Abstract
Avian influenza viruses continue to cross the species barrier, and if such viruses become transmissible among humans, it would pose a great threat to public health. Since its emergence in China in 2013, H7N9 has caused considerable morbidity and mortality. In the absence of a universal influenza vaccine, preparedness includes development of subtype-specific vaccines. In this study, we developed and evaluated in ferrets an intranasal live attenuated influenza vaccine (LAIV) against H7N9 based on the A/Leningrad/134/17/57 (H2N2) cold-adapted master donor virus. We demonstrate that the LAIV is attenuated and safe in ferrets and induces high hemagglutination- and neuraminidase-inhibiting and virus-neutralizing titers. The antibodies against hemagglutinin were also cross-reactive with divergent H7 strains. To assess efficacy, we used an intratracheal challenge ferret model in which an acute severe viral pneumonia is induced that closely resembles viral pneumonia observed in severe human cases. A single- and two-dose strategy provided complete protection against severe pneumonia and prevented virus replication. The protective effect of the two-dose strategy appeared better than the single dose only on the microscopic level in the lungs. We observed, however, an increased lymphocytic infiltration after challenge in single-vaccinated animals and hypothesize that this a side effect of the model.
Introduction
Avian influenza viruses continue to cross the species barrier, and although infections detected in humans are incidental, they often have a severe outcome. In February 2013, a novel influenza virus of the subtype H7N9 emerged in China and has since continued to infect humans in a seasonal-like fashion. As of May 2015, 657 cases were confirmed to have been infected with H7N9 of which 261 have perished.1 Infections were mostly observed in people who visited live bird markets. Since the virus is mainly detected in poultry at these markets, it is assumed that H7N9 is transmitted from poultry to humans.2 The disease starts with typical influenza-like illness including fever and cough. Severe cases present viral pneumonia, acute respiratory distress syndrome (ARDS), and multiorgan failure. However, these cases usually have underlying chronic conditions.3
Despite the fact that H7N9 has not yet been capable of establishing a sustained transmission among humans, the virus is considered to have potential to become pandemic in humans for several reasons.2,4 Firstly, as the virus is low pathogenic in birds,5 it slips under the radar unless active monitoring is performed. Secondly, since the first detection, the virus continues to emerge, expands geographically, and has diverged into different clades,6 increasing the chance of acquiring the ability to transmit between humans. Furthermore, H7N9 viruses bind both avian-type (α2,3-linked sialic acid) and, to a lesser extent, human-type (α2,6-linked sialic acid) receptors.7,8 Regardless of the limited respiratory droplet transmission between ferrets, this dual-receptor specificity can be a critical feature for the acquisition of sustained human-to-human transmission in case more adaptive changes occur in the receptor-binding site.9,10 Moreover, several isolates have been shown to be resistant to antivirals, which impedes preventive measures and treatment.11,12 Obviously, the human population is naive to H7N9,4 which allows the virus to spread easily without an immunological barrier once it becomes transmissible between humans.
To anticipate possible future pandemics, the World Health Organization (WHO) has initiated several programs, the Global Action Plan (GAP) for influenza vaccines is one among them.13,14 This program includes the development of vaccines against potentially pandemic influenza viruses. Since for influenza, a universal vaccine is not yet available, development of subtype-specific vaccines up to clinical phase 1/2 is a manner to be able to rapidly act in the event of a pandemic. Careful consideration of which influenza viruses have a potency to initiate a pandemic is however required and still provides no guarantee. Based on the arguments above, H7N9 was recently included. Within the GAP program, we developed and evaluated a live attenuated influenza vaccine (LAIV) against H7N9 based on the A/Leningrad/134/17/57 (H2N2) cold-adapted master donor virus.
Prior to the H7N9 emergence, a number of H7 candidate vaccine viruses had been developed and tested in clinical trials, however, unfortunately none of them contained N9 neuraminidase (NA).15,16,17 Antibodies against NA block the sialidase activity of neuraminidase and, as a consequence, impair viral budding and limit the infection.18 Existing H7 vaccines contain N1, N3, or N7 NA genes; however, since their amino acid homology with N9 is less than 50%, there will be most likely no cross-reactive antibodies to N9. Although some studies show that previously developed vaccines against divergent H7 provide cross protection against H7N9,19,20,21 this would only be a temporal solution in the absence of specific H7N9 vaccines. Vaccines that induce cross-reactive antibodies against H7 only and no antibodies against neuraminidase are likely less effective and therefore urge development of vaccines specific for the current H7N9 outbreak.
Inactivated influenza vaccines (IIVs) provide strain-specific humoral immunity that does not protect against antigenic variations of the influenza virus. Moreover, IIVs do not induce mucosal immunity and thus fail to protect at the gate. Inactivated vaccines against H7N9 prove to be weak immunogenic, and adjuvation is required for sero-conversion,22 which makes the vaccine more complex. LAIVs are believed to be immunologically superior vaccines due to their potential to induce all arms of the adaptive immune responses, including induction of serum antibodies, mucosal immunity, and cytotoxic T lymphocytes targeted to conserved virus epitopes.23,24,25 Other advantages of LAIVs over traditional IIVs are a much cheaper and quicker manufacturing process and easier administration by intranasal spray.14
We developed an intranasal H7N9 live attenuated vaccine and evaluated safety, immunogenicity, and efficacy in an intratracheally induced severe pneumonia ferret model. We show that the vaccine is safe and raises high hemagglutination-inhibiting (HI) and neuraminidase-inhibiting (NI) titers and virus-neutralizing (VN) titers, which provide sterilizing immunity in challenged ferrets. Importantly, the vaccine protects against weight loss, fever, clinical disease, (severe) pneumonia, and death.
Results
Generation and in vitro characterization of the H7N9 LAIV candidate
To prepare for the event that H7N9 becomes transmissible between humans and a pandemic is on the verge, an H7N9 LAIV based on the A/Anhui/1/2013 isolate representative for currently circulating H7N9 influenza in China was generated. The LAIV was constructed by classical reassortment in developing chicken embryos using the cold-adapted and temperature-sensitive master donor virus (MDV) A/Leningrad/134/17/57 (H2N2) as a backbone.26 The resulting A/17/Anhui/2013/61 (H7N9 LAIV) vaccine strain inherited HA and NA genes from wild-type (WT) H7N9 A/Anhui/1/2013 parental virus and six internal genes from the MDV as confirmed by partial sequencing.27 Full-length genome sequencing confirmed nucleotide sequences of six internal genes identical to the internal genes of the MDV. However, the HA gene of A/17/Anhui/2013/61 reassortant virus revealed two nucleotide changes of A to G at positions 421 and 499 (ORF numbering) compared with WT H7N9 (GISAID: EPI439507). These changes resulted in changes at the amino acid level of Asn-123-Asp and Asn-149-Asp by H7 numbering (Figure 1). In addition, the NA gene of the reassortant also contained a nucleotide change of C to T, which resulted in Thr-10-Ile change compared with NA of WT virus (GISAID: EPI439509).
Figure 1.
Mutations as a result of egg adaptation. Sequencing diagrams of HA (nt 421 and 499) and NA (nt 29) of WT A/Anhui/1/2013 (H7N9) influenza after multiple passaging on eggs and a single passage on MDCKs.
To reveal whether these mutations were spontaneous or result from egg adaptation, we sequenced WT A/Anhui/1/2013 passaged three, four, and five times in eggs and three times in eggs followed by one passage on Madin–Darby canine kidney (MDCK) cells (Figure 1). Interestingly, mutation at position 421 (Asn-123-Asp) was appearing gradually during virus passaging in eggs, and after four sequential passages, the original nucleotide was detected at a very low level (approximately 5%) and almost disappeared by fifth egg passage. Noteworthy, one passage in MDCK cells could significantly restore the original population of WT virus, and the heterogeneity was seen at approximately 45% level. Similarly, the second mutation at position 499 could be partially restored after single passage in MDCK cells, whereas all egg-grown variants shared Asn-149-Asp mutation without quasi species. The Thr-10-Ile change in the NA protein was also appearing gradually during egg passaging and could be partially restored by a single passage in a mammalian host (Figure 1). These data demonstrate a strong connection between egg passaging of WT H7N9 virus with the appearance of Asn-123-Asp and Asn-149-Asp mutations in HA and Thr-10-Ile mutation in NA, i.e., these mutations are egg adapted. Passaging in mammalian cell culture can restore these mutations to the original residues. Therefore, during the generation of LAIV reassortant virus using the egg-based system, it is impossible to maintain the original mammalian cell–adapted component of WT virus HA and NA, and the presence of egg-adapted component is inevitable.
H7N9 LAIV has a cold-adapted and temperature-sensitive phenotype
LAIVs are characterized by their temperature-sensitive (ts) and cold-adapted (ca) phenotype. The first phenotype is characterized by the inability to grow at temperatures that represent fever in humans and the latter by the ability to grow at low (26 °C) temperatures. In chicken embryos, H7N9 LAIV (A/17/Anhui/2013/61) exhibited high reproductive activity at an optimum incubation temperature of 33 °C (Supplementary Table S1). Similar to the MDV (A/Leningrad/134/17/57 (H2N2)), the H7N9 LAIV acquired the ts and ca phenotypes. It reproduced well at a reduced temperature of 26 °C and practically lost the ability to reproduce at temperature elevated to 38–39 °C. The WT parent virus A/Anhui/1/2013 (H7N9), in contrast, was distinguished by the ability to reproduce to high titers at 38–39 °C and low reproduction at 26 °C.
H7N9 LAIV is attenuated in ferrets
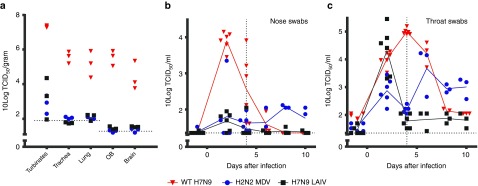
Next, attenuation of the H7N9 LAIV was evaluated in a ferret safety test performed according to the WHO biosafety guidelines on production and quality control of H7N9 vaccines.28 When ferrets were intranasally infected with WT H7N9 (A/Anhui/1/2013, 107 TCID50), they did not develop any clear clinical signs of influenza illness. They did develop a mild fever and showed limited weight loss over a period of 14 days (Supplementary Table S2). In contrast, H7N9 LAIV and MDV did not induce fever or weight loss. High viral titers were detected 4 days after infection with WT H7N9 in the upper (nasal turbinates) and lower (trachea and lung) respiratory tract and exterior of the respiratory system in the olfactory bulb (OB) and brain (Figure 2a), but not systemically (spleen and intestine—data not shown). In the H7N9 LAIV and MDV infected ferrets, viral titers were only detected in the upper respiratory tract (nasal turbinates). WT H7N9 replicated at high levels in the nose and throat and continued until 6 and 8 days after infection, respectively, whereas only low levels in the nose and high levels in the throat were detected in the H7N9 LAIV infected ferrets until 4 days after infection (Figure 2b,c). Infection with the MDV resulted in low levels of virus replication in the nose and substantial virus replication in the throat, however, this continued until day 10. Gross pathology performed on the lungs of ferrets sacrificed 4 and 14 days after infection showed only minor lesions in the lungs of WT H7N9-infected ferrets, which were approximately equal in MDV infected ferrets and less in the H7N9 LAIV infected ferrets (data not shown). Thus, the absence of clinical signs of disease, limitation of virus replication to only the upper respiratory tract, a reduced time span of viral replication, and reduced gross pathology confirmed the attenuated phenotype of the H7N9 LAIV.
Figure 2.
Virus replication in the respiratory tract and brain. (a) Virus titers in the nasal turbinates, trachea, lung, olfactory bulb (OB), and brain of ferrets sacrificed 4 days after intranasal infection with either WT H7N9 (red triangles), H2N2 master donor virus (MDV; blue circles), or H7N9 live attenuated influenza vaccine (LAIV) (black squares). No virus replication was detected in the intestines or the spleen (not shown). (b) Nose and (c) throat swabs were performed on all animals (six per group) 1 day prior to challenge and 2 and 4 days after infection. After termination of three animals per group 4 days post infection (d.p.i.), sampling was continued in the remaining three animals on days 6, 8, and 10. The virus titers in the homogenized tissue samples or transport buffer of the swabs were determined by end-point titration on Madin–Darby canine kidney cells using a fivefold serial dilution. WT H7N9 titrations were incubated at 37 °C whereas H2N2 MD and H7N9 LAIV titrations were performed at 32 °C. Presented are the individual 10 log transformed titers connected with a line through the averages (b and c only). Dotted horizontal lines indicate limit of detection. Dotted vertical lines indicate day of termination of first three ferrets.
Single and booster vaccination with H7N9 LAIV induce high HI, NI, and VN antibody titers
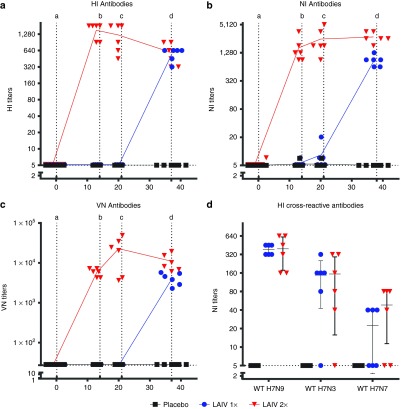
To investigate the immunogenicity of H7N9 LAIV, ferrets were intranasally vaccinated according to a single (1×LAIV) and booster (2×LAIV) strategy. As observed in the safety study (Figure 2), 2 days after first vaccination, high virus titers were detected in the throat of H7N9 LAIV-vaccinated ferrets. However, after booster vaccination (21 days after primary vaccination), no virus was detected, indicating that the booster had been neutralized by the antibodies induced at primary vaccination. H7N9 LAIV induced high HI, NI, and VN antibody titers as detected in serum collected on days 13, 20, and 23 after single vaccination and on day 17 after booster vaccination (Figure 3). Seven days after first vaccination, no relevant titers could be detected, but the responses peak at 14 (HI) and 21 days after first vaccination (NI and VN). Despite booster vaccination, HI and VN titers declined but NI titers increased; however, none of the effects were significant.
Figure 3.
Functional and neutralizing antibody responses. (a) Hemagglutination inhibition, (b) neuraminidase inhibition, and (c) virus neutralization antibody titers detected in sera during the course of the study in serum of placebo (black squares), 1×LAIV (blue circles), or 2×LAIV (red triangles) intranasally vaccinated ferrets. (d) Cross-reactive hemagglutination inhibition titers at the day before challenge. Titers were measured against four hemagglutinating units of H7N9 LAIV as a representative of the hemagglutinin of (a) WT virus, (d) WT H7N9, H7N3, or H7N7 using a concentration of 1% horse erythrocytes (a), an H6N9 (RN19/13-human reassortant strain) dilution with a neuraminidase activity resulting in ~OD 1 after incubation of 1 hour at 37 °C (b), and against 100 TCID50 of WT H7N9 (c). Inhibition of neuraminidase was considered at an OD of less than three times the SD below the maximal OD (standard H6N9). VN titer was determined at 50% virus neutralization according to Reed and Muench.46 Presented are the individual titers and a line connecting the averages. Vertical dotted lines indicate the following: a. 1st vaccination 2×LAIV; b. 1st vaccination 1×LAIV; c. 2nd vaccination 2×LAIV; d. challenge. LAIV, live attenuated influenza vaccine.
We next evaluated the cross-reactive potency of the vaccine-induced H7 antibodies against A/Netherlands/33/03 (H7N7) and A/mallard/Netherlands/12/00 (H7N3) strains in sera obtained 23 days after single vaccination and 17 days after booster vaccination (all obtained the day before challenge). The H7N7 strain was isolated from a human case during the 2003 outbreak in the Netherlands, and the H7N3 is a low-pathogenic avian influenza strain also used in a vaccine strain.15 The H7N7 and H7N3 strains share 96.3% and 97.1% homology with A/Anhui/1/2013 (H7N9), respectively. The H7N9 LAIV vaccine induces cross-reactive antibodies against both H7 variants (Figure 3d), although higher responses were observed for the H7N3 strains, which also reflects the higher homology with H7N9.
Thus, H7N9 is highly immunogenic and induces functional antibodies against hemagglutinin and neuraminidase surface proteins that result in high VN antibody titers. Moreover, the H7-directed antibodies cross react with divergent H7.
H7N9 LAIV protects against severe disease and death
Since intranasal infection only induced mild disease in ferrets (Supplementary Table S2),9,29 which does not represent the severe disease observed in H7N9-infected humans, the intratracheal challenge model was chosen to establish the protective efficacy of H7N9 LAIV. In a dose-escalation infection study, a dose of 107 TCID50 WT H7N9 influenza consistently induced severe pneumonia (data not shown), similar to as previously reported.30 Since at this dose, the disease progressed rapidly, termination was performed 3 days post challenge (d.p.c.) to be able to compare end-point measurements. When placebo-vaccinated ferrets were challenged accordingly, they became less active (score 1) that progressed to inactive behavior (score 2) after 2–3 days. They initially presented faster breathing (score 1) which developed to heavy stomach breathing (score 2) by 2–3 d.p.c. (Supplementary Table S3). Finally, three out of six animals succumbed to the infection prior to termination of the study. Ferrets vaccinated with H7N9 LAIV, on the other hand, only developed mild disease and showed none to some inactivity (score 1) and few animals displayed faster breathing (score 1), and none of the animals died.
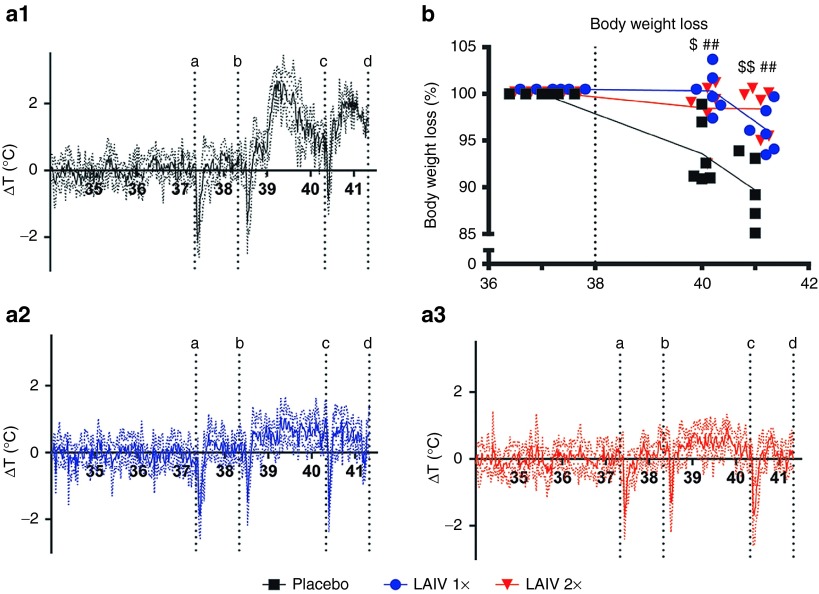
Less than 1 d.p.c., placebo-vaccinated ferrets developed a substantial fever, which lasted for approximately 1 day and after a small dip returned for another day (Figure 4a.1). During the 3 d.p.c., the average temperature was ~1.2 °C higher than normal, and the highest recorded fever was 3 °C above baseline (Supplementary Table S4). In H7N9 LAIV-vaccinated ferrets, only a very mild induction of fever was registered after challenge (Figure 4a.2, a.3). Vaccination significantly reduced the average temperature increase with 0.7–0.8 °C and the highest recorded fever with ~1.5 °C (Supplementary Table S4). Placebo-vaccinated ferrets lost between 5% and 15% of their body weight by 3 d.p.c. (Figure 4b), the average weight was 8% lower than baseline and the average maximum recorded weight loss was 10% (Supplementary Table S4). These effects were significantly (P < 0.05 for 1×LAIV and P < 0.01 for 2×LAIV) reduced in vaccinated ferrets (Supplementary Table S4), although the weight of single-vaccinated ferrets started to descend slightly at 3 d.p.c., while ferrets that had received a booster remained stable (Figure 4b). Hence, vaccination with H7N9 LAIV significantly reduced fever and weight loss. The booster strategy appeared more effective but was not significantly different from a single vaccination.
Figure 4.
Fever and weight loss. Body temperature was recorded every 30 minutes by transponders that were placed in the intraperitoneal cavity: (a.1) placebo; (a.2) 1×LAIV; (a.3) 2×LAIV. To analyze fever development, the temperature difference compared with baseline was calculated and the average ΔT per group was plotted. The dotted lines indicate the SD. The several sharp decreases in temperature are a result of sedation indicated by vertical dotted lines: a. baseline swabs and weight; b. challenge; c. swabs and weight; d. section. The temperature decrease of animals that succumbed to the infection started a couple of hours prior to death, and therefore in the placebo group five animals (one transponder had a power down) were included until day 39 (2.00 p.m.); four animals until day 40 (8.00 a.m.), and the remaining two animals until section. (b) Body weight prior to challenge was set at 100%, and percentage weight loss was subtracted from baseline. Presented are the individual body weight loss and lines connecting the averages. Placebo (black squares), 1×LAIV (blue circles) or 2×LAIV–vaccinated (red triangles)–vaccinated ferrets. ##P < 0.01(1×LAIV,) $P < 0.05 (2×LAIV), and $$P < 0.01(2×LAIV) indicate difference compared with placebo by the Mann–Whitney test. LAIV, live attenuated influenza vaccine.
H7N9 LAIV vaccination inhibits virus replication
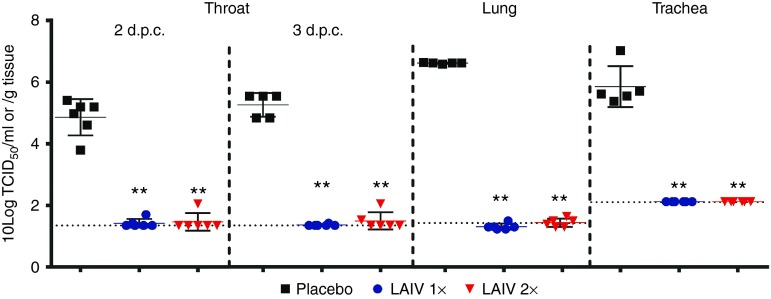
Upon intratracheal challenge, H7N9 WT virus replicated to high titers in the throat (2 and 3 d.p.c.) and in the lung and trachea (3 d.p.c.; Figure 5) of placebo-vaccinated ferrets. In the previous dose-escalation study, no virus replication was detected in the nasal turbinates or brain (data not shown) and were therefore not analyzed here. Vaccination with either a single or a booster strategy inhibited virus replication, since the challenge virus could not be detected in the lung and trachea in any of the vaccinated ferrets and was also absent in the throat of almost all animals. In the throat, some virus replication just above the detection limit was observed in three vaccinated animals.
Figure 5.
Virus replication in the respiratory tract. Virus titers in the throat 2 and 3 days post challenge (d.p.c.) and in the trachea and lung 3 d.p.c. in placebo (black squares), 1×LAIV–vaccinated (blue circles), or 2×LAIV (red triangles)–vaccinated ferrets. The virus titers in the transport buffer of the swabs or homogenized tissue samples were determined by end-point titration on Madin–Darby canine kidney cells using a fivefold serial dilution. Presented are the individual 10 log transformed titers, their averages (bar), and SD (error bar). Dotted horizontal lines indicate limit of detection. Throat swabs taken prior to challenge were negative for virus replication (data not shown). LAIV, live attenuated influenza vaccine.
H7N9 LAIV vaccination prevents severe pneumonia
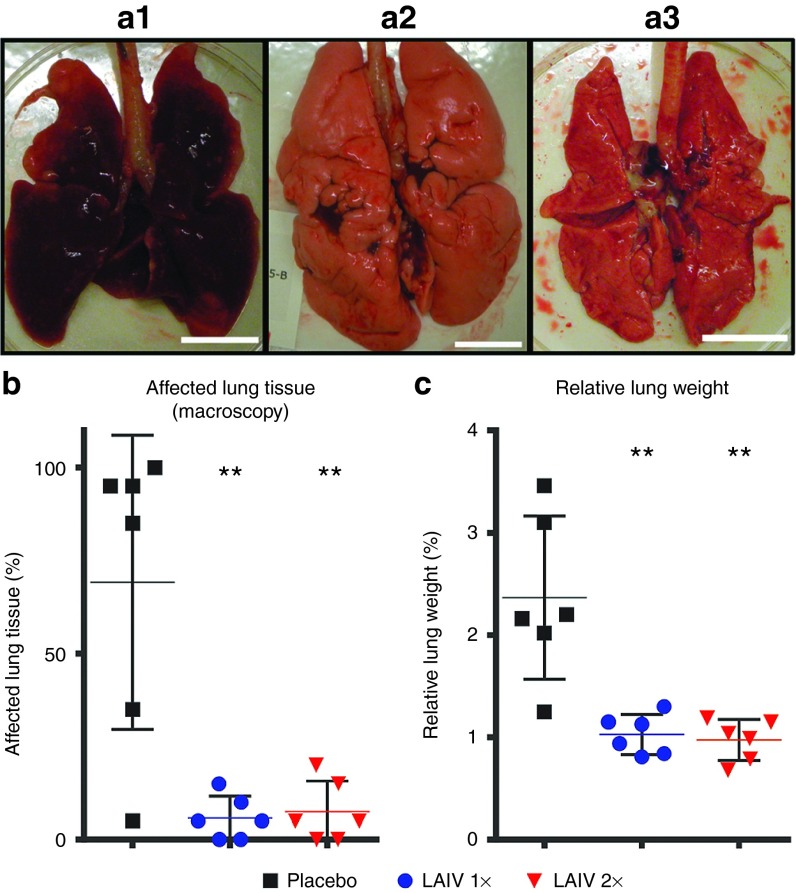
Three days after challenge, the lungs of four out of six placebo-immunized ferrets were swollen, appeared opalescent, and showed dark red lesions, which in extreme cases almost affected the whole lung (Figure 6a.1,b). One ferret showed a relatively normal macroscopic appearance and one ferret had a moderately affected lung. This difference may be an effect of timing; since animals were sacrificed relatively short after challenge, severe pneumonia may not yet have been developed to such an extent in these animals to be macroscopically visible. Regardless, similar virus titers were detected in the respiratory tract as compared with the macroscopically severely affected ferrets, confirming successful challenge. Edema was observed flowing from the trachea of placebo-vaccinated ferrets, and due to presence of edema in the lungs, the weight of the lungs was increased up to three times compared with historic data. To rule out the differences in size of the ferrets, the ratio of the lung and body weight (relative lung weight (RLW)) was determined to be able to compare groups (Figure 6). Interestingly, the lung of the placebo-vaccinated ferret which had normal appearance was clearly increased in weight (RLW of 2.2) indicating initiation of pneumonia.
Figure 6.
Affected lung tissue at the macroscopic level and edema formation. Representative images of inflated lungs (dorsal view) of (a.1) placebo, (a.2) 1×LAIV–vaccinated, and (a.3) 2×LAIV–vaccinated ferrets. Bar = 2 cm. (b) During section, the lung lobes were macroscopically examined and an estimate of the percentage tissue that was affected (dark red) was recorded. (c) The relative lung weight (RLW) was calculated as the percentage of the lung weight 3 days post challenge (d.p.c.) relative to the body weight at the day of challenge. An increase of the RLW is caused by edema formation and is a measure for pneumonia. Presented are the individual values, their means (bar), and SDs (error bar). LAIV, live attenuated influenza vaccine.
The severe effects observed in placebo-vaccinated ferrets were absent in H7N9 LAIV-vaccinated ferrets. Macroscopically only a few small spots (<10%) were visible (Figure 6a.2, a.3,b) on the lung surface, and the RLW of either vaccinated groups was significantly lower than the RLW of the placebo group (Figure 6c). Moreover, the LAIV-vaccinated ferrets had normal lung weights compared with historic data. Therefore, H7N9 LAIV vaccination with either strategy protects against severe lung damage and edema formation.
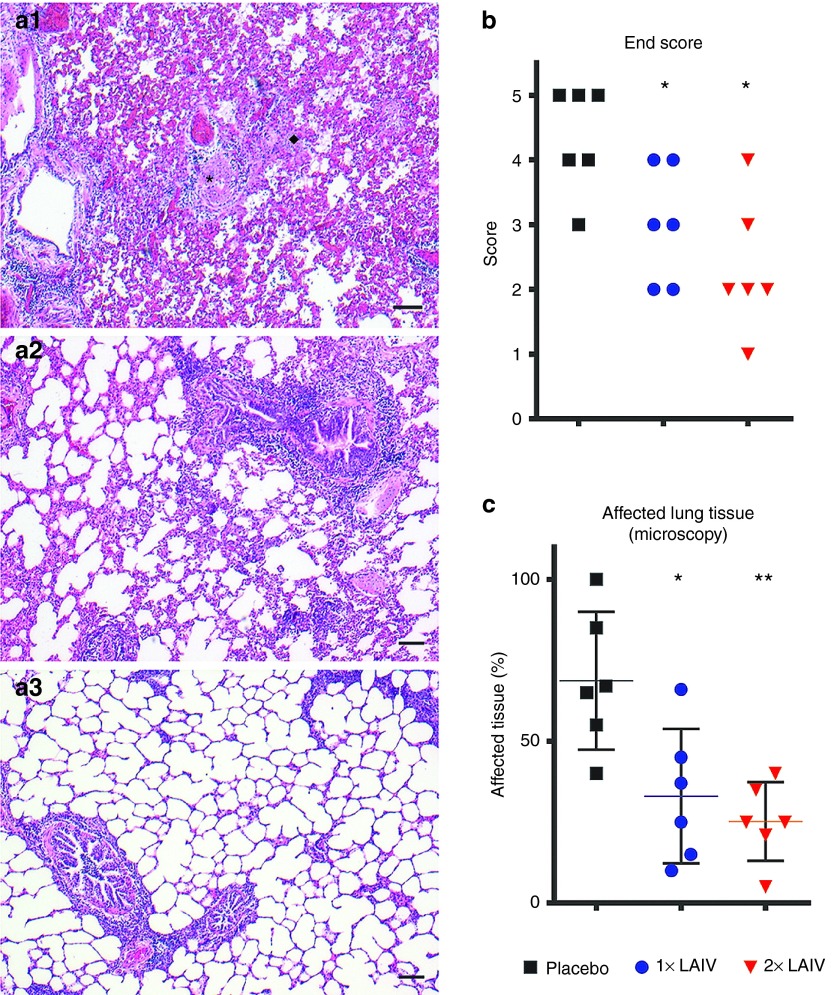
Histopathological analysis of the lungs showed that after intratracheal challenge with WT H7N9, five out of six placebo-vaccinated ferrets developed a severe lung pathology characterized by a moderate-to-strong bronchopneumonia and a moderate interstitial and alveolar pneumonia (Figure 7a). Bronchiolar lumens were regularly obstructed by exudates, and bronchioli were inflamed. Necrosis and denudation of bronchiolar epithelium were frequently observed. Hyperemia is strongly present, and the alveolar space is regularly filled with edema, fibrin, debris, and large amounts of polymorphonuclear (PMN) cells. Because of the severe pathology, examination of hypertrophy of alveolar type II cells was not possible. Part of the blood vessels and large bronchi were minimally involved in this inflammation. The severity and the amount of lung pathology were greatly decreased in the single-vaccinated group and further diminished in the booster group, and most of the above-described effects were either absent or minimally present. The remaining pathology was characterized by a slight-to-strong peribronchiolitis and perivasculitis, which was less pronounced in the 2×LAIV group than in the 1×LAIV group (Figure 7). A moderate hypertrophy of alveolar type II cells was observed in the 1×LAIV group and to a lesser extent in the 2×LAIV group. The overall pathology was expressed as the end score (Figure 7b) and determined based on the degree of damage and inflammation and the amount of tissue involved, using a scoring range from 0 (no aberrations) to 5 (severely affected). These scores show that both vaccination strategies significantly reduce lung pathology and that a booster vaccination is likely more effective. This is also reflected by the percentage of tissue that is involved in the infection pathology, which is clearly reduced compared with placebo-vaccinated ferrets (Figure 7c).
Figure 7.
Microscopic analysis of lung pathology. Representative hematoxylin and eosin (HE) stained lung sections of (a.1) placebo, (a.2) 1×LAIV–vaccinated, and (a.3) 2×LAIV–vaccinated ferrets. Panel (a.1) shows necrotizing and denudation of inflamed bronchioli with luminal plugs of inflammatory exudate (indicated by *), accompanied by a small peribronchiolar rim of lymphocytes and some polymorphonuclear (PMN) cells. Hyperemia is strongly present, and the alveolar space is partly filled with edema, fibrin, and debris; PMN cells are regularly present (indicated by ♦). Panels (a.2) and (a.3) show peribronchiolitis and (a.2) also an enhanced cellularity of the interstitium. Alveolar inflammation is minimal or absent. Single-vaccinated ferrets (a.2) reveal a moderate hyperplasia of the bronchiolar epithelial lining, some luminal exudate of PMN cells, and a strong peribronchiolar accumulation of lymphocytes. Double-vaccinated animals (a.3) present a slight peribronchiolar infiltrate and slight hyperplasia of the epithelial lining and a minimal luminal exudate. Bar = 100 µm. (b) The end score is an overall pathology score which is determined based on the amount of tissue affected, the degree of inflammation, and the amount of damage in the left caudal and cranial lung lobe. The scores range from 0 (no aberrations) to 5 (severely affected). (c) The estimated percentage of lung parenchyma involved in the infection pathology. Bars represent the mean and the error bars represent the SD. Statistical analysis was performed using (b) an adapted version of the Mann–Whitney test for ordinal data with a small sample size and (c) the classical Mann–Whitney test. LAIV, live attenuated influenza vaccine.
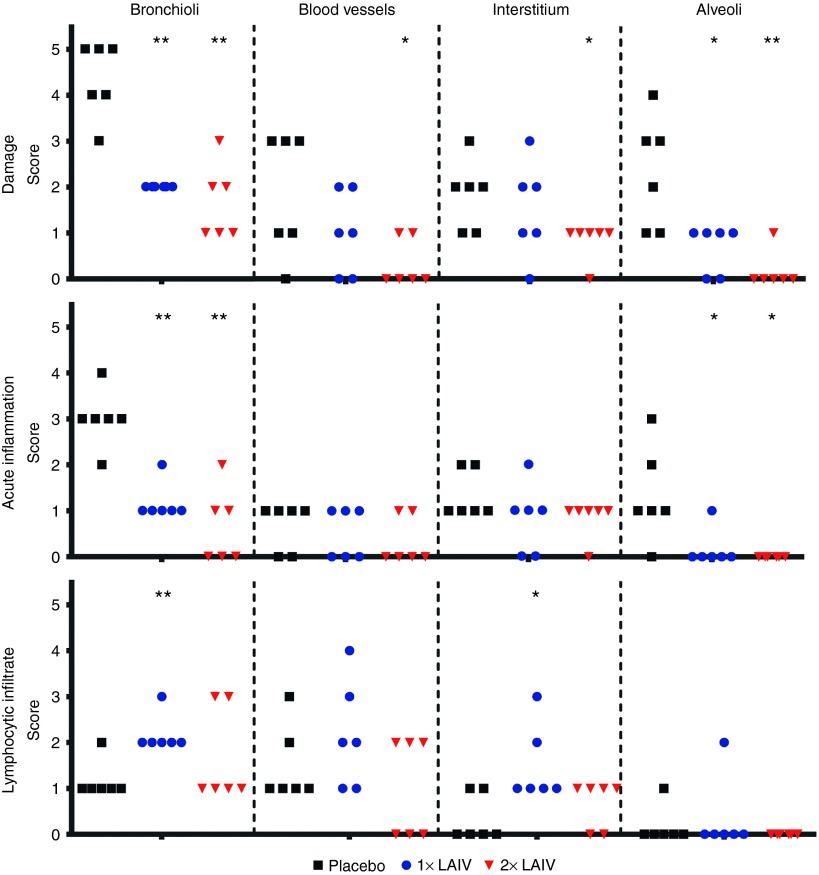
To further examine the nature of the infection pathology, we categorized pathology parameters that related to damage, acute inflammation and lymphocytic infiltrate. This allowed distinguishing between directly virus-induced effects (damage) and immune system–related effects (the latter two categories). The category acute inflammation included infiltrates of PMN cells and macrophages and the category lymphocytic infiltrate included presence of lymphocytes, lymphoblasts, and plasma cells. These two categories allowed to discriminate between cells that relate to the innate and adaptive host response, respectively, although the latter category does not exclude natural killer cells. The three categories were separately assessed for the bronchi, bronchioli, blood vessels, interstitium, and alveoli (Figure 8). In the bronchi, only minor effects were observed and are not presented.
Figure 8.
Detailed histopathological analysis. Pathology parameters were categorized according to their relation to damage (upper panel), acute inflammation (middle panel), and lymphocytic infiltration (lower panel), and these were assessed for the different lung compartments: bronchioli, blood vessels, interstitium, and alveoli. For each category, a final score was determined ranging from 0 (no aberrations) to 5 (severely affected). The category damage includes directly virus-induced effects, the category acute inflammation includes innate effects, and the category lymphocytic infiltrate includes mostly adaptive immune effects. The bronchi only showed minimal effects (not presented). Statistical analysis was performed using an adapted version of the Mann–Whitney test for ordinal data with a small sample size.
Damage induced by the intratracheal infection is most pronounced in the bronchioli and to a lesser extent in the alveoli, and these are significantly reduced in the single and booster approach (Figure 8; upper panel). Significant reduction of damage in the interstitium and blood vessels was only observed for the booster strategy. Acute inflammation of the bronchioli is moderately to strong in the lungs of placebo-vaccinated ferrets (Figure 8, middle panel) and significantly reduced by either the single or booster vaccination approach. Acute inflammation in the alveoli is also significantly reduced in the vaccinated animals. Strikingly, the infiltration of lymphocytes including activated large lymphocytes and a few plasma cells is enhanced in single-vaccinated ferrets compared with placebo-vaccinated ferrets (Figure 8; lower panel). This effect is greatest and significant around the bronchioli and to a lesser extent around blood vessels and in the interstitium. The booster strategy does not show this significantly enhanced infiltration of lymphocytes, although lymphocytes are still present. Hence, single and booster vaccination with H7N9 LAIV reduce the severe damage and acute inflammation at the various levels in the lung induced by an intratracheal challenge. This effect appears more pronounced in the booster strategy, although there were no significant differences between the two vaccination strategies. However, a single vaccination enhances infiltration of lymphocytes.
Discussion
Avian influenza A viruses keep crossing the species barrier, and would such a virus become transmissible between humans, this would pose a serious risk for the world population. The GAP for Influenza vaccines is one of WHO's programs to anticipate on such an event. Within this program, we developed a LAIV against potentially pandemic H7N9 influenza by classical reassortment26 based on the backbone of the master donor strain A/Leningrad/134/17/57 (H2N2). H7N9 LAIV showed a shift of the replication optimum toward lower temperatures in embryonated chicken eggs, replicated only in the upper respiratory tract of ferrets, and did not induce clinical signs of influenza disease including fever or weight loss. As such, the vaccine showed an attenuated phenotype as compared with WT H7N9. Administration of a single dose induced high functional antibody titers against HA and NA and high VN titers in ferrets. A booster vaccination did not further enhance the antibody response. Both strategies induced cross-reactive HI titers against two divergent H7 strains (A/Netherlands/33/03 (H7N7) and A/mallard/Netherlands/12/00 (H7N3)), indicating that the vaccine may also protect against variants of the H7N9 virus. Single- and a two-dose vaccination strategies both induced sterile immunity since virus could not be detected in the throat, trachea, and lungs after intratracheal challenge. The vaccine was highly effective as it protected ferrets against fever, weight loss, severe pneumonia, and death when challenged with H7N9. The protective efficacy appeared to be better in the two-dose strategy, although there were no significant differences between single- and double-vaccinated ferrets. In view of the limited production capacity during a pandemic outbreak, a single dose would be preferable and we here show that, at least in ferrets, this strategy provides ample protection.
An ever-difficult obstacle in preclinical research is extrapolation to the human situation. The better the human situation is mimicked the more likely the results will predict the outcome in humans. For influenza disease, mainly mice and ferrets have been used as a model.31,32,33 Although many vaccine efficacy studies are currently performed in mice, this model does not reflect influenza disease in humans and often requires adaptation of the challenge virus, which altogether impacts the extrapolation. Partly by the similar lung physiology and sialic acid receptor distribution in the respiratory tract, the ferret is a model that closely resembles human influenza pathogenesis. The severity of disease, however, is greatly influenced by the route of inoculation. We observed that when H7N9 was inoculated via the intranasal route in the safety study, a mild disease was induced with the only observations a mild fever and moderate weight loss which was in line with previous studies.9,29 The lungs of the animals showed minor deviations, but nowhere near the viral pneumonia observed in humans. Previously Van den Brand et al. induced severe viral pneumonia with an H5N1 infection in ferrets using the intratracheal route,34 and recently this method was also applied for H7N9.30 Despite the unnatural route of infection, this method induced disease similar to severe cases of H5N1- or H7N9-infected humans. Intratracheal inoculation is therefore the better model to study vaccine efficacy in relation to protection from disease.
Thus far, the efficacy of H7N9 LAIVs has only been evaluated in the intranasal challenge ferret model.35,36 This is mainly a model in which reduction of virus replication can be measured, which is important in the light of containing a pandemic. We here show, however, that in an H7N9 intratracheal challenge ferret model, H7N9 LAIV protects against severe pneumonia and death and also provides sterilizing immunity.
Pathological examination 3 days after challenge showed that at the macroscopic level on average, the percentage of severely affected lungs was reduced with 60% to almost normal appearance in the LAIV-vaccinated ferrets. In addition, relative lung weight, which is a measure for the presence of edema, was maintained at baseline in immunized animals. These observations cohere with the absence of severely impaired breathing and protection against death as a consequence of ARDS. For histopathological analysis, we categorized parameters associated with damage, acute inflammation and lymphocytic infiltration, which allowed distinguishing between primarily virus-induced effects, effects related to the innate and to the adaptive host response, respectively. LAIV vaccination significantly reduced bronchiolar and alveolar damage that was observed in placebo-vaccinated ferrets. Acute inflammation characterized by PMN cells and macrophages in the bronchioli and alveoli was also significantly reduced in both LAIV strategies. However, an increase in infiltrate of (large) lymphocytes around the bronchioli, blood vessels and in the interstitium, was observed for the single LAIV-vaccinated ferrets.
LAIVs have been shown to induce cellular responses,37,38 and therefore, the increased lymphocytic infiltrates may include specific T cells induced by LAIV vaccination; however, the observation of larger lymphocytes also indicates the presence of B cells, and also some plasma cells were found. Since the challenge was performed relatively shortly after vaccination (24 days), the immune response is not yet in the memory phase, and lymphocytes have not yet fully contracted.39,40,41 A high challenge dose (107 TCID50) in combination with the presence of a high number of lymphocytes may lead to the aberrant increased infiltrates, which may not have been induced when challenged with a lower dose or at a later stage. Hence, the increased infiltration may be a side effect of the model with respect to timing and dose of the challenge. The identity of the lymphocytes and whether they are indeed vaccine specific and why these effects are less prominent in boosted ferrets remain to be further elucidated.
During the development of the H7N9 LAIV, the vaccine was passaged multiple times in eggs, which resulted in amino acid changes in HA (Asn-123-Asp and Asn-149-Asp) and NA (Thr-10-Ile) compared with the human wt isolate. These mutations could partly be restored after one passage on MDCK cells. The changes may result from host system–dependent positive selection and apparently were required for optimizing growth in eggs since the final vaccine reassortant is characterized by high yield in eggs (> 10 logEID50). This is supported by a study by Chen et al. in which they found that a reverse genetically derived LAIV candidate with the human H7N9 WT HA sequence grew poorly in eggs.36 A number of egg-adapted mutations had to be introduced into the H7 protein to generate LAIV candidates based on the A/Ann Arbor/6/60 MDV with high-yield characteristics. Interestingly, one of these candidates had the same two mutations in HA as the Len/17-based H7N9 LAIV virus (Asn-123-Asp and Asn-149-Asp) underlining that the mutations indeed are characteristic for optimized replication.
We developed an H7N9 LAIV by classical reassortment that is attenuated and highly immunogenic in ferrets. The vaccine prevented virus replication and protected against severe pneumonia and death after an intratracheal challenge in ferrets. Based on these encouraging results, the vaccine candidate is currently under clinical evaluation.
Materials and Methods
Ethical statement. The animal experiments were approved by the Committee on Animal Experimentation of the Antonie van Leeuwenhoek terrain (DEC-Alt, Bilthoven, the Netherlands) under permit numbers 201300088 and 2013002215. Animal handling was carried out in accordance with relevant Dutch national legislation, including the 1997 Dutch Act on Animal Experimentation. Animals were examined for general health on a daily basis. If animals show severe disease according to the defined end points prior to scheduled termination, they were euthanized by cardiac bleeding under anesthesia with ketamine (5 mg/kg; Ketamine 10%, Alfasan BV, Woerden, NL) and medetomidine (0.1 mg/kg; Sedator, Eurovet Animal Health BV, Bladel, NL).
Viruses. H7N9 A/Anhui/1/2013 influenza virus, an isolate from a human fatal case, was obtained from the National Institute for Biological Standards and Control under the conditions of the Pandemic Influenza Preparedness (PIP) framework. Cold-adapted A/Leningrad/134/17/57 (H2N2) MDV was obtained from the influenza strain repository of the Institute of Experimental Medicine (Saint Petersburg, Russia). A/Netherlands/33/03 (H7N7) and A/mallard/Netherlands/12/00 (H7N3) were obtained from the repository of the Centre for Infectious Disease Control of the RIVM (Bilthoven, the Netherlands).
Live attenuated influenza virus vaccine. The A/17/Anhui/2013/61 (H7N9) live attenuated influenza virus vaccine strain was obtained by a classical genetic reassortment method as previously described.26 Briefly, parental virus A/Anhui/1/2013 (H7N9) and MDV A/Leningrad/134/17/57 (H2N2) were mixed in equal doses (106 EID50) and administered together into embryonated eggs followed by 48 hours of incubation at optimal temperature (33 °C). The resulting mixture of reassortants was subjected to several cloning passages by limiting dilutions at low temperature (26 °C) in the presence of hyper immune serum against MDV. Subsequently, two additional cloning procedures at optimal temperature without antiserum were performed. The genome composition of multiple resulting reassortant viruses was determined by partial sequencing of all eight viral genes using universal primers as previously described.27,42 After the reassortant with required genome composition was selected, full-genome sequencing and phenotypic analyses were performed.
Generation of H6N9 reassortant virus. To avoid nonspecific inhibition of HA antibodies in the enzyme-linked lectin NA inhibition assay, RN9/13-human A(H6N9) was generated as described above. For this purpose, nonpathogenic avian virus A/herring gull/Sarma/51/2006 (H6N1) kindly provided by the Institute of Influenza (Ministry of Healthcare of the Russian Federation, Saint Petersburg, Russia) and vaccine strain A/17/Anhui/2013/61 (H7N9) were used as donor for HA and NA, respectively. By means of low temperature selection and selection in the presence of rat anti-sera against А/17/mallard/Netherlands/00/95 (H7N3) and А/17/Solomon Islands/06/9 (H1N1) viruses, reassortant strain of A(H6N9) subtype on the cold-adapted master donor strain backbone was obtained. The inheritance of HA and NA from A/herring gull/Sarma/51/2006 (H6N1) and A/17/Anhui/2013/61 (H7N9) strains, respectively, was confirmed by HAI test and/or RT-PCR analysis.
Phenotypic and genetic analysis. The ts and ca phenotypes of parental and reassortant viruses were assessed by evaluating viral replication in eggs at permissive (33 °C) and restrictive temperatures (26, 37, 38, 39 and 40 °C). Eggs were incubated for 48 hours for temperatures 33–40 °C and 6 days for 26 °C before detection of virus by hemagglutination assay. Genetic analysis of reassortant viruses was performed either by partial- or full-genome sequencing using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and a 3130xl Genetic Analyzer (Applied Biosystems) according to the instructions of the manufacturer.
Ferrets and handling. Female ferrets (Mustela putorius furo), aged 6–12 months were obtained from Schimmel B.V. (Uddel, The Netherlands). Ferrets were confirmed to be negative for previous infection with circulating influenza virus and Aleutian disease. A temperature transponder (DST micro T, Star-Oddi, Gardabaer, Iceland) was implanted intra-abdominally 14 days prior to commencement of the experiment, which recorded the temperature every 30 minutes. For this purpose, sevoflurane (Sevorane, Aesica Queenborough Ltd, Kent, UK) was used as an anesthetic and buprenorphine (Buprecare, AST Farma BV, Oudewater, NL, 0.2 ml subcutaneously) as a post-operative analgesic. Blood sampling, immunizations, swabs, and challenge were performed under anesthesia by ketamine (5 mg/kg) and medetomidine (0.1 mg/kg), and the anesthesia was antagonized with atipamezole (0.25 mg/kg). After intranasal vaccination and infection or intratracheal challenge, ferrets were antagonized after half an hour to avoid sneezing and coughing reflexes. All animals had ad libitum access to pelleted standard ferret food and tap water.
Safety study in ferrets. Three groups of six ferrets were placed in BSL-3 isolators and were intranasally infected with either WT H7N9, H2N2 MDV, and H7N9 LAIV at a dose of 107 TCID50 or EID50 in 0.5 ml. Animals were weighed every other day, and clinical signs were recorded daily. Nose and throat swabs were taken every other day until 10 days post infection. Swabs were vortexed in 2-ml transport medium (15% sucrose (Sigma, Zwijndrecht, The Netherlands), 2.5 μg/ml fungizone (Gibco, Bleiswijk, the Netherlands), 100 U/ml penicillin, 100 μg/ml streptomycin (Invitrogen, Bleiswijk, the Netherlands), 250 μg/ml gentamicin (Sigma) in phosphate-buffered saline (PBS, Gibco) and were stored at less than −70 °C until further virological analysis. Four days after infection, three animals of each group were sacrificed, and gross pathology was performed on the lungs. Longitudinal sections of the right and accessory lung lobes, the middle section of the trachea, nasal turbinates, a cross section of the spleen, a cross section of the cerebrum and cerebellum, the OB, and cross sections of the intestine were isolated and stored at less than −70 °C for further virological analysis. Fourteen days post infection (d.p.i.), the remaining three animals were sacrificed, and gross pathology was performed on the lungs.
Immunization and challenge of ferrets. Groups of six ferrets were intranasally vaccinated (0.5 ml) twice on days 0 and 21 with a placebo or with H7N9 LAIV or once only on day 14 with H7N9 LAIV. Groups were housed in open pens in different animal rooms to avoid transmission of the LAIV between the groups. Serum samples were collected at regular time intervals (on days 1, 13, 20, 37, and 41) and stored at −20 °C for serological analysis. Thirty-eight days after start of the study, the animals were challenged intratracheally with WT influenza A/Anhui/1/2013 (H7N9) with a virus load of 107 TCID50 in 3 ml. This dose was obtained from a previously performed dose-titration infection experiment and resulted in the most consistent induction of lung pathology and disease. Prior to challenge and 2 and 3 days after challenge, throat swabs were taken and ferrets were weighed. Ferrets were scored twice daily for the clinical parameters activity and impaired breathing according to the following scoring system: 0 = active; 1 = active when stimulated; 2 = inactive, and 3 = lethargic and 0 = normal breathing; 1 = fast breathing, and 2 = heavy and stomach breathing, respectively.
On day 41, three days after challenge, animals were sacrificed by cardiac bleeding. The timing of section was based on a previous infection study in which some animals died between 3 and 4 days after infection when a dose of 107 TCID50 was administered. Scheduling section at a later time point than 3 days after challenge would imply that some animals would die prior to scheduled section. This difference in timing would impair comparison of the pathological results.
During section, the trachea was clamped off after inhalation, and the inflated lungs were isolated, examined for gross pathology, and photographed. The middle section of the trachea (~1 cm) and a section of the three right lung lobes and the accessory lobe along the proximodistal axis (~1 cm by 3 mm) were isolated and stored at −70 °C for virological analysis. The left cranial and caudal lung lobes were fixed in 10% buffered formalin for histopathological analysis.
Serological analysis. HAI titers in ferret sera were determined according to standard methods with a modification specifically published for H7N9 antibody responses.43 Briefly, sera were inactivated (30 minutes at 56 °C), treated with receptor destroying enzyme (Denka-Seiken, Tokyo, Japan), twofold serially diluted, and tested in duplicate against four hemagglutinating units (HAUs) of H7N9 WT or H7N9 LAIV using 1% horse red blood cells.
VN titers were determined as described elsewhere.44 Briefly, sera were heat inactivated for 30 minutes at 56 °C and twofold serially diluted in virus growth medium (MEM medium (Gibco, 31095) containing: 40 µg/ml gentamycin (Sigma), 0.01 M Tricin (Sigma) and 2 µg/ml L-1-Tosylamide-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (Sigma)) in a 96-well plate using a starting dilution of 1:8. An equal volume of virus at a concentration of 100 TCID50 was added to each well. Wells containing only medium or medium with only virus served as a cell viability and negative control, respectively. In addition, a back titration of the virus stock used for incubation with the serum dilutions was prepared by ½log10 serial dilutions. All the preparations were subsequently incubated for 2 hours at 37 °C. Following, the virus-serum mixture, the controls and the back-titration samples were transferred to 96-well plates containing a confluent monolayer of MDCK cells and incubated for another 2 hours at 37 °C, 5% CO2 after which the medium was refreshed. Plates were incubated until the back-titration plate reached cytopathic effect (CPE) at a titer of 100 TCID50 (4–5 days).
In the neuraminidase inhibition assay, homologous H7N9 virus could not be used since antibodies directed against H7 could impede NA activity through sterical hindrance. To resolve this issue, the RN9/13-human A(H6N9) reassortant, which contains an HA against which ferrets are naïve was prepared as described above. The assay was performed as described previously.45 Briefly, 96-well plates (Immulon 2HB, Thermo Scientific, Bleiswijk, the Netherlands) were coated overnight with 100 µl of 50 µg/ml fetuin (Sigma). Twofold serial dilutions of serum were prepared in duplicates and incubated with a RN9/13-human A(H6N9) dilution of which the neuraminidase activity was predetermined at OD450 = 1 after 1-hour incubation at 37 °C. Following this, 100 µl of the virus/serum dilutions were transferred to the fetuin-coated wells. After 4 hours of incubation at 37 °C, the plates were washed with PBS/Tween20 (0.5%), supplemented with 100 µl of peroxidase-labeled peanut lectin (2.5 µg/ml; Sigma), incubated for 1 hour at 37 °C, and washed again. Peroxidase substrate (TMB, 100 µl) was added, and the reaction was stopped after 10 minutes with 100 µl of 2 M sulfuric acid. The OD450 was read using the EL808, Bio-Tek Instruments, Winooski, VT. The NI titer was determined from the last dilution with an OD450 ≤ (OD450 of the virus control – 3 × SD).
Virus quantification. Lungs were collected in lysing A matrix tubes (2 ml, MP Biomedicals, Santa Ana, CA), homogenized using a FastPrep homogenizer (MP Biomedicals) and clarified by low speed centrifugation. Transport medium from the swabs was used directly. Virus titers were determined by end-point titration on MDCK cells as previously described.44 In short, MDCK cells were seeded in 96-well plates at a density of 1–5 × 104 cells/well and incubated at 37 °C until 90–100% confluence. The cells were inoculated in sextuplets with 100 µl in virus growth medium (MEM medium (Gibco, 31095) containing: 40 µg/ml gentamycin (Sigma), 0.01 M Tricin (Sigma) and 2 µg/ml TPCK-treated trypsin (Sigma)) and diluted fivefold serially. After 6 days of incubation at 37 °C, wells were scored for CPEs. The TCID50 titer was determined by the Reed and Muench method.46
Pathology. After fixation, the left cranial and caudal lung lobes were embedded in paraffin. Sections were cut at a thickness of 5 µm and stained with hematoxylin and eosin. Slides were examined microscopically with an objective of 4× or 10×. For evaluation of the infection pathology, three pathology categories were defined that included parameters indicating damage of the epithelial lining, acute inflammation, or lymphocytic infiltration. Damage-related parameters included hypertrophy, hyperplasia, flattened or pseudo squamous, epithelia, necrosis and denudation of bronchi(oli) epithelium, hyperemia of septa, and alveolar emphysema and hemorrhages. Acute inflammatory–related parameters included (peri)bronchi(oli)tis, interstitial infiltrate, alveolitis, and (peri)vasculitis characterized by PMN cells and macrophages. Lymphocytic infiltrate–related parameters included lymphocytes, lymphoblasts, and plasma cells. These categories were semiquantitatively scored on a scale of 0 (no aberrations) to 5 (severe damage) resulting in three scores: one for damage, one for acute inflammation, and one for lymphocytic infiltrate each for the bronchi(oli), interstitium, alveoli, and blood vessels. Both lung lobes were completely examined, and at least six microscopic fields were scored for each lobe. The affected percentage lung surface was estimated with an objective of 2× by examining the whole lung lobe. Finally, an end score was determined by taking into account the severity and percentage of tissue affected of the various parameters. Microscopic slides were randomized and scored blindly.
Statistical analysis and calculations. Data were analyzed with GraphPad Prism (v6.04) or R (v3.1.1). Virus titers and antibody titers were log transformed, and significant differences were determined by the Mann–Whitney U test (one-tailed). The pathology scores were analyzed by an adapted version of the Wilcoxon–Mann–Whitney test using mid P-values validated for ordinal data with a small sample size (manuscript in preparation). Unless otherwise indicated, P < 0.05 (*) and P < 0.01 (**) were considered significantly different compared with the placebo group.
Temperature data were retrieved from the implanted temperature loggers and consisted of measurements taken every 30 minutes. Body weight data consisted of a single measurement maximally once per day at the indicated days. Body weight and temperature baselines were calculated as the average body weight and temperature over a period of 2 (body weight) and 7 days (temperature) prior to infection or challenge. The change in temperature and body weight (ΔT and Δweight) was calculated by subtracting the baseline from the measurement after infection or challenge. The sum of all these measurement divided by the number of measurements is the average change in body weight or temperature. The maximum change in body weight or temperature was the maximum change compared with baseline recorded during any of the measurement after infection or challenge.
SUPPLEMENTARY MATERIAL Table S1. Reproductive activity of reassortant vaccine strain A/17/Anhui/2013/61 (H7N9), and Parent viruses in developing chicken embryos at various incubation temperatures. Table S2. Fever and weight loss analysis after intranasal infection. Table S3. Clinical score and survival rates. Table S4. Fever and weight loss analysis after challenge with WT H7N9.
Acknowledgments
This study was funded by the World Health Organization (WHO) under grant 2013/342684. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We thank Geert van Amerongen and Johan Meijer for their excellent biotechnical assistance and we thank Adam Meijer and Marcel Jonges for kindly providing the H7N7 and H7N3 strains
Supplementary Material
References
- WHO (2015). Influenza at the human-animal interface. Summary and assessment as of 23 June 2015.
- Watanabe, T, Watanabe, S, Maher, EA, Neumann, G and Kawaoka, Y (2014). Pandemic potential of avian influenza A (H7N9) viruses. Trends Microbiol 22: 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, HN, Lu, HZ, Cao, B, Du, B, Shang, H, Gan, JH et al. (2013). Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med 368: 2277–2285. [DOI] [PubMed] [Google Scholar]
- Jernigan, DB and Cox, NJ (2015). H7N9: preparing for the unexpected in influenza. Annu Rev Med 66: 361–371. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood, MJ, Miller, PJ, Spackman, E, Swayne, DE, Susta, L, Costa-Hurtado, M et al. (2014). Role of poultry in the spread of novel H7N9 influenza virus in China. J Virol 88: 5381–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, TT, Zhou, B, Wang, J, Chai, Y, Shen, Y, Chen, X et al. (2015). Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature 522: 102–105. [DOI] [PubMed] [Google Scholar]
- Shi, Y, Zhang, W, Wang, F, Qi, J, Wu, Y, Song, H et al. (2013). Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342: 243–247. [DOI] [PubMed] [Google Scholar]
- Zhou, J, Wang, D, Gao, R, Zhao, B, Song, J, Qi, X et al. (2013). Biological features of novel avian influenza A (H7N9) virus. Nature 499: 500–503. [DOI] [PubMed] [Google Scholar]
- Belser, JA, Gustin, KM, Pearce, MB, Maines, TR, Zeng, H, Pappas, C et al. (2013). Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501: 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q, Shi, J, Deng, G, Guo, J, Zeng, X, He, X et al. (2013). H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341: 410–414. [DOI] [PubMed] [Google Scholar]
- Hai, R, Schmolke, M, Leyva-Grado, VH, Thangavel, RR, Margine, I, Jaffe, EL et al. (2013). Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat Commun 4: 2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, Y, Shichinohe, S, Nakayama, M, Igarashi, M, Ishii, A, Ishigaki, H et al. (2015). Emergence of H7N9 influenza a virus resistant to neuraminidase inhibitors in nonhuman primates. Antimicrob Agents Chemother 59: 4962–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieny, MP, Costa, A, Hombach, J, Carrasco, P, Pervikov, Y, Salisbury, D et al. (2006). A global pandemic influenza vaccine action plan. Vaccine 24: 6367–6370. [DOI] [PubMed] [Google Scholar]
- WHO (2006). Global pandemic influenza action plan to increase vaccine supply WHO/IVB/06.13; WHO/CDS/EPR/GIP/2006.1.
- Rudenko, L, Kiseleva, I, Naykhin, AN, Erofeeva, M, Stukova, M, Donina, S et al. (2014). Assessment of human immune responses to H7 avian influenza virus of pandemic potential: results from a placebo-controlled, randomized double-blind phase I study of live attenuated H7N3 influenza vaccine. PLoS One 9: e87962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu, TM, Levine, M, Fitzgerald, T, Luke, C, Sangster, MY, Jin, H et al. (2014). Live attenuated H7N7 influenza vaccine primes for a vigorous antibody response to inactivated H7N7 influenza vaccine. Vaccine 32: 6798–6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, RJ, Madhun, AS, Hauge, S, Sjursen, H, Major, D, Kuhne, M et al. (2009). A phase I clinical trial of a PER.C6 cell grown influenza H7 virus vaccine. Vaccine 27: 1889–1897. [DOI] [PubMed] [Google Scholar]
- Eichelberger, MC and Wan, H (2015). Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol 386: 275–299. [DOI] [PubMed] [Google Scholar]
- Rudenko, L, Isakova-Sivak, I and Donina, S (2013). H7N3 live attenuated influenza vaccine has a potential to protect against new H7N9 avian influenza virus. Vaccine 31: 4702–4705. [DOI] [PubMed] [Google Scholar]
- Krammer, F, Albrecht, RA, Tan, GS, Margine, I, Hai, R, Schmolke, M et al. (2014). Divergent H7 immunogens offer protection from H7N9 virus challenge. J Virol 88: 3976–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q, Chen, Z, Cheng, X, Xu, L and Jin, H (2013). Evaluation of live attenuated H7N3 and H7N7 vaccine viruses for their receptor binding preferences, immunogenicity in ferrets and cross reactivity to the novel H7N9 virus. PLoS One 8: e76884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan, MJ, Bernstein, DI, Winokur, P, Rupp, R, Anderson, E, Rouphael, N et al.; DMID 13-0032 H7N9 Vaccine Study Group. (2014). Serological responses to an avian influenza A/H7N9 vaccine mixed at the point-of-use with MF59 adjuvant: a randomized clinical trial. JAMA 312: 1409–1419. [DOI] [PubMed] [Google Scholar]
- Hayden, FG, Howard, WA, Palkonyay, L and Kieny, MP (2009). Report of the 5th meeting on the evaluation of pandemic influenza prototype vaccines in clinical trials: World Health Organization, Geneva, Switzerland, 12–13 February 2009. Vaccine 27: 4079–4089. [DOI] [PubMed] [Google Scholar]
- Bardiya, N and Bae, JH (2005). Influenza vaccines: recent advances in production technologies. Appl Microbiol Biotechnol 67: 299–305. [DOI] [PubMed] [Google Scholar]
- Jin, H, and Subbarao, K (2015). Live attenuated influenza vaccine. In: Oldstone, MBA and Compans RW (eds.). Influenza Pathogenesis and Control—Volume II, vol. 386. Springer, Cham, Switzerland. pp. 181–204. [DOI] [PubMed] [Google Scholar]
- Aleksandrova, GI (1977). [Use of the genetic recombination method for obtaining vaccinal strains of the influenza virus]. Vopr Virusol: 387–395. [PubMed]
- Isakova-Sivak, I, de Jonge, J, Smolonogina, T, Rekstin, A, van Amerongen, G, van Dijken, H et al. (2014). Development and pre-clinical evaluation of two LAIV strains against potentially pandemic H2N2 influenza virus. PLoS One 9: e102339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. (2013). Update of WHO biosafety risk assessment and guidelines for the production and quality control of human influenza vaccines against avian influenza A(H7N9) virus. http://www.who.int/biologicals/BIOSAFETY_RISK_ASSESSMENT_21_MAY_2013.pdf?ua=1 (accessed 10 February 2016).
- Zhu, H, Wang, D, Kelvin, DJ, Li, L, Zheng, Z, Yoon, SW, et al. (2013). Infectivity, transmission, and pathology of human H7N9 influenza in ferrets and pigs. Science 341: 183–186. [DOI] [PubMed] [Google Scholar]
- Kreijtz, JH, Kroeze, EJ, Stittelaar, KJ, de Waal, L, van Amerongen, G, van Trierum, S et al. (2013). Low pathogenic avian influenza A(H7N9) virus causes high mortality in ferrets upon intratracheal challenge: a model to study intervention strategies. Vaccine 31: 4995–4999. [DOI] [PubMed] [Google Scholar]
- Belser, JA, Katz, JM and Tumpey, TM (2011). The ferret as a model organism to study influenza A virus infection. Dis Model Mech 4: 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margine, I and Krammer, F (2014). Animal models for influenza viruses: implications for universal vaccine development. Pathogens 3: 845–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes, R, Rimmelzwaan, GF and Osterhaus, AD (2010). Animal models for the preclinical evaluation of candidate influenza vaccines. Expert Rev Vaccines 9: 59–72. [DOI] [PubMed] [Google Scholar]
- van den Brand, JM, Stittelaar, KJ, van Amerongen, G, Rimmelzwaan, GF, Simon, J, de Wit, E et al. (2010). Severity of pneumonia due to new H1N1 influenza virus in ferrets is intermediate between that due to seasonal H1N1 virus and highly pathogenic avian influenza H5N1 virus. J Infect Dis 201: 993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, H, Zhang, Q, Gu, C, Shi, J, Deng, G, Ma, S et al. (2015). A live attenuated vaccine prevents replication and transmission of H7N9 virus in mammals. Sci Rep 5: 11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z, Baz, M, Lu, J, Paskel, M, Santos, C, Subbarao, K et al. (2014). Development of a high-yield live attenuated H7N9 influenza virus vaccine that provides protection against homologous and heterologous H7 wild-type viruses in ferrets. J Virol 88: 7016–7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirkova, TV, Naykhin, AN, Petukhova, GD, Korenkov, DA, Donina, SA, Mironov, AN et al. (2011). Memory T-cell immune response in healthy young adults vaccinated with live attenuated influenza A (H5N2) vaccine. Clin Vaccine Immunol 18: 1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn, KG, Bredholt, G, Brokstad, KA, Pathirana, RD, Aarstad, HJ, Tøndel, C et al. (2015). Longevity of B-cell and T-cell responses after live attenuated influenza vaccination in children. J Infect Dis 211: 1541–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, C, Linderman, JJ and Kirschner, D (2014). Harnessing the heterogeneity of T cell differentiation fate to fine-tune generation of effector and memory T cells. Front Immunol 5: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry, EJ, Teichgräber, V, Becker, TC, Masopust, D, Kaech, SM, Antia, R et al. (2003). Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol 4: 225–234. [DOI] [PubMed] [Google Scholar]
- Kaech, SM, Wherry, EJ and Ahmed, R (2002). Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol 2: 251–262. [DOI] [PubMed] [Google Scholar]
- Hoffmann, E, Stech, J, Guan, Y, Webster, RG and Perez, DR (2001). Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146: 2275–2289. [DOI] [PubMed] [Google Scholar]
- WHO. (2013). Laboratory procedures: Serological detection of avian influenza A(H7N9) virus infections by modified horse red blood cells haemagglutination-inhibition assay. http://www.who.int/influenza/gisrs_laboratory/cnic_serological_diagnosis_hai_a_h7n9_20131220.pdf accessed 10 Februari 2016).
- WHO. (2002). WHO Manual on Animal Influenza Diagnosis and Surveillance; WHO/CDS/CSR/NCS/2002.5 Rev. 1.
- Lambré, CR, Terzidis, H, Greffard, A and Webster, RG (1990). Measurement of anti-influenza neuraminidase antibody using a peroxidase-linked lectin and microtitre plates coated with natural substrates. J Immunol Methods 135: 49–57. [DOI] [PubMed] [Google Scholar]
- Reed, LJ, Muench, H. (1938). A simple method of estimating fifty per cent endpoints. Am J Hyg 27: 493–497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.