Abstract
MicroRNAs (miRNAs) are small noncoding transcripts that regulate gene expression. Aberrant expression of miRNAs can affect development of cancer and other diseases. Synthetic miRNA mimics can modulate gene expression and offer an approach to therapy. Inside cells, mature miRNAs are produced as double-stranded RNAs and miRNA mimics typically retain both strands. This need for two strands has the potential to complicate drug development. Recently, synthetic chemically modified single-stranded silencing RNAs (ss-siRNA) have been shown to function through the RNAi pathway to induce gene silencing in cell culture and animals. Here, we test the hypothesis that single-stranded miRNA (ss-miRNA) can also mimic the function of miRNAs. We show that ss-miRNAs can act as miRNA mimics to silence the expression of target genes. Gene silencing requires expression of argonaute 2 (AGO2) protein and involves recruitment of AGO2 to the target transcripts. Chemically modified ss-miRNAs function effectively inside cells through endogenous RNAi pathways and broaden the options for miRNA-based oligonucleotide drug development.
Introduction
RNAi is associated with gene silencing by duplex RNAs.1,2,3 miRNAs are endogenously-expressed gene-silencing agents that regulate cellular gene expression. miRNAs are synthesized as long single-stranded RNAs that can fold into hairpin loop structure. These hairpins are processed by the enzymes drosha and dicer into double-stranded mature species that can bind to argonaute (AGO) protein.3 The guide strand complementary to target transcripts is loaded into AGO proteins while the passenger strand is removed. The guide strand:AGO complex then binds sequence-specifically to target sequences that are typically located within 3'-untranslated regions (3'-UTR) of mRNAs.
miRNAs contribute to the regulation of many cellular processes and levels of miRNA expression can affect normal physiology and diseases.3 Synthetic duplex RNAs that mimic miRNAs increase the effective concentration of miRNAs within cells and enhance silencing of target genes. This ability to control networks of genes involved in disease makes miRNA mimics a promising approach for therapy.
While the use of duplex miRNA mimics is promising, RNA duplexes have potential drawbacks as therapeutic agents. The need for a passenger strand increases the complexity of the molecule. Both strands must be synthesized and the passenger strand has the potential to cause off-target effects.4 Duplex RNAs are less able to enter tissues in vivo than are single-stranded oligonucleotides, necessitating use of targeting ligands or nanoparticles.5,6 Single-stranded miRNA (ss-miRNA) mimics might combine the power of function through the RNAi pathway with the more favorable pharmacological properties of single stranded oligonucleotides.
Recently, synthetic single-stranded silencing RNAs (ss-siRNA) that are fully complementary to their targets have been demonstrated to function through the RNAi pathway and silence gene expression in cell culture and in animals.7,8 These ss-siRNAs can be successfully loaded into RNA-induced silencing complex and the AGO2/ss-siRNA complex can cleave target transcripts in vitro and in vivo. ss-siRNAs that target CAG repeats were shown to allele-selectively inhibit the expression of mutant huntingtin (HTT),9 ataxin-3 (ATXN3),10 and atrophin-1 (ATN1).11 Inhibition of expression for these trinucleotide repeat genes was dependent on expression of AGO2. An ss-siRNA targeting nuclear antisense transcripts for progesterone receptor gene (PR) can cause transcriptional gene silencing and was also AGO2-dependent.12
These experiments established that ss-siRNAs could function through the RNAi pathway and control gene expression inside animals when delivered in a saline solution. In vivo effects in the liver were achieved after systemic administration.8 For a variety of tissues within the brain, inhibition of HTT gene expression was achieved after intraventricular administration.7,9
One previous study examined chemically modified single-stranded miRNAs as mimics for miR-124 and miR-122.13 These synthetic miRNAs were fully or partially modified with 2'-fluoro or 2'-O-methyl groups and appeared to function as miRNA mimics. While these findings were promising, potency was substantially reduced relative to unmodified RNA mimics and involvement of the RNAi pathway was not demonstrated. The findings were not followed up and the potential for single-stranded mimics to efficiently act through the RNAi pathway remained obscure.
Here, we report the design of chemically modified single-stranded miRNA mimics for miR-34a and let-7a. The mimics contain 2'-fluoro, 2'-O-methyl, and 2'-O-methoxyethyl nucleotides and a 5'-phosphate group, with phosphorothioate substitutions at the internucleotide linkages. We show that the mimics suppress expression of their direct target genes. The characteristics of inhibition by the ss-miRNA mimics are similar to unmodified duplex miRNA mimics. Inhibition of gene expression by the ss-miRNA mimic is dependent on expression of AGO2 and AGO2 is associated with target transcripts, demonstrating involvement of the RNAi pathway. We find that silencing of specific target genes depends on the nature of the chemical modification (type and position of modification), or whether a duplex or single-stranded RNA is used.
Results
Design of miRNA mimics
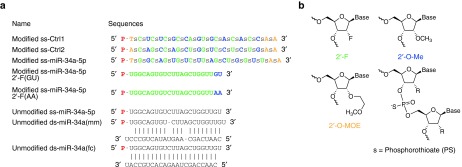
ss-miRNAs are designed to contain chemical modifications to stabilize the RNA strand against digestion by cellular nucleases while still permitting efficient entry into the RNA-induced silencing complex.8 We chose to focus most attention on designing ss-miRNAs to mimic the action of miR-34a because target genes and double-stranded miRNA mimics for that miRNA had been characterized previously.14 The ss-miRNA mimic consists of 2'-methyl, 2'-fluoro, and 2'-methoxyethyl modified nucleotides and a phosphate group at the 5' end (Figure 1). Two thirds of the backbone linkages are substituted with phosphorothioate (PS) bonds, and the remaining linkages are phosphodiester.
Figure 1.
Design of chemically modified single-stranded and unmodified single-stranded/double-stranded miR-34a mimics. (a) Sequences and chemical modifications for miR-34a mimics. Modifications: 2-fluoro (2'-F, green), 2'-O-methyl (2'-O-Me, blue), 2'-O-methoxyethyl (2'-O-MOE, orange), phosphorothioate (PS) linkage (s, black), 5'-phosphate (P, red), unmodified RNA (black). (b) Structure of modified nucleotides.
In parallel with testing the ss-miRNA, we also tested a corresponding 5'-phosphorylated single-stranded RNA with no nucleotide modifications (Unmodified ss-miR-34a-5p) and two types of unmodified double-stranded miRNA mimics for miR-34a (Figure 1a). One mimic is a double-stranded RNA consisting of a 5'-phosphorylated miR-34a-5p guide strand and an unmodified miR-34a-3p passenger strand (Unmodified ds-miR-34a(mm)), which contains internal bulges or mismatch bases in the duplex. The other is a double-stranded RNA that consists of a 5'-phosphorylated miR-34a-5p guide strand and a perfectly complementary passenger strand (Unmodified ds-miR-34a(fc)) except for the 3' overhang.
For comparison, we also tested single-stranded modified RNAs based on the design for single-strand miRNA mimics described by Lim and coworkers.13 These RNAs (modified ss-miR-34a-5p 2'-F(GU)/2'-F(AA)) were primarily composed of 2'-F nucleotides but also contained two 3' terminal 2'-O-methyl modifications (Figure 1a). All linkages were phosphodiester. 2'-F(AA) contained 3' adenosine bases so as to be directly comparable to the other ss-siRNAs used in this study. 2'-F(GU) retains the original sequences of miR-34a-5p at the 3' end (GU) like the miRNA mimics reported by Lim and coworkers.13
Chemically modified ss-miR-34a-5p inhibits SIRT1 gene expression
We tested ss-miRNA and ds-miRNA mimics of miR-34a in HeLa cells. HeLa cells were used for this assay because it was previously reported that the miR-34a level is low,14 enabling us to see effects of miRNA mimics more definitively. We chose SIRT1 as a target gene for monitoring effects on expression because SIRT1 gene is one of the direct targets for miR-34a14 and there are at least two potential binding sites for miR-34a within the 3'-UTR of SIRT1 mRNA (Supplementary Figure S1). We delivered the mimics using a cationic lipid into HeLa cells and evaluated inhibition of SIRT1 gene expression by western blot.
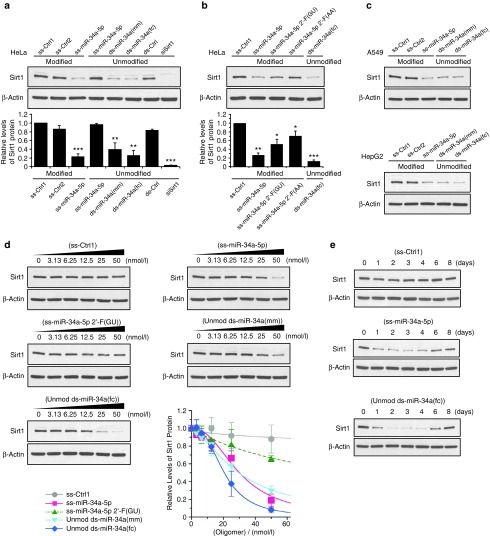
We found that chemically modified ss-miR-34a-5p mimic can knock down SIRT1 expression in HeLa cells to a level similar to unmodified double-stranded miRNA mimics ds-miR-34a(mm) and ds-miR-34a(fc) (Figure 2a). Unmodified ss-miR-34a-5p did not inhibit SIRT1 expression, consistent with the expectation that an unmodified single-stranded RNA would not be stable upon addition to cell culture media or within cells. Non-complementary control ssRNAs (ss-Ctrl1, ss-Ctrl2) (Figure 1a) containing a similar chemical modification pattern as ss-miR-34a-5p did not affect SIRT1 expression, nor did a non-complementary duplex RNA (ds-Ctrl). Single-stranded mimics 2'-F(GU) and 2'-F(AA) were also active but less potent when tested at 50 nmol/l (Figure 2b). Similar SIRT1 inhibition by the mimics was observed also in A549 and HepG2 cells (Figure 2c).
Figure 2.
Inhibition of SIRT1 gene expression by modified ss-miR-34a-5p and unmodified ds-/ss-miR-34a mimics. (a, b) Western blot analysis showing the effect of ss- and ds-miRNA mimics (50 nmol/l) on Sirt1 expression in HeLa cells. An siRNA specific for SIRT1 mRNA (siSirt1) was used as a positive control. Cells were harvested 3 days after transfection for western blot analysis. Error bars are mean ± SD. n = 4 (a), n = 3 (b). (c) Inhibition of Sirt1 by single stranded and double-stranded miR-34a mimics in A549 and HepG2 cells. The oligomers were transfected at 50 nmol/l into A549 cells and at 30 nmol/l into HepG2 cells. (d) Dose response profiles for Sirt1 inhibition by modified ss-Ctrl1, modified ss-miR-34a-5p, modified ss-miR-34a-5p 2'-F(GU) and unmodified ds-miR-34a(mm)/(fc). Western blots and their quantitation from three to four independent experiments are presented. Error bars are mean ± SD. (e) Time course profiles of Sirt1 protein levels in HeLa cells treated with modified ss-Ctrl1, modified ss-miR-34a-5p, or unmodified ds-miR-34a(fc) (50 nmol/l). Western blots are representative from two independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (relative to ss-Ctrl1, paired t-test).
We then examined knockdown of SIRT1 as a function of oligonucleotide concentration (Figure 2d). Noncomplementary control ssRNA (ss-Ctrl1) did not affect SIRT1 expression at any concentration. Fully complementary duplex RNA ds-miR-34a(fc) and mismatch-containing duplex ds-miR-34a(mm) inhibited protein expression with IC50 values of 21 ± 0.3 and 30 ± 1 nmol/l, respectively. Single-stranded ss-miR-34a-5p also inhibited with an IC50 value of 30 ± 3 nmol/l, while ss-miR-34a-5p 2'-F(GU) based on the design of Lim et al. possessed an IC50 > 50 nmol/l.
We also examined inhibition of SIRT1 expression as a function of time. We did not observe inhibition by a noncomplementary control ss-Ctrl1 at any time point (Figure 2e). The time course assay showed that SIRT1 inhibition by modified ss-miR-34a-5p or unmodified ds-miR-34a(fc) was similar. Inhibition of SIRT1 expression was evident after one day and was still observable 6 days after transfection, after which expression returned to the basal level. These results suggest that ss-miRNAs can mimic the activity of endogenous miRNAs in repressing gene expression.
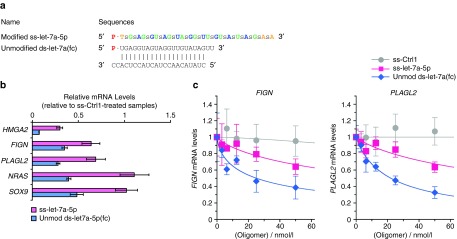
After examining inhibition of SIRT1 by mimics of miR-34a-5p, we analyzed inhibition of a second miRNA, let-7a-5p (Figure 3a). We examined expression of five known let-7a-5p targets using quantitative PCR (qPCR) and observed significant inhibition of HMGA2, FIGN, and PLAGL2 (Figure 3b). Dose response analyses revealed IC50 values for inhibition of FIGN and PLAGL2 by unmodified ds-let-7a(fc) are 25 ± 7 and 22 ± 2 nmol/l, respectively (Figure 3c). Modified ss-let-7-5p also inhibited expression of these genes, but with a lower efficacy, > 50 nmol/l.
Figure 3.
Single-stranded and double-stranded let-7a mimics reduce mRNA levels of let-7a target genes. (a) Sequences and chemical modifications for modified ss-let-7a-5p and unmodified ds-let-7a(fc). (b) Quantitative polymerase chain reaction (qPCR) data showing the effect of modified ss-let-7a-5p and unmodified ds-let-7a(fc) (50 nmol/l) on expression of let-7a target genes (HMGA2, FIGN, PLAGL2, NRAS, SOX9) relative to ss-Ctrl1 treatment. Cells were harvested 2 days after transfection for qPCR analysis. Error bars are mean ± SD. n = 3. (c) Dose response profiles for FIGN and PLAGL2 expression by modified ss-let-7a-5p and unmodified ds-let-7a(fc) mimics. Cells were harvested 2 days after transfection for qPCR analysis. n = 3, Error bars are mean ± SD.
Modified ss- and unmodified ds-miR-34a share targets for gene expression regulation
Unlike fully complementary siRNAs that are used as laboratory tools for knocking down the expression of a single chosen gene, miRNAs generally are not fully complementary to their targets and have the potential to control the expression of multiple genes based on recognition of complementary seed sequences. The phenotypes miRNAs produce, therefore, are derived from individual effects on many genes. It is important, therefore, to determine how single-stranded and double stranded miRNA mimics compare for their ability to knock down expression of a range of potential target genes.
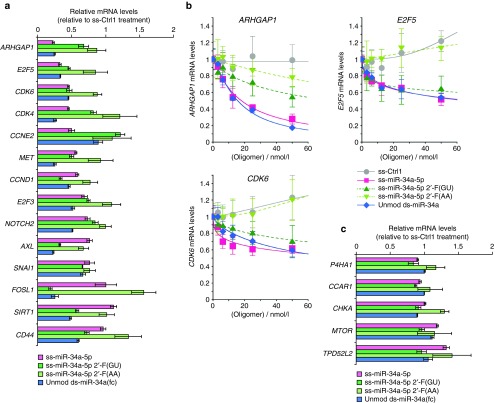
To address this question, we evaluated expression levels of SIRT1 and 19 other miR-34a target genes for modified ss-miR-34a-5p- and unmodified ds-miR-34a(fc)-treated samples by RT-qPCR. These genes had been identified in previous reports of miR-34 activity15 and it is important to note that some target genes have been more thoroughly validated than others and the miRNA activity is dependent on cell context.
We evaluated knockdown relative to effects on gene expression of two noncomplementary ss-miRNA controls (Figure 4a, Supplementary Figure S2). We found that 10 of the genes (ARHGAP1, E2F5, CDK6, CDK4, MET, CCND1, E2F3, NOTCH2, AXL, SNAI1) were inhibited by both modified ss-miR-34a-5p and unmodified ds-miR-34a(fc) (24–77%, Figure 4a). Little or no inhibition was shown for six genes (MYC, CREB1, AXIN2, BIRC5, BCL2, YY1) by either single or duplex mimics in HeLa cells under the conditions used in this assay (Supplementary Figure S2). These results indicate that many of the target genes for the unmodified duplex miR-34a mimic can be targeted by modified ss-miR-34a-5p, suggesting that the rules governing target recognition by single-stranded and double-stranded miRNA mimics are similar.
Figure 4.
Single-stranded and double-stranded miR-34a mimics reduce mRNA levels of miR-34a target genes. (a) Quantitative polymerase chain reaction (qPCR) data showing the effect of modified ss-mR-34a-5p, ss-miR-34a 2'-F(GU), ss-miR-34a 2'-F(AA), and unmodified ds-miR-34a(fc) (50 nmol/l) on expression of miR-34a target genes relative to ss-Ctrl1 treatment. Cells were harvested 2 days after transfection for qPCR analysis. Error bars are mean ± SD. n = 4. Complete qPCR data for 20 potential miR-34a target genes are presented in Supplementary Figure S2. (b) Dose response profiles for ARHGAP1, E2F5, and CDK6 inhibition by ss- or ds-miR-34a mimics. Cells were harvested 2 days after transfection for qPCR analysis. Error bars are mean ± SD. n = 4. (c) qPCR data showing expression of genes that lack target sites for miR-34a after treating HeLa cells with miR-34a mimics (50 nmol/l). Cells were harvested 2 days after transfection for qPCR analysis. Error bars are mean ± SD. n = 4.
We note, however, that mRNA levels for FOSL1, SIRT1, and CD44 were significantly reduced by duplex RNA ds-miR-34a(fc) but not by single-stranded mimic ss-miR-34a-5p, while CCNE2 mRNA level was significantly reduced only by modified ss-miR-34a-5p. These observations suggest that although the targets are mostly shared between the modified ss- and unmodified ds-miRNA mimics, the target preference could be changed depending on the chemical modifications.
Consistent with this suggestion, although we observed significant reduction of Sirt1 protein level for both modified ss-miR-34a-5p- and unmodified ds-miR-34a(fc)-treated samples (Figure 2), we observed significant reduction of SIRT1 mRNA level only for unmodified ds-miR-34a(fc)-treated samples. This finding suggests that the mechanism of gene knockdown may also be affected by the chemical modifications necessary to achieve ss-miRNA activity. This conclusion is supported from data describing inhibition by ss-miRNAs patterned after the designs of Lim and coworkers. In general, both ss-miR-34a-5p 2'-F(AA) and 2'-F(GU) were less active than ss-miR-34a-5p with regards to blocking gene expression (Figure 4a, Supplementary Figure S2). For several gene targets (e.g., CCND1, AXL, FOSL1), however, ss-miR-34a-5p 2'-F(GU) exhibited greater potency than the other miRNA mimics.
We compared dose responses for target genes ARHGAP, E2F5, and CDK6 (Figure 4b). In all cases, the AA variant performed poorly (IC50 value > 50 nmol/l), similar to the noncomplementary control (Figure 4b). By contrast, modified ss-miR-34a-5p and unmodified ds-miR-34a(fc) showed similar activities for ARHGAP1, E2F5, and CDK6 mRNA inhibitions (modified ss-miR-34a-5p: IC50(ARHGAP1) = 18 ± 3 nmol/l; unmodified ds-miR-34a(fc): IC50(ARHGAP1) = 16 ± 1 nmol/l). The GU variant has similar or a little less potency for silencing E2F5 and CDK6. Inhibition of ARHGAP expression by the GU variant was at an intermediate level (IC50(ARHGAP1) = ~75 nmol/l).
As a control, we examined expression of a panel of genes that lack target sequences complementary to the seed sequence of miR-34a-5p (Figure 4c). Expression of these genes was unchanged upon addition of ss-miR-34-5p variants or the duplex miRNA mimic. These data support the conclusion that inhibition is seed-sequence dependent.
The action of ss-miRNA mimic is dependent on AGO2
Argonaute (AGO) is a critical protein factor involved in RNAi.16,17,18 There are four argonaute proteins (AGO1–4) in human cells. AGO1 and AGO2 can be involved in gene regulation by endogenous miRNAs, but functional differences between AGO proteins remain unclear. ss-siRNA/miRNAs contain chemical modifications that are similar to those found within antisense oligonucleotides that do not silence gene expression through RNAi. Because it is plausible that the single strands tested in these studies might function through a mechanism that is independent of RNAi, it was essential to establish the involvement of RNAi factors in gene silencing mediated by ss-miRNAs.
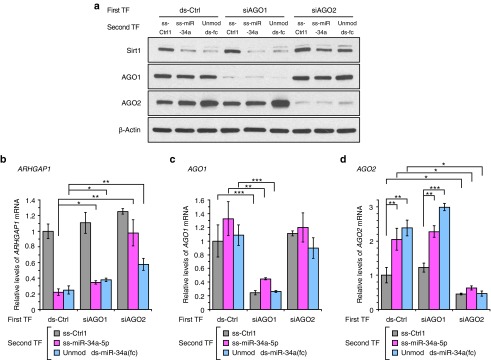
To gain mechanistic insights into action of ss-miRNA mimics, we performed AGO1 and AGO2 knockdown experiments using siRNAs designed to reduce expression of either AGO. In the first transfection, HeLa cells were treated with ds-Ctrl or anti-AGO1 (siAGO1) or anti-AGO2 (siAGO2) siRNA to reduce AGO1 or AGO2 expression. Two days after the first transfection, cells were treated with noncomplementary control ss-Ctrl1, ss-miRNA ss-miR-34a-5p, or unmodified RNA duplex ds-miR-34a(fc). We then analyzed Sirt1, AGO1, and AGO2 protein levels by western blot to investigate how SIRT1 inhibition is affected by knockdown of AGO1 or AGO2.
We observed efficient silencing of AGO1 or AGO2 expression by siRNAs delivered in the initial transfection (Figure 5a,c,d). Reduced AGO1 expression did not affect silencing of SIRT1 by modified ss-miR-34a-5p or unmodified ds-miR-34a(fc) (Fig 5a). By contrast, silencing AGO2 reversed silencing of SIRT1 expression by both the ss-miRNA and duplex miRNA mimics. We also checked ARHGAP1 mRNA levels by RT-qPCR. Inhibition of ARHGAP1 expression by the mimics was also reversed by knockdown of AGO2 relative to knockdown of AGO1 (Figure 5b). These results are consistent with silencing by the ss-miRNA mimic being dependent on expression of AGO2. We also noticed that treatment with miR-34a mimics significantly increase AGO2 expression (Figure 5a,d). Increased AGO2 might cause positive feedback and enhance activity of miR-34a.
Figure 5.
Involvement of AGO2 during inhibition of SIRT1 and ARHGAP1 expression by single-stranded and double-stranded miR-34a mimics. (a) Western blot data showing involvement of AGO2 in SIRT1 inhibition by miR-34a mimics. Data are representative from three independent experiments. (b) qPCR data showing involvement of AGO2 in ARHGAP1 inhibition by miR-34a mimics. (c) qPCR data confirming that AGO1 mRNA levels were reduced after the double transfection with anti-AGO1 siRNA. (d) qPCR data conforming that AGO2 mRNA levels were reduced after the double transfection with anti-AGO2 siRNA. ds-Ctrl, siAGO1, or siAGO2 was transfected into HeLa cells at 25 nmol/l (Day 0). Two days after the first transfection, cell was treated with ss-Ctrl1, modified ss-miR-34a-5p, or unmodified ds-miR-34a(fc) (second transfection, 50 nmol/l) (Day 2) and then harvested for qPCR (Day 4) and western blot (Day 5) analysis. Error bars in panel b–d are mean ± SD. n = 3. *P < 0.05; **P < 0.01; ***P < 0.001 (relative to ds-Ctrl/ss-Ctrl1, unpaired t-test).
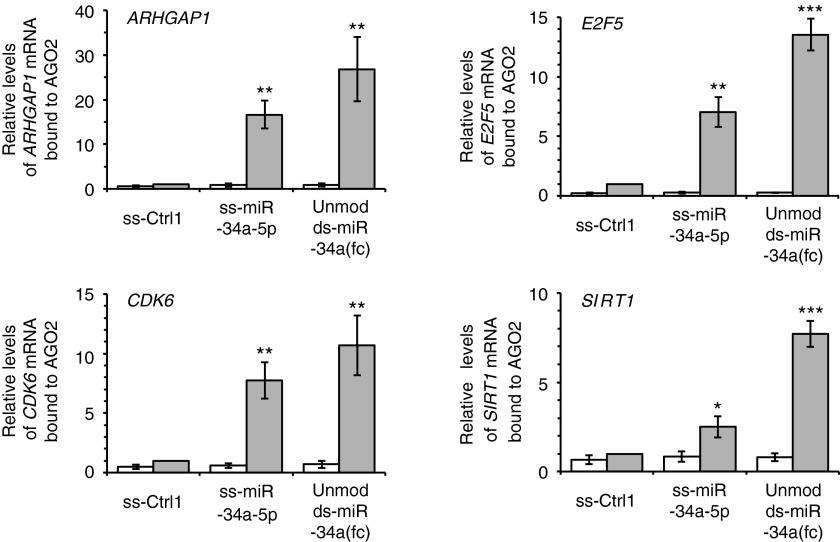
To further test the potential for the involvement of AGO2 in the action of ss-miRNA mimics, we used RNA immunoprecipitation to examine whether AGO2 was recruited to transcripts targeted by miR-34a including ARHGAP1, E2F5, CDK6, and SIRT1 mRNAs (Figure 6). These genes were chosen for analysis because they are among those that can be knocked down by addition of modified ss-miR-34a-5p (Figures 2 and 4). After treatment with duplex miRNA or ss-miRNA mimics, AGO2-associated transcripts were immunoprecipitated using anti-AGO2 antibody and then quantified by RT-qPCR.
Figure 6.
RNA immunoprecipitation (RIP) after treatment with miR-34a mimics reveals recruitment of AGO2. The oligomers were transfected into HeLa cells at 50 nmol/l. Three days after transfection, cells were harvested for AGO2-RIP experiments. Error bars are mean ± SD. n = 4. *P < 0.05; **P < 0.01; ***P < 0.001 (relative to ss-Ctrl1, paired t-test).
Analysis of RNA immunoprecipitation results revealed that AGO2 was recruited to the ARHGAP1, E2F5, CDK6, and SIRT1 mRNA at an enrichment of from 2.5- to 27-fold relative to noncomplementary control ss-miRNA (Figure 6; Supplementary Figure S3). In all cases, enrichment by the duplex RNA mimic was higher than that observed for the ss-miRNA mimic.
Destabilizing effect of ss-miRNA mimics on target mRNAs
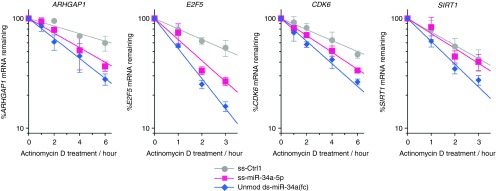
miRNAs inside cells can trigger either or both translational repression or/and mRNA destabilization for gene regulation.19 To investigate how chemically modified ss-miRNA mimics can regulate expression of each target gene, we examined the effects of miRNA mimics on target mRNA stability. After transfection of ss- or ds-miRNA mimics or a control oligomer (ss-Ctrl1) into HeLa cells, cells were treated with actinomycin D to stop transcription. Cells were then harvested at the different time points and mRNA levels of miR-34a targets, ARHGAP1, E2F5, CDK6, and SIRT1, were analyzed by RT-qPCR to reveal decay profiles and half-lives of each transcript.
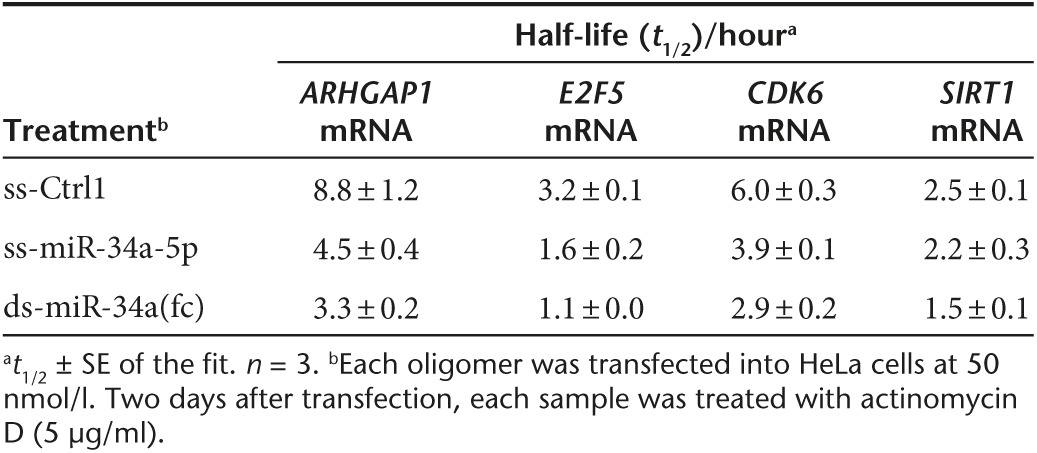
Unmodified ds-miR-34a mimic yielded the greatest reduction on mRNA stability of all of the four genes (Figure 7) and half-lives of the mRNAs were 1.7–2.9-fold shorter relative to those in ss-Ctrl1-treated cells (Table 1). These results are consistent with significant reductions of target transcripts upon unmodified ds-miR-34a(fc) transfection (Figure 3). Modified ss-miR-34a-5p also destabilized ARHGAP1, E2F5, and CDK6 mRNAs but less efficiently than unmodified ds-miR-34a(fc) mimic. Consistent with the lack of reduction of SIRT1 mRNA observed in qPCR analysis (Figure 3a), SIRT1 mRNA stability was not decreased upon treatment with modified ss-miR-34a-5p (Figure 7, Table 1). This finding may be suggesting that modified ss-miR-34a-5p regulates SIRT1 expression through translational repression with little reduction of SIRT1 mRNA rather than mRNA destabilization.
Figure 7.
Effect of miR-34a mimics on stability of ARHGAP1, E2F5, CDK6, and SIRT1 mRNAs. Single-stranded and double-stranded miR-34a mimics were transfected into HeLa cells at 50 nmol/l. Two days after transfection, cells were treated with actinomycin D (5 μg/ml) for 0–6 hours (ARHGAP1, CDK6) or 0–3 hours (E2F5, SIRT1). Cells were harvested at the indicated time points and each mRNA level was measured by RT-qPCR. Error bars are mean ± SD. n = 3.
Table 1. Half-lives of miR-34a target mRNAs after treatment with miR-34a mimics in HeLa cells.
These data suggest that, chemically modified ss-miRNA mimics have the potential to decrease stability of target transcripts like unmodified ds-miRNA mimics. However, the data also reinforce the conclusion that the exact mechanism of gene expression regulation by each miRNA mimic can be altered depending on targets and chemical modifications incorporated into mimics.
Discussion
Oligonucleotide-based therapeutics provides a promising approach for managing unmet medical needs. The use of oligonucleotides began to draw broad attention during the 1970s20,21 with commercial developing beginning in the late 1980's and 1990's. Progress was slow because of the need to develop insights into new fields of scientific investigation involving nucleic acid chemistry, in vivo nucleic acid drug delivery, and mechanisms of action of oligonucleotide drugs. After three decades, steady experimental progress is leading to promising clinical successes. Several oligonucleotide drugs are offering encouraging signs of efficacy in clinical trials and one drug, Kynamro (mipomersen sodium), was approved by the United States Food and Drug Administration in 2013 for systemic administration to patients with familial hypercholesterolemia.22
One of the strengths inherent in nucleic acids as an approach to drug development is that they can act through many different mechanisms. Many clinical trials use antisense oligonucleotides that recognize sequences within mRNA and block translation. The mechanism of these antisense oligonucleotides generally involves inducing cleavage of the target mRNA by RNase H. Other trials employ duplex RNAs that induce cleavage of the mRNA target by RNA-induced silencing complex.
The primary strength of duplex RNAs as a tool for controlling gene expression is their potent gene silencing. The weakness of duplex RNAs for drug development is that they are not as readily delivered into target tissues in vivo as are analogous single-stranded antisense oligonucleotides.5,6 A nucleic acid that combines the favorable delivery properties of single-stranded antisense oligonucleotides with the robustness of function through an RNAi mechanism would be a valuable addition to the options available for controlling gene expression.
Previous studies have shown that fully complementary ss-siRNAs can mimic the action of duplex RNAs and inhibit the expression of target genes in both cell culture and animals.7,8 In animals, gene knockdown was observed both in the liver and throughout the central nervous system. Inhibition by ss-siRNAs generally proceeded through an RNAi mechanism, although in at least one case it was shown that ss-siRNAs could function in parallel as antisense oligonucleotides. This latter result emphasizes the need to always investigate the mechanism of action. One previous report had described the action of single-stranded miRNA mimics consisting of a different pattern of chemical modifications.13 These mimics repressed expression of target genes but mechanistic characterization was limited.
MRX34 (Mirna Therapeutics) is a double-stranded miR-34 mimic that was developed for treatment of primary liver cancer and entered in phase 1 clinical trial to evaluate its safety, tolerability, pharmacokinetic profile, biological activity, and clinical outcomes.15 MRX34 is formulated by liposome for delivery and is intravenously administered in patients. It is possible that an ss-miRNA might have some advantages and provide a useful option for future development.
In this study, we tested the effect of ss-miRNA mimics of miR-34a. We have shown that ss-miRNA mimics of miR-34a reduce expression of SIRT1 (Figure 2) and other target genes (Figure 4). ss-miRNA mimic of let-7a inhibits expression of the let-7a target genes FIGN, PLAGL2, and HMGA2 (Figure 3). This inhibition requires AGO2 expression (Figure 5) and AGO2 is recruited to target transcripts (Figure 6). AGO1 expression does not appear to be required. Similar to the unmodified ds-miR-34a mimic, the modified ss-miR-34a-5p mimic reduces stability of ARHGAP1, E2F5, and CDK6 mRNA (Figure 7). Taken together, these data indicate that the ss-miRNA mimic can function through the RNAi pathway and possess potencies similar to those of analogous duplex RNA mimics.
While the actions of ss-miRNA and duplex miRNA mimics are similar, they are not identical. This conclusion is consistent with previous comparisons of ss-siRNAs and duplex RNAs that were complementary to CAG trinucleotide repeats.11 Our data show that although target genes are mostly common between modified ss- and unmodified ds-miR-34a mimics, the target preference could be changed depending on the chemical modifications (Figure 4). FOSL1, SIRT1, and CD44 mRNA levels were reduced more preferentially by the unmodified ds-miR-34a mimic, while CCNE2 mRNA level was reduced only by the modified ss-miR-34a-5p mimic. Target specificity of miRNAs is primarily determined by seed sequence complementarity to target transcripts, but our results suggest that chemical modification incorporated into miRNA sequences can alter target specificity depending on the local sequences and structures surrounding target sites.
A previous study had examined inhibition of gene expression by single-stranded miRNA mimics but provided only indirect evidence for involvement of RNAi.13 We have tested ss-miRNAs patterned after these previous designs. ss-miR-34a-5p 2'-F(AA) consistently performed poorly. ss-miR-34a-5p 2'-F(GU) was more active. For most genes, ss-miR-34a-5p 2'-F(GU) was less active than ss-miR-34a-5p, but for a few it was more active. This variation reinforces the conclusion that the identity of the chemical modification affects the exact potency and efficacy of inhibition in a target dependent manner.
There are four mammalian AGO proteins (AGO1-4).16 We had previously examined the importance of AGO1-4 for transcriptional silencing by duplex RNAs that target a noncoding transcript overlapping the progesterone promoter and found that AGO2 was the primary AGO variant involved in modulating expression.23 We now examine the relative roles for AGO1 and AGO2 in the action of ss-miRNAs and find, once again, that AGO2 is the primary AGO variant. Why AGO2 appears more critical is not fully defined, but may be partially due to the fact that AGO2 is expressed fivefold highly than AGO1 in HeLa cells (unpublished result). More broadly, the relative roles of AGO1–AGO4 and biological reasons for cells possessing multiple AGO variants remain a general question for the RNAi field. It is also important to realize that non-AGO dependent mechanisms may also contribute to the action of ss-siRNAs in some cases, a result that we have observed previously for inhibition of ataxin-3 by ss-siRNAs that target CAG trinucleotide repeats.10
Our studies suggest several potentially significant insights into the use of ss-miRNA mimics as a therapeutic platform. Our data confirm the earlier conclusion that ss-miRNAs could act as gene-silencing agents13 and substantiate their role as mimics that act through the RNAi pathway. The choice of chemical modification pattern is critical, as is whether the mimic contains one or two strands. Not only do the mimics vary in potency, they also possess different profiles for inhibition of target genes.
In the future, development programs that employ ss-miRNAs are likely to benefit from extensive structure activity analysis to identify compounds that are both potent and modulate a beneficial range of physiologic target genes. For example, in this study, we focused on 2'-O-methyl, 2'-O-methoxyethyl, and 2'-fluoro modifications, with phosphorothioate internucleotide substitutions. Many other substitutions are available for tailoring ss-miRNAs,24 providing wide scope for developing improved agents.
ss-siRNAs have been used to affect expression of several different classes of RNA targets including mRNA coding regions,8 expanded trinucleotide repeats,7,9,10,11 and promoter RNAs.12 Our data show that they can also act as miRNA mimics. In all these studies, activity has been robust and dependent on AGO2 expression, suggest that chemically modified single-stranded RNAs will often be valuable molecular tools and have the potential to be useful starting points for therapeutic development. More broadly, RNAi-active single stranded RNAs continue to be a robust technology for modulating diverse cellular processes and their sensitivity to chemical modifications suggests that more effort developing their basic recognition properties may be rewarded with even higher biological activities.
Materials and Methods
Cell culture and ssRNA/dsRNA transfection. HeLa and HepG2 cells (ATCC, Manassas, VA) were cultured in minimum essential medium eagle (MEM; Sigma-Aldrich, St. Louis, MO) supplemented with 1% MEM nonessential amino acids (Sigma-Aldrich) and 10% FBS (Sigma-Aldrich). A549 cells (ATCC) were cultured in F-12K media containing 10% FBS. ss-miRNA mimics and dsRNAs were reverse transfected into cells at 50 nmol/l (HeLa and A549) or 30 nmol/l (HepG2) using Lipofectamine RNAiMAX (Thermo Fisher Scientific (Invitrogen), Waltham, MA). Three days after transfection, cells were harvested for western blot analysis. For AGO1 or AGO2 knockdown experiments, ds-Ctrl, siAGO1, and siAGO2 were reverse transfected into HeLa cells at 25 nmol/l. Two days after the first transfection, cells were trypsinized, split at the ratio of 1:2 in another six-well plate with ss-/ds-miRNA mimics or a control (ss-Ctrl1) (50 nmol/l). Cells were harvested 2 days after the second transfection for RT-qPCR and 3 days after the second transfection for western blot. Sequences of the ss-miRNAs and ss/dsRNAs are listed in Figures 1a and 2a and Supplementary Table S1.
Western blot Analysis. Three days after transfection, cells were harvested using lysis buffer (50 mmol/l Tris-HCl, 120 mmol/l NaCl, 0.5% NP-40, 1 mmol/l ethylenediaminetetraacetic acid (EDTA) (Thermo Fisher Scientific (Ambion)), 1 mmol/l dithiothreitol (Sigma-Aldrich), and 1× protease inhibitor cocktail set I (EMD Millipore, Billerica, MA)). Protein concentrations in each sample were determined using micro BCA protein assay kit (Thermo Fisher Scientific (Thermo Scientific)). Twenty micrograms of total protein were analyzed by SDS-PAGE (4–20% TGX gels (Bio-Rad, Hercules, CA)). Gels were run at 110 V for 80 minutes. After gel electrophoresis, proteins were transferred to nitrocellulose membrane (Amersham Protran Supported 0.45 μm NC (GE Healthcare Life Science, Pittsburgh, PA) at 100 V for 1 hour. After blocking the membrane with 5% non-fat dry milk (Lab Scientific, Livingston, NJ)/Tris-buffered saline with Tween 20 (50 mmol/l Tris-HCl pH 7.4, 150 mmol/l NaCl, 0.1% Tween 20) at room temperature for 1 hour, the membrane was incubated with primary antibodies at the following dilution ratio: anti-Sirt1 antibody (EMD Millipore; 07-131; 1:5,000), anti-AGO1 antibody (Cell Signaling Technology, Danvers, MA; #5053; 1:2,000), anti-AGO2 antibody (Wako Chemicals USA, Richmond, VA; 015-22411; 1:2,000), anti-β-actin antibody (Sigma-Aldrich; A5441; 1:15,000). Horseradish peroxidase (HRP)-conjugated anti-mouse IgG (#715-035-150) or anti-rabbit IgG (#711-035-152) (Jackson ImmunoResearch, West Grove, PA; 1:1,000–10,000) secondary antibody was used for visualizing proteins using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific (Thermo Scientific)). Protein bands were quantified using ImageJ software (U.S. National Institutes of Health, Bethesda, MD).
Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR). Two days after transfection, cells were harvested using TRI reagent (Sigma-Aldrich) to extract total RNAs. Total RNAs were treated with DNase I (Worthington Biochemical, Lakewood, NJ) at 25 °C for 20 minutes, followed by heating for deactivation at 80 °C for 15 minutes. cDNAs were prepared using High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific (ABI)). qPCR was performed using iTaq Universal SYBR Green Supermix or iTaq Supermix (for TaqMan primers) (Bio-rad) and specific primer sets. 18S ribosomal RNA levels were measured for normalization. The sequences of the qPCR primers are listed in Supplementary Table S2.
RNA Immunoprecipitation. ss- and ds-miRNA mimics were reverse transfected into HeLa cells (3.5 million cells/15-cm dishes) at 50 nmol/l using Lipofectamine RNAiMAX. Three days after transfection, cells were harvested by scraping in Dulbecco's phosphate-buffered saline (PBS) (Sigma-Aldrich) and centrifuged at 500 × g for 10 minutes. The supernatant was removed from each sample and then resuspended in 1 ml of lysis buffer (20 mmol/l Tris-HCl (pH 7.4), 3 mmol/l MgCl2, 150 mmol/l KCl, 0.5% NP-40) containing 1× EDTA-free protease inhibitor cocktail (Roche, Basel) and RNasin Plus RNase inhibitor (Promega, Madison, WI). Each sample was sonicated (20% power, 20 seconds, 1 pulse) and then centrifuged at 13,000 rpm (18,928 × g) for 10 minutes. The supernatant was kept as whole cell lysates and used for the subsequent RNA immunoprecipitation. To 500 μl of lysis buffer, 60 μl of whole cell lysates and 2 μg of anti-AGO2 antibody (Wako) or normal mouse IgG (EMD Millipore) were added and then incubated at 4 °C overnight. After that, 50 μl of Protein G Plus/Protein A Agarose Beads (EMD Millipore) were added to each sample and the samples were incubated at 4 °C for 2 hours. The beads were washed with wash buffer I (50 mmol/l Tris-HCl (pH 7.4), 2 mmol/l MgCl2, 300 mmol/l NaCl, 0.05% NP-40) and wash buffer II (50 mmol/l Tris-HCl (pH 7.4), 2 mmol/l MgCl2, 500 mmol/l NaCl, 0.05% NP-40) twice.
Antibody-protein-DNA/RNA complexes were eluted from the beads with 250 μl of elution buffer (1% SDS, 0.1 mol/l NaHCO3) containing RNasin Plus RNase inhibitor twice (500 μl in total). For input samples, 10 μl of whole cell lysates were mixed with 500 μl of elution buffer. To each sample, 20 μl of 5 mol/l NaCl, 20 μl of 1 mol/l Tris-HCl (pH 7.4), 10 μl of 0.5 mol/l EDTA, and 20 μg of proteinase K (Thermo Fisher Scientific (Ambion)) were added and then the samples were incubated at 42 °C for 50 minutes, followed by phenol extraction to remove proteins. Nucleic acids in each sample were further purified by ethanol precipitation. Each pellet was resuspended in nuclease-free water. Each sample was treated with DNase I for 20 minutes to remove genomic DNA. cDNAs were made through reverse transcription using High Capacity cDNA Reverse Transcription Kit. Samples without reverse transcription were also prepared as negative (no RT) controls. qPCR was performed for each sample using primers specific for SIRT1, ARHGAP1, E2F5, or CDK6 mRNA. qPCR data for each sample were normalized by qPCR data for input. qPCR products were also analyzed by agarose gel electrophoresis to check specificity in amplification (Supplementary Figure S3).
Measuring mRNA stability. ss- and ds-miRNA mimics were transfected into HeLa cells at 50 nmol/l using Lipofectamine RNAiMAX. Two days after transfection, cells were treated with actinomycin D (stock solution: 4 mg/ml in DMSO; final concentration: 5 μg/ml in 2 ml of MEM). At the indicated time points (0, 1, 2, 4, and 6 hours for ARHGAP1 and CDK6 mRNA; 0, 1, 2, and 3 hours for E2F5 and SIRT1 mRNA), cells were harvested using TRI reagent for RT-qPCR analysis. mRNA decay profiles were shown as %mRNA remaining as a function of the treatment time of actinomycin D.
Statistical analysis and curve fitting. Data were analyzed using KaleidaGraph Ver4.1 software (Synergy Software, Reading, PA). Data for dose response were fitted to the following equation (only when inhibition was observed): y = 1−xn/(IC50n+xn), where x is the oligomer concentration and y is the ratio of remaining proteins or transcripts. n (Hill co-efficient) and IC50 are fitting parameters. IC50 values are reported as IC50 ± SE of the fit. Student's two-tailed unpaired (Figure 5) or paired t-test (Figures 2a,b and 6) was performed to evaluate statistical significance relative to ss-Ctrl1 or ds-Ctrl treatment. Data for mRNA decay (Figure 7) were fitted to the following equation: y = 100*exp(-ln(2)/t1/2*t), where y is %mRNA remaining and t is the treatment time of actinomycin D. t1/2 (half-life) is a fitting parameter. Half-lives of transcripts are reported as t1/2 ± SE of the fit. P values less than 0.05 were considered statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Potential target sites for miR-34a-5p on the 3' UTRs of SIRT1, ARHGAP1, E2F5, and CDK6 mRNAs. Figure S2. qPCR data showing the effect of miR-34a mimics on mRNA expression of 20 different miR-34a target genes. Figure S3. Analysis of qPCR products from AGO2-RIP experiments by agarose gel electrophoresis. Table S1. Sequences of duplex RNAs used in this study. Table S2. Sequences of qPCR primers used in this study.
Acknowledgments
This work was supported by the National Institutes of Health (GM106151, GM73042) and the Robert Welch Foundation (I-1244). We thank Roya Kalantari for preliminary planning of this project. We also thank Stanley Crooke and Walt Lima for their contribution to the foundation of this work and their helpful comments on this manuscript. D.R.C. holds the Rusty Kelley Professorship in Medical Science. T.P. is an employee of Ionis Pharmaceutical.
Supplementary Material
References
- Bartel, DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Kim, VN (2005). MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385. [DOI] [PubMed] [Google Scholar]
- Ha, M and Kim, VN (2014). Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524. [DOI] [PubMed] [Google Scholar]
- Jackson, AL, Bartz, SR, Schelter, J, Kobayashi, SV, Burchard, J, Mao, M et al. (2003). Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21: 635–637. [DOI] [PubMed] [Google Scholar]
- Koller, E, Vincent, TM, Chappell, A, De, S, Manoharan, M and Bennett, CF (2011). Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res 39: 4795–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, JK and Corey, DR (2012). Silencing disease genes in the laboratory and the clinic. J Pathol 226: 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D, Pendergraff, H, Liu, J, Kordasiewicz, HB, Cleveland, DW, Swayze, EE et al. (2012). Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell 150: 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, WF, Prakash, TP, Murray, HM, Kinberger, GA, Li, W, Chappell, AE et al. (2012). Single-stranded siRNAs activate RNAi in animals. Cell 150: 883–894. [DOI] [PubMed] [Google Scholar]
- Hu, J, Liu, J, Yu, D, Aiba, Y, Lee, S, Pendergraff, H et al. (2014). Exploring the effect of sequence length and composition on allele-selective inhibition of human huntingtin expression by single-stranded silencing RNAs. Nucleic Acid Ther 24: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J, Yu, D, Aiba, Y, Pendergraff, H, Swayze, EE, Lima, WF et al. (2013). ss-siRNAs allele selectively inhibit ataxin-3 expression: multiple mechanisms for an alternative gene silencing strategy. Nucleic Acids Res 41: 9570–9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J, Liu, J, Narayanannair, KJ, Lackey, JG, Kuchimanchi, S, Rajeev, KG et al. (2014). Allele-selective inhibition of mutant atrophin-1 expression by duplex and single-stranded RNAs. Biochemistry 53: 4510–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, M, Prakash, TP and Corey, DR (2013). Transcriptional silencing by single-stranded RNAs targeting a noncoding RNA that overlaps a gene promoter. ACS Chem Biol 8: 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorn, G, Klein-McDowell, M, Zhao, L, Saunders, MA, Flanagan, WM, Willingham, AT et al. (2012). Single-stranded microRNA mimics. RNA 18: 1796–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi, M, Ferlito, M and Lowenstein, CJ (2008). miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA 105: 13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini, M and Knight, RA (2014). miR-34: from bench to bedside. Oncotarget 5: 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J, Carmell, MA, Rivas, FV, Marsden, CG, Thomson, JM, Song, JJ et al. (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- Meister, G, Landthaler, M, Patkaniowska, A, Dorsett, Y, Teng, G and Tuschl, T (2004). Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197. [DOI] [PubMed] [Google Scholar]
- Rand, TA, Ginalski, K, Grishin, NV and Wang, X (2004). Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc Natl Acad Sci USA 101: 14385–14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger, E and Izaurralde, E (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110. [DOI] [PubMed] [Google Scholar]
- Zamecnik, PC and Stephenson, ML (1978). Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci USA 75: 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson, ML and Zamecnik, PC (1978). Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc Natl Acad Sci USA 75: 285–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouni-Berthold, I and Berthold, HK (2015). Mipomersen and lomitapide: Two new drugs for the treatment of homozygous familial hypercholesterolemia. Atheroscler Suppl 18: 28–34. [DOI] [PubMed] [Google Scholar]
- Chu, Y, Yue, X, Younger, ST, Janowski, BA and Corey, DR (2010). Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res 38: 7736–7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleavey, GF and Damha, MJ (2012). Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol 19: 937–954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.