to identify additional miR-16 mimic targets in the brain, and (ii) to assess potential indirect effects associated with miR-16 mimic delivery. For these studies, we chose the hippocampus and brainstem, two functionally and temporally distinct regions related to AD.53,54,55 iTRAQ (isobaric tags for relative and absolute quantification) analysis56 identified a total of 4,058 proteins in the adult mouse brain (data not shown). Compared to the control group, a total of 16 proteins were significantly misregulated in the hippocampus, including 5 upregulated and 11 downregulated proteins (fold change <0.8 and >1.2, P < 0.05) (see Supplementary Table S1). In the brainstem, a total of 102 proteins were changed using similar cut-off values, including 47 upregulated and 55 downregulated proteins. Using the miRWalk algorithm, we identified 7/11 (64%) and 31/55 (56%) of downregulated proteins with at least one predicted miR-16 target site in their 3'UTR (see Supplementary Table S1). Furthermore, 14/55 (25%) of proteins misregulated in the brainstem had at least one miR-16 site within the coding sequence (open reading frame).
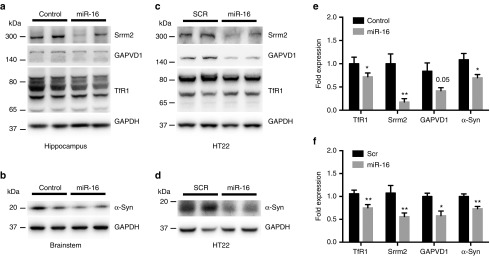
We selected four proteins for further validation, including α-Synuclein (α-Syn) (fold 0.766, P = 0.001), serine/arginine repetitive matrix protein 2 (Srrm2) (fold 0.798, P = 0.023), GTPase-activating protein, VPS9 domain-containing protein 1 (GAPVD1) (fold 0.771, P = 0.038), and Transferrin receptor protein 1 (TfR1) (fold 0.666, P = 0.037). By western blot, we could confirm the downregulation of α-Syn, Srrm2, GAPVD1, and TfR1 in mimic-treated mice when compared to controls (Figure 5). To determine whether these effects were a direct consequence of miR-16 overexpression, we performed gain-of-function studies in HT22 cells. These experiments confirmed the regulation of identified genes by miR-16 in neuronal cells.
Figure 5.
Proteomics validation in vivo and in neuronal cells. (a,b) Representative western blot analysis of selected proteins in the hippocampus and brainstem of miR-16 mimic-treated mice. (c,d) Results on HT22 cells treated with 50 nmol/l final concentration of miR-16 mimics (n = 3 in triplicate). Statistical significance between control- and miRNA-treated mice was determined using unpaired t-test with Bonferroni multiple comparison test as a post-test. Data are shown as mean ± standard error of the mean. (e,f) Quantification of protein levels. Statistical significance was calculated by parametric unpaired t-test with Welch's correction, where *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as mean ± standard error of the mean. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as normalization control.
We next explored the functional relationship of misregulated proteins (up- and downregulated) using the Genemania online tool.57 This analysis identified a significant (physical) interaction map between α-Syn and various other affected proteins in the brainstem (see Supplementary Figure S9). Unfortunately, the relatively low number of affected proteins in the hippocampus made similar predictions impossible. We also performed an enrichment analysis of genes encoding all top ranked proteins in the brainstem using the DAVID software.58 The highest-ranking network associated with miR-16 delivery was AD (P = 5.1E−10). Other relevant networks and pathways included Parkinson's disease (PD) (P = 6.3E−9), oxidative phosphorylation (P = 7.5E−9), and cytoskeleton protein binding (P = 3.6E−7) (see Supplementary Table S2).
Discussion
This study examines the potential application of miRNA mimics as therapeutic agents in AD and provides new information about the role of miR-16 in the brain. Overexpression studies in vitro, in cells and in mice suggest that miR-16 could target simultaneously (or a combination of) endogenous regulators of Aβ, Tau, inflammation, and oxidative stress. Together, these results suggest that selected miR-15/107 family members can function as promising multifactorial drug targets for AD.
The results presented herein support the notion that miR-16 and its homologues are involved in the physiological regulation of AD genes across species. It is noteworthy that miR-16 itself is highly conserved.23 The observation that miR-103 did not regulate BACE1 in most conditions tested herein might reflect a different mode of regulation of miR-103/107 close homologues (i.e., via coding sequence or other regulatory elements).34,59 Our overexpression paradigms cannot discriminate if identified miR-16 targets in vivo are regulated mainly during development, adult maintenance, and/or disease conditions. Also, this study does not address the question of whether miR-16 regulates these genes simultaneously, and in the same cell populations. Obviously, these questions are biologically meaningful, and should be addressed in future experiments, for instance using loss-of-function paradigms. The overexpression conditions used herein remain, however, a method of choice for therapeutic applications, particularly in miRNA replacement therapy. It is interesting to note that the mimics could effectively target genes expressed mainly in neurons (i.e., APP, BACE1, and Tau), consistent with recent observations45 (although some non-neuronal genes may also be regulated—see below). The fact that central nervous system-delivered mimics can distribute throughout the brain can be viewed as an advantage in targeting widespread diseases such as AD; however, further preclinical studies are required to ascertain this hypothesis. The delivery of mimics to specific brain regions and/or cell types is also feasible, for instance using receptor-specific peptides.60 As shown here, brain cells adapt well to “high” levels of exogenous miRNA mimics, thus opening an interesting therapeutic window.61,62,63,64
Most AD mouse models used in preclinical studies express only the coding sequence of mutant genes of interest (e.g., APP, BACE1, PSEN), therefore excluding partially or entirely the 3'UTR. The use of wild-type mice is therefore essential to address our questions, since harboring all physiological regulatory elements. For instance, the use of wild-type mice allowed us to investigate in more detail the role of miR-16 in the regulation of endogenous Tau phosphorylation. In addition, and importantly, we identified protein networks downstream of miR-16 overexpression in the adult brain. These effects are independent of mutant transgenes, known to have pleiotropic effects,65 and thus provide a more accurate view of regulated pathways. We hypothesize that miRNA-based therapies will benefit mostly patients with sporadic AD, thus with no causative mutations in APP or PSEN genes.66
Continuous delivery of miR-16 mimics in the brain did not induce overt inflammation, another significant advantage when developing central nervous system-based drugs. We actually noticed a downregulation of Gfap and Aif1 in the treated mice. All members of the miR-15/107 family are predicted to target the human GFAP 3'UTR (targetscan.org). Whether this phenomenon is conserved in mice remains an interesting possibility, and would indicate that a pool of miR-16 mimics could effectively target non-neuronal genes. Interestingly, previous studies have shown that Gfap deficiency in mice protects neurons against metabolic and excitotoxic insults,67 whereas interference with glial activation in AD mice results in improved cognitive and synaptic function.68 In AD brain, a large number of GFAP-positive astrocytes are colocalized with amyloid plaques.69 While the role of inflammation in AD remains under debate,70 our results suggest a potential neuroprotective role for miR-16 upregulation in AD. The underlying mechanisms involved in this process remain to be determined, but might involve previously identified miR-16 downstream effectors (e.g., TNF, IL-8).51,71 Of course, longer treatments are required to determine if these effects are maintained over time.
Our quantitative proteomics analyses identified various putative miR-16 targets in vivo with potential important functions in AD and neurodegeneration. For instance, α-Synuclein, the major component of Lewy bodies in PD,72 can induce the fibrillation of Tau.73 TfR1 is a major iron binding protein, with high affinity for transferrin. Recent evidence suggests that ferritin iron accumulation in the hippocampus of AD patients concurs with decreased tissue integrity.74 The reduction of TfR1 in the hippocampus is also thought to be protective against oxidative stress in AD.75 GAPVD1, also known as RAP6, is a regulator of endocytosis76 and regulates Glut4 trafficking mainly in adipocytes.77 Although the role of GAPVD1 in the brain is unknown, profiling studies suggest the GAPVD1 mRNA is upregulated in AD patients (NextBio databank: http://www.nextbio.com). Srrm2/SRm300 plays an important role in pre-mRNA splicing as a spliceosome component,78 and is a candidate gene for PD.79 Again, mRNA expression studies suggest an upregulation of Srrm2 in AD patients when compared to nondemented controls (NextBio databank, data sets GSE48350 and GSE5281).80,81 Interestingly, miR-16 mimics also induced a downregulation of various mitochondrial respiration components, including Ndufb5, Ndufb9, ATP5j, ATP6V0, COX4i, Cox5a,b,-6C in the brainstem. In the rat brain, it has been suggested that aging elicits elevates metabolic activity by regulating in part these genes.82 Whether miR-16 introduction (or reintroduction) could prevent or attenuate oxidative stress associated with ageing and/or disease is an interesting possibility.
Considering that a single miRNA can modulate a large number of genes, miRNA-based therapeutics have their own challenges that must be overcome before assessing their efficacy in humans, like stability, delivery, and safety.83 One should keep in mind that certain miRNAs can, however, function through specific “master switches”, thus limiting the number of affected genes.84,85 In this context, our study provides important new information with regard to the efficiency of miRNA mimics for AD therapy, by showing the combined action of miR-16 on APP, BACE1, and Tau. Although bioinformatics predict a large number of miR-16 targets (e.g., >1,000 using miRWalk), our in vivo studies in the brain show that relatively few genes (~100 in total) are affected by miR-16 overexpression, and in a region-specific manner. These observations suggest that miRNA replacement therapy could be safe with minimal side effects in humans. While previous studies have linked miR-16/15a misregulation to cancer,86,87,88,89 it is not expected that proposed target genes (e.g., Bcl-2, Mcl1, Ccnd1, and Wnt3a) are regulated in postmitotic neurons. Such cell and tissue specificity is well documented, for instance, with transcription factors. Consistent with this notion, Bcl-2 protein levels remained unchanged following miR-16 mimic overexpression (data not shown), consistent with our proteomics analysis. In addition, there is no clear indication that major cancer-related networks and pathways are affected in the treated mice.
Considering that miR-16 dysregulation is associated with various other neurodegenerative and psychiatric diseases, these results set the stage to explore in more detail the role of this superfamily in brain disorders in general. Future experiments include testing the effects of mimics in animal models of neurodegeneration (taking into account their limitations), as well as performing detailed pharmacokinetics analyses of mimics in the brain. Finally, our research suggests that miR-16 replacement therapy can specifically be used for AD and possibly PD.
Materials and methods
Cell culture. Mouse neuroblastoma Neuro2a cells, mouse Neuro2a cells expressing the Swedish mutant of APP and Δ9 mutant of PSEN1 (Neuro2a APPSwe/Δ9) (Dr. Gopal Thinakaran, University of Chicago, USA), mouse hippocampal-derived HT22 cells (Dr. Schubert, Salk institute, USA), human HEK293T cells, and human HEK293 cells expressing the Swedish mutant of APP (HEK293-APPSwe) were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum (ThermoFischer Scientific, Waltham, MA).
Cell transfection. Cells were seeded into six-well plates at the concentration of 1.5 × 105 cells per well the day before transfection. All miRNA mimics used for vitro studies were purchased from Ambion (Life Technologies, Burlington, Canada). These were transfected at various concentrations (see text) using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions.
Western blotting. Total proteins from cells were extracted from cells using RIPA buffer (25 mmol/l Tris-HCl (pH 7.6), 150 mmol/l NaCl, 1% NP-40, 1% sodium deoxycholate) supplemented with 0.01% Protease and phosphatase inhibitor cocktail and 0.025% Na-deoxycholate10%. Brain proteins and total RNA was extracted using the mirVana PARIS kit (Life Technologies). Protein lysates were separated by electrophoresis using 4–12% NuPAGE precast gels and Tris-Acetate3-8% for protein more than 200kD (Life Technologies) and wet transferred onto a 0.45 μm Nitrocellulose membrane (Bio-Rad, Mississauga, Canada). GAPVD1 (k-22, cat#sc-133607) and SRM300 (H111, cat#sc-292291) antibodies were purchased from Santa Cruz Biotechnology. (Dallas, TX) Phospho-ERK1/2 (cat#9101), ERK1/2 (cat #9102), Nicastrin (D38F9, cat#5665), BACE1 (D10E5, cat#5606), and PEN2 (D2G6, cat#8502) antibodies were purchased from Cell Signaling (Danvers, MA). TAU1 (cat#MAB3420), goat anti-rat IgG-HRP (#catAP136P), PRESENILIN-1 (PS1-loop, cat#MAB5232), AMYLOID-β (WO-2, cat#MABN10), and glyceraldehyde 3-phosphate dehydrogenase (cat#MAB374) antibodies were purchased from Millipore (Etobicoke, Canada). Other antibodies included: TAU total (cat#A0024, Dako, Denmark), TAU PHF13 (cat# ab24716, Abcam, Toronto, Canada), APP (cat#A8717, Sigma), ALPHA-SYNUCLEIN (cat#PA1-18264, Thermo Fisher Scientifics, Burlington, Canada), and Peroxidase-conjugated affinipure goat-anti mouse IgG (Jackson immuno research, West Grove, PA). Images were acquired using Immobilon Western Chemiluminescent HRP Substrate (cat#WBKLS0050, Millipore) and Fusion FX (Vilber Lourmat, Eberhardzell, Germany) imaging system.
qRT-PCR. Total RNA was reverse-transcribed to cDNA using iScript Reverse Transcription Supermix (Bio-Rad) according to the manufacturer's instructions. cDNA was used as template in qRT-PCR reaction performed using soFast EvaGreen Supermix (Bio-Rad). Primer sequences used were: Aif1 Forward: ATCAACAAGCAATTCCTCGATGA, Aif1 Reverse: CAGCATTCGCTTCAAGGACATA (Primer Bank ID 9506379a1); Tlr2 Forward: ACAACTTACCGAAACCTCAGAC, Tlr2 Reverse: ACCCCAGAAGCATCACATG; Bad Forward: TGAGCCGAGTGAGCAGGAA, Bad Reverse: GCCTCCATGATGACTGTTGGT; Gfap Forward: AGAGGGACAACTTTGCACAG (Primer Bank ID 6671610a1), Gfap Reverse: TCCAAATCCACACGAGCC; CD86 Forward: CTGGACTCTACGACTTCACAATG, CD86 Reverse: AGTTGGCGATCACTGACAGTT; Tau Forward: TGACACGGACGCTGGCCTGAA, Tau Reverse: CACTTGGAGGTCACCTTGCTC; APP Forward: CGAGAGAGAATGTCCCAGGT, APP Reverse: AGTTCTTGGCTTGACGCTCT; Bace1 Forward: CGTGTGGAAATCAATGGTCAAG, Bace1 Reverse: GACGGCAGCTTCAAATACTTTC; Ncstn Forward: TCCGTGGTACTGGCAGGATT Ncstn Reverse: CCCCTGTATCCCCACTAATTGA (Primer Bank ID 31981205a1). Relative expression was calculated by using the ΔΔCt method using a LightCycler 480 II (Roche, Laval, Canada). Glyceraldehyde 3-phosphate dehydrogenase was used as normalization control. For miRNA quantification, TaqMan miRNA assays (Applied Biosystem, Burlington, Canada) for miR-16 was used, and relative levels were calculated using the ΔΔCt method against RNU19 as reference control.90
Luciferase assays. The full-length hAPP, hBACE1, and hNicastrin 3'UTR luciferase constructs were described previously.28,50,90 Mutagenesis was performed by TOP gene Technologies (Montréal, Canada) and validated by sequencing. miRNA mimics (pre-miRs) with concentrations between 0–25 nmol/l, pRL Renilla (10 ng) and pGL3 plasmids harboring 3'UTR of interest (500 ng) were cotransfected using LipofectAMINE 2000 into HEK293 cells. Twenty-four hours after cotransfection, luciferase activities were measured by using a Dual-Glo Luciferase Assay System (Promega, Madison, WI) according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
ELISA. Supernatants of Neuro2a-APPSwe/Δ9 cells were collected 48 hours post-transfection with miR-16 mimics or scrambled control. Soluble (secreted) human Aβ1–40 and Aβ1–42 levels were measured by ELISA (cat #KHB3481 and KHB3441, Invitrogen, Burlington, Canada) following the manufacture's protocol.
In vivo administration of mimics. Mouse miR-16 mimics (CONmir mimics) used for in vivo studies were purchased from Riboxx (Redebul, Germany). All animal protocols were approved by the animal protection committee of the CHU de Québec. Wild-type mice (C57BL/6, female, 2 months old) were used in all experiments. Mice were maintained in a 12-hour light/12-hour dark cycle and received routine veterinary monitoring. The mini-pumps (ALZET model 1007D) and brain infusion kits (cat#8663) were purchased from Durect (Cupertina, CA). Preoperative procedure included 30 µl of Anafen (1 mg/ml), 100 µl Marcaine (5.0 mg/ml), and 500 µl saline (0.9%). ALZET mini-osmotic pumps were implanted subcutaneously. Mimics were administrated into the brain using coordinates: ventricle A/P = −0.22 M/L = 0.5 D/v = 3.5) (12 hours per day) (0.5 µl per hour with concentration of 4.2 µg/μl). During the postoperative procedure, mice were treated with 50 µl Anafen (1 mg/ml) and 500 µl saline (0.9%).
Proteomics. Proteomics was performed by the proteomics platform of the Centre de recherche du CHU de Québec on two selected regions of brainstem and hippocampus with n = 4 mice/region. Frozen tissues were disrupted using a mortar and pestle. Samples were kept frozen on dry-ice, and grind to fine powder. Then lysis buffer (50 mmol/l ammonium bicarbonate, 50 mmol/l dithiothreitol, 0.5% sodium deoxycholate) containing protease inhibitors cocktail (Roche Diagnostics, Indianapolis, IN) was added, and the sample preparation was homogenized on ice by sonication with a Sonic Dismembrator (Fisher, Pittsburgh, PA) with 1 second pulse (20 times). Samples were centrifuged 10 minutes at 16,000 g. The supernatants were mixed with five volumes of acetone (stored at −20 °C) and incubated overnight at −20 °C. Precipitated proteins were centrifuged 15 minutes at 16,000 g. Protein pellets were air dried and then resuspended in 0.5 M triethylammonium bicarbonate—0.5% sodium deoxycholate. Fifteen micrograms of protein for each group were used for iTRAQ labeling. Triethylammonium bicarbonate and sodium deoxycholate were added to each sample to reach a final concentration of 0.5 M and 0.5%, respectively. Proteins were then reduced and alkylated according to the iTRAQ kit manufacturer's instruction (Applied Biosystems, Burlington, Canada). Samples were digested with trypsin (Sequence grade Modified, Promega) using 1:30 ratio overnight at 37 °C. After digestion, peptides were acidified to precipitate deoxycholate, and then purified with an oasis HLB cartridge (1 cc, 10 mg, Waters) and lyophilized. Dried peptides were dissolved in 30 µl 0.5 M triethylammonium bicarbonate and labeled with iTRAQ label reagent (Applied Biosystems). Four-plex labeling was performed for 2 hours at room temperature in the dark. Labeled peptides were combined in one tube and dried with the SpeedVac. Samples were cleaned up using HLB cartridge. Samples were dried and reconstituted 200 µl HPLC H2O and 1/100 ampholytes pH 3–10 (Biorad). Then peptides were fractionated with 7 cm IPG strips pH 3–10 using an isoelectric focusing method, and performed for 10,000 V h. IPG strips were cut in 14 fractions and peptides were extracted in 2% ACN −0.1%FA solution followed by 50% ACN-1% FA. Finally, fractions were dried with the SpeedVac. The proteins listed were the one considered to be differentially expressed and they were identified at a false discovery rate less than 1% as estimated by a Protein Pilot tool using reverse database search strategy and their iTRAQ ratios were <0.8 and >1.2 with a P value lower than 0.05 as calculated by Protein Pilot based on two-tailed t-tests where the degree of freedom is equal to the number of distinct peptide minus one.
RNA immunoprecipitation (RIP-Chip). RIP immunoprecipitations were performed as described previously.91,92 Briefly, anti-AGO2 (2A8, cat# MABE56, Millipore) and control mouse IgGs were coupled to Protein G Sepharose (GE Healthcare Bio-science, Mississauga, Canada). Brainstem and cortex tissues were homogenized in a lysis buffer (25 mmol/l Tris-HCl pH8, 150 mmol/l NaCl, 2 mmol/l MgCl2, 0.5% Triton X-100, 5 mmol/l dithiothreitol, 250 U/ml RNAsin, and protease inhibitors). Proteins were transferred to a clean tube after high-speed centrifugation. Total lysate was precleared by incubating with protein G alone and then separated into two fractions. These were incubated with either the antibody (AGO2) or control IgG-coupled beads. Following washes (high salt buffer = lysis buffer at 900 mmol/l NaCl and low Triton X-100 buffer = lysis buffer at 0.05% Triton X-100), proteins, including RNA-binding proteins, were eluted with sample buffer. Immunoprecipitated total RNAs were extracted directly from the beads using Trizol (Invitrogen). MiR-16 was subjected to qRT-PCR analysis. Following the immunoprecipitation, the protein fraction was subjected to western blot analysis (anti-AGO2 C34C6, cat#2897, Cell Signalling) in order to validate the efficiency of Ago2 immunoprecipitation (data not shown).
Statistics. Statistical significance and normality was calculated using GraphPad version 6.0d software (La Jolla, CA). Western blot images were analyzed by ImageJ V1.47 software. Statistical significance was calculated by parametric unpaired t-test with Welch's correction (P < 0.05 considered as significant) and multiple comparisons was done using the Bonferroni method.
SUPPLEMENTARY MATERIAL Figure S1. Experimental overview of current study. Figure S2. Characterization of miR-16/15a family members in cells. Figure S3. Regulation of endogenous APP in native HEK293 cells. Figure S4. Downregulation of the miR-16/15a cluster in AD. Figure S5. Validation of miR-16 mimic specificity in vivo. Figure S6. Characterization of miR-16/15a family members in the brain. Figure S7. Analysis of APP, BACE1, and Tau mRNA in vivo. Figure S8. Nicastrin is directly regulated by miR-16. Figure S9. Physical interaction networks between putative miR-16 targets in vivo. Table S1. Protein changes in the brainstem and hippocampus of treated mice versus controls (n=4/group). Table S2. DAVID gene enrichment analysis of top ranked targets identified in brainstem using Homo sapiens background.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (CIHR), the Alzheimer Society of Canada, the Fonds de recherche du Québec Santé (FRQS), and Université Laval/Bourse Ven-Huguette-Anil-Murthy (to S.P.). The authors declare no conflict of interest.
Supplementary Material
References
- Prince, M, Bryce, R, Albanese, E, Wimo, A, Ribeiro, W and Ferri, CP (2013). The global prevalence of dementia: a systematic review and metaanalysis. Alzheimers Dement 9: 63–75.e2. [DOI] [PubMed] [Google Scholar]
- Campion, D, Dumanchin, C, Hannequin, D, Dubois, B, Belliard, S, Puel, M et al. (1999). Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet 65: 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling, RA, Aisen, PS, Beckett, LA, Bennett, DA, Craft, S, Fagan, AM et al. (2011). Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 7: 280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper, B, Iwatsubo, T and Wolfe, MS (2012). Presenilins and γ-secretase: structure, function, and role in Alzheimer Disease. Cold Spring Harb Perspect Med 2: a006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, J and Selkoe, DJ (2002). The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 297: 353–356. [DOI] [PubMed] [Google Scholar]
- Jia, Q, Deng, Y and Qing, H (2014). Potential therapeutic strategies for Alzheimer's disease targeting or beyond β-amyloid: insights from clinical trials. Biomed Res Int 2014: 837157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutajangout, A and Wisniewski, T (2014). Tau-based therapeutic approaches for Alzheimer's disease - a mini-review. Gerontology 60: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum, WI (2014). Why Alzheimer trials fail: removing soluble oligomeric beta amyloid is essential, inconsistent, and difficult. Neurobiol Aging 35: 969–974. [DOI] [PubMed] [Google Scholar]
- Jaturapatporn, D, Isaac, MG, McCleery, J and Tabet, N (2012). Aspirin, steroidal and non-steroidal anti-inflammatory drugs for the treatment of Alzheimer's disease. Cochrane Database Syst Rev 2: CD006378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreiras, MC, Mendes, E, Perry, MJ, Francisco, AP and Marco-Contelles, J (2013). The multifactorial nature of Alzheimer's disease for developing potential therapeutics. Curr Top Med Chem 13: 1745–1770. [DOI] [PubMed] [Google Scholar]
- Hébert, SS, Wang, WX, Zhu, Q and Nelson, PT (2013). A study of small RNAs from cerebral neocortex of pathology-verified Alzheimer's disease, dementia with lewy bodies, hippocampal sclerosis, frontotemporal lobar dementia, and non-demented human controls. J Alzheimers Dis 35: 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros, V (2004). The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- He, L and Hannon, GJ (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531. [DOI] [PubMed] [Google Scholar]
- Bartel, DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- van Rooij, E and Kauppinen, S (2014). Development of microRNA therapeutics is coming of age. EMBO Mol Med 6: 851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, H, Fabbri, M and Calin, GA (2013). MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 12: 847–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, AL and Levin, AA (2012). Developing microRNA therapeutics: approaching the unique complexities. Nucleic Acid Ther 22: 213–225. [DOI] [PubMed] [Google Scholar]
- Hammond, SM (2007). MicroRNAs as tumor suppressors. Nat Genet 39: 582–583. [DOI] [PubMed] [Google Scholar]
- Bader, AG, Brown, D and Winkler, M (2010). The promise of microRNA replacement therapy. Cancer Res 70: 7027–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson, B and Das, S (2015). MicroRNA Therapeutics: the Next Magic Bullet? Mini Rev Med Chem 15: 467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader, AG (2012). miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet 3: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli, AE (2012). MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13: 271–282. [DOI] [PubMed] [Google Scholar]
- Finnerty, JR, Wang, WX, Hébert, SS, Wilfred, BR, Mao, G and Nelson, PT (2010). The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol 402: 491–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley, PS, Schelter, J, Burchard, J, Kibukawa, M, Martin, MM, Bartz, SR et al. (2007). Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27: 2240–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, M, Kuiperij, HB, Claassen, JA, Küsters, B and Verbeek, MM (2014). MicroRNAs in Alzheimer's disease: differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol Aging 35: 152–158. [DOI] [PubMed] [Google Scholar]
- Liu, W, Liu, C, Zhu, J, Shu, P, Yin, B, Gong, Y et al. (2012). MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer's-associated pathogenesis in SAMP8 mice. Neurobiol Aging 33: 522–534. [DOI] [PubMed] [Google Scholar]
- Wang, WX, Huang, Q, Hu, Y, Stromberg, AJ and Nelson, PT (2011). Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta Neuropathol 121: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert, SS, Horré, K, Nicolaï, L, Papadopoulou, AS, Mandemakers, W, Silahtaroglu, AN et al. (2008). Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci USA 105: 6415–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioya, M, Obayashi, S, Tabunoki, H, Arima, K, Saito, Y, Ishida, T et al. (2010). Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol Appl Neurobiol 36: 320–330. [DOI] [PubMed] [Google Scholar]
- Nunez-Iglesias, J, Liu, CC, Morgan, TE, Finch, CE and Zhou, XJ (2010). Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer's disease cortex reveals altered miRNA regulation. PLoS One 5: e8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert, SS, Papadopoulou, AS, Smith, P, Galas, MC, Planel, E, Silahtaroglu, AN et al. (2010). Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet 19: 3959–3969. [DOI] [PubMed] [Google Scholar]
- Cogswell, JP, Ward, J, Taylor, IA, Waters, M, Shi, Y, Cannon, B et al. (2008). Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis 14: 27–41. [DOI] [PubMed] [Google Scholar]
- Ai, J, Sun, LH, Che, H, Zhang, R, Zhang, TZ, Wu, WC et al. (2013). MicroRNA-195 protects against dementia induced by chronic brain hypoperfusion via its anti-amyloidogenic effect in rats. J Neurosci 33: 3989–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, WX, Rajeev, BW, Stromberg, AJ, Ren, N, Tang, G, Huang, Q et al. (2008). The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci 28: 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, PT and Wang, WX (2010). MiR-107 is reduced in Alzheimer's disease brain neocortex: validation study. J Alzheimers Dis 21: 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, P, Al Hashimi, A, Girard, J, Delay, C and Hébert, SS (2011). In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J Neurochem 116: 240–247. [DOI] [PubMed] [Google Scholar]
- Zhu, HC, Wang, LM, Wang, M, Song, B, Tan, S, Teng, JF et al. (2012). MicroRNA-195 downregulates Alzheimer's disease amyloid-β production by targeting BACE1. Brain Res Bull 88: 596–601. [DOI] [PubMed] [Google Scholar]
- van Rooij, E, Purcell, AL and Levin, AA (2012). Developing microRNA therapeutics. Circ Res 110: 496–507. [DOI] [PubMed] [Google Scholar]
- Jopling, CL (2010). Targeting microRNA-122 to Treat Hepatitis C Virus Infection. Viruses 2: 1382–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, N, Hoang, D, Miller, N, Ansaloni, S, Huang, Q, Rogers, JT et al. (2008). MicroRNAs can regulate human APP levels. Mol Neurodegener 3: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Absalon, S, Kochanek, DM, Raghavan, V and Krichevsky, AM (2013). MiR-26b, upregulated in Alzheimer's disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J Neurosci 33: 14645–14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancolio, K, Dumanchin, C, Barelli, H, Warter, JM, Brice, A, Campion, D et al. (1999). Unusual phenotypic alteration of beta amyloid precursor protein (betaAPP) maturation by a new Val-715 –> Met betaAPP-770 mutation responsible for probable early-onset Alzheimer's disease. Proc Natl Acad Sci USA 96: 4119–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planel, E, Krishnamurthy, P, Miyasaka, T, Liu, L, Herman, M, Kumar, A et al. (2008). Anesthesia-induced hyperphosphorylation detaches 3-repeat tau from microtubules without affecting their stability in vivo. J Neurosci 28: 12798–12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, GA, Dumitru, CD, Shimizu, M, Bichi, R, Zupo, S, Noch, E et al. (2002). Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99: 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koval, ED, Shaner, C, Zhang, P, du Maine, X, Fischer, K, Tay, J et al. (2013). Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Hum Mol Genet 22: 4127–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes, D (2014). Programmable intrathecal pumps for the management of chronic pain: recommendations for improved efficiency. J Pain Res 7: 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin, R, Ge, P, Chen, Q, Sutherland, JE, Zhang, Z, Gash, DM et al. (2015). Onset Time and Durability of Huntingtin Suppression in Rhesus Putamen After Direct Infusion of Antihuntingtin siRNA. Mol Ther Nucleic Acids 4: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesemann, K, Coffey, RJ, Wallace, MS, Tan, Y, Broste, S and Buvanendran, A (2014). Clinical accuracy and safety using the SynchroMed II intrathecal drug infusion pump. Reg Anesth Pain Med 39: 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep, H, Sticht, C, Pandey, P and Gretz, N (2011). miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44: 839–847. [DOI] [PubMed] [Google Scholar]
- Delay, C, Dorval, V, Fok, A, Grenier-Boley, B, Lambert, JC, Hsiung, GY et al. (2014). MicroRNAs targeting Nicastrin regulate Aβ production and are affected by target site polymorphisms. Front Mol Neurosci 7: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, R, Li, X, Hu, G, Gong, AY, Drescher, KM and Chen, XM (2012). miR-16 targets transcriptional corepressor SMRT and modulates NF-kappaB-regulated transactivation of interleukin-8 gene. PLoS One 7: e30772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca, D, Diaz, A, Romero, M, Ferracci, F and Blomberg, BB (2015). MicroRNAs miR-155 and miR-16 Decrease AID and E47 in B Cells from Elderly Individuals. J Immunol 195: 2134–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padurariu, M, Ciobica, A, Mavroudis, I, Fotiou, D and Baloyannis, S (2012). Hippocampal neuronal loss in the CA1 and CA3 areas of Alzheimer's disease patients. Psychiatr Danub 24: 152–158. [PubMed] [Google Scholar]
- Grinberg, LT, Rueb, U and Heinsen, H (2011). Brainstem: neglected locus in neurodegenerative diseases. Front Neurol 2: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic, G, Stanic, G, Mladinov, M, Jovanov-Milosevic, N, Kostovic, I and Hof, PR (2009). Does Alzheimer's disease begin in the brainstem? Neuropathol Appl Neurobiol 35: 532–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese, S, Reidegeld, KA, Meyer, HE and Warscheid, B (2007). Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7: 340–350. [DOI] [PubMed] [Google Scholar]
- Warde-Farley, D, Donaldson, SL, Comes, O, Zuberi, K, Badrawi, R, Chao, P et al. (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38(Web Server issue): W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, da W, Sherman, BT and Lempicki, RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Wang, WX, Wilfred, BR, Madathil, SK, Tang, G, Hu, Y, Dimayuga, J et al. (2010). miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol 177: 334–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y, Hauert, S, Lo, JH and Bhatia, SN (2012). Identification and characterization of receptor-specific peptides for siRNA delivery. ACS Nano 6: 8620–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang, P, Wiggins, JF, Daige, CL, Cho, C, Omotola, M, Brown, D et al. (2011). Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther 19: 1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino, MT, Campani, V, Misso, G, Gallo Cantafio, ME, Gullà, A, Foresta, U et al. (2014). In vivo activity of miR-34a mimics delivered by stable nucleic acid lipid particles (SNALPs) against multiple myeloma. PLoS One 9: e90005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, DW, Bracken, CP, Szubert, JM and Goodall, GJ (2013). On measuring miRNAs after transient transfection of mimics or antisense inhibitors. PLoS One 8: e55214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y, Wang, Z and Gemeinhart, RA (2013). Progress in microRNA delivery. J Control Release 172: 962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robakis, NK (2014). Cell signaling abnormalities may drive neurodegeneration in familial Alzheimer disease. Neurochem Res 39: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magen, I and Hornstein, E (2014). Oligonucleotide-based therapy for neurodegenerative diseases. Brain Res 1584: 116–128. [DOI] [PubMed] [Google Scholar]
- Hanbury, R, Ling, ZD, Wuu, J and Kordower, JH (2003). GFAP knockout mice have increased levels of GDNF that protect striatal neurons from metabolic and excitotoxic insults. J Comp Neurol 461: 307–316. [DOI] [PubMed] [Google Scholar]
- Furman, JL, Sama, DM, Gant, JC, Beckett, TL, Murphy, MP, Bachstetter, AD et al. (2012). Targeting astrocytes ameliorates neurologic changes in a mouse model of Alzheimer's disease. J Neurosci 32: 16129–16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hol, EM, Roelofs, RF, Moraal, E, Sonnemans, MA, Sluijs, JA, Proper, EA et al. (2003). Neuronal expression of GFAP in patients with Alzheimer pathology and identification of novel GFAP splice forms. Mol Psychiatry 8: 786–796. [DOI] [PubMed] [Google Scholar]
- Meraz-Ríos, MA, Toral-Rios, D, Franco-Bocanegra, D, Villeda-Hernández, J and Campos-Peña, V (2013). Inflammatory process in Alzheimer's Disease. Front Integr Neurosci 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, F, Gong, B, Wang, H, Yu, D, Zhao, G, Lin, L et al. (2012). miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis 17: 702–716. [DOI] [PubMed] [Google Scholar]
- Spillantini, MG, Schmidt, ML, Lee, VM, Trojanowski, JQ, Jakes, R and Goedert, M (1997). Alpha-synuclein in Lewy bodies. Nature 388: 839–840. [DOI] [PubMed] [Google Scholar]
- Nonaka, T, Watanabe, ST, Iwatsubo, T and Hasegawa, M (2010). Seeded aggregation and toxicity of {alpha}-synuclein and tau: cellular models of neurodegenerative diseases. J Biol Chem 285: 34885–34898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven, EP, Lu, PH, Tishler, TA, Heydari, P and Bartzokis, G (2013). Increased iron levels and decreased tissue integrity in hippocampus of Alzheimer's disease detected in vivo with magnetic resonance imaging. J Alzheimers Dis 37: 127–136. [DOI] [PubMed] [Google Scholar]
- Huang, XT, Qian, ZM, He, X, Gong, Q, Wu, KC, Jiang, LR et al. (2014). Reducing iron in the brain: a novel pharmacologic mechanism of huperzine A in the treatment of Alzheimer's disease. Neurobiol Aging 35: 1045–1054. [DOI] [PubMed] [Google Scholar]
- Hunker, CM, Galvis, A, Kruk, I, Giambini, H, Veisaga, ML and Barbieri, MA (2006). Rab5-activating protein 6, a novel endosomal protein with a role in endocytosis. Biochem Biophys Res Commun 340: 967–975. [DOI] [PubMed] [Google Scholar]
- Leto, D and Saltiel, AR (2012). Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol 13: 383–396. [DOI] [PubMed] [Google Scholar]
- Blencowe, BJ, Issner, R, Nickerson, JA and Sharp, PA (1998). A coactivator of pre-mRNA splicing. Genes Dev 12: 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehadeh, LA, Yu, K, Wang, L, Guevara, A, Singer, C, Vance, J et al. (2010). SRRM2, a potential blood biomarker revealing high alternative splicing in Parkinson's disease. PLoS One 5: e9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair, LJ, Nordhues, BA, Hill, SE, Scaglione, KM, O'Leary, JC 3rd, Fontaine, SN et al. (2013). Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J Clin Invest 123: 4158–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, WS, Dunckley, T, Beach, TG, Grover, A, Mastroeni, D, Walker, DG et al. (2007). Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics 28: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville, KA, Kent, C, Personett, D, Lai, WR, Park, PJ, Coleman, P et al. (2008). Aging elevates metabolic gene expression in brain cholinergic neurons. Neurobiol Aging 29: 1874–1893. [DOI] [PubMed] [Google Scholar]
- Junn, E and Mouradian, MM (2012). MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol Ther 133: 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, CY, Choi, YS and McManus, MT (2010). Analysis of microRNA knockouts in mice. Hum Mol Genet 19(R2): R169–R175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidigal, JA and Ventura, A (2015). The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol 25: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqeilan, RI, Calin, GA and Croce, CM (2010). miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ 17: 215–220. [DOI] [PubMed] [Google Scholar]
- Yang, TQ, Lu, XJ, Wu, TF, Ding, DD, Zhao, ZH, Chen, GL et al. (2014). MicroRNA-16 inhibits glioma cell growth and invasion through suppression of BCL2 and the nuclear factor-κB1/MMP9 signaling pathway. Cancer Sci 105: 265–271. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen, L, Wang, Q, Wang, GD, Wang, HS, Huang, Y, Liu, XM et al. (2013). miR-16 inhibits cell proliferation by targeting IGF1R and the Raf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS Lett 587: 1366–1372. [DOI] [PubMed] [Google Scholar]
- Calin, GA, Cimmino, A, Fabbri, M, Ferracin, M, Wojcik, SE, Shimizu, M et al. (2008). MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA 105: 5166–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert, SS, Horré, K, Nicolaï, L, Bergmans, B, Papadopoulou, AS, Delacourte, A et al. (2009). MicroRNA regulation of Alzheimer's Amyloid precursor protein expression. Neurobiol Dis 33: 422–428. [DOI] [PubMed] [Google Scholar]
- Dorval, V, Mandemakers, W, Jolivette, F, Coudert, L, Mazroui, R, De Strooper, B et al. (2014). Gene and MicroRNA transcriptome analysis of Parkinson's related LRRK2 mouse models. PLoS One 9: e85510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, WX, Wilfred, BR, Hu, Y, Stromberg, AJ and Nelson, PT (2010). Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA 16: 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.