Abstract
Nanochannel electroporation (NEP) was applied to deliver precise dosages of myeloid cell leukemia-1 (Mcl-1)-specific siRNA and molecular beacons to two types of acute myeloid leukemia (AML) cells, FMS-like tyrosine kinase-3 wild-type (WT) and internal tandem duplications (ITD) type at the single-cell level. NEP, together with single-cell quantitative reverse transcription PCR, led to an observation showing nearly 20-folds more Mcl-1 siRNA than MCL1 mRNA were required to induce cell death for both cell lines and patient blasts, i.e., ~8,800 siRNAs for ~500 ± 50 mRNAs in ITD cells and ~6,000 siRNAs for ~300 ± 50 mRNAs in WT cells. A time-lapse study revealed that >75% MCL1 mRNA was downregulated within 1 hour after delivery of a small amount of siRNA. However, additional siRNA was required to inhibit the newly transcribed mRNA for >12 hours until the cell lost its ability of self-protection recovery. A multidelivery strategy of low doses and short delivery interval, which require 77% less siRNA and has the potential of lower side effects and clinical cost, was as effective as a single high-dose siRNA delivery. Our method provides a viable analytical tool to investigate gene silencing at the single-cell level for oligonucleotide-based therapy.
Introduction
Acute myeloid leukemia (AML) is one of the most common acute leukemia. Currently, 60% of the patients achieve complete remission; however, the majority of them experience relapse and die.1,2,3 Therefore, novel therapeutic strategies are needed.
The elevated expression of the B-cell lymphoma 2 (BCL2) proto-oncogene contributes to apoptotic-resistance.4,5,6,7A BCL2 family member, myeloid cell leukemia-1 (MCL1), serves the role of apoptosis delay in hematopoietic cells. MCL1 was found overexpressed in AML cells compared to normal bone marrow cells.8,9 Emerging evidence supports that aberrant activation of a receptor tyrosine kinase, FMS-like tyrosine kinase-3 (FLT3) may lead to MCL1 upregulation in AML.10,11 The FLT3-internal tandem duplication (ITD) is a genetic aberration leading to constitutive kinase activation of FLT3, which occurs in 20–30% of AML patients and is associated with poor prognosis.12,13Aberrant kinase activation in this molecular subset of AML patients may increase MCL1 and partly explain the dismal clinical outcome. Thus, we hypothesized that specific knockdown of MCL1 by RNA-induced silencing (RNA interference, RNAi) could be a viable treatment strategy for AML patients, including those harboring a FLT3 activating mutation.
Short interfering RNA (siRNA) has been widely used to silence its targets through complementary-binding to their target mRNAs.14,15,16,17,18,19,20 The conventional delivery methods such as bulk electroporation and nanocarriers (e.g., lipoplex and polyplex) are widely applied; however, they cannot provide uniform delivery and are stochastic in nature.21,22,23,24,25 The variation in delivery to each cell may cause different downstream consequences, making it difficult to understand the fundamentals of cell response to different drug dosages. The authors have recently developed a nanochannel electroporation (NEP) method capable of single-cell-based DNA/RNA delivery with excellent dosage control and minimal cell damage.25,26 The empirical data demonstrated by Gao et al.25 showed that the dosage in single cells delivered by bulk electroporation varied greatly while NEP provided more uniform dosage and a sharp distribution.
Herein, we demonstrated for the first time the time-lapse effect of Mcl-1 siRNA on MCL1 mRNA and cell apoptosis in AML cells at the single-cell level by integration of NEP with single-cell qRT-PCR27 and fluorescence resonance energy transfer-based molecular beacons (MBs) detection.28,29,30,31 We also demonstrated at a single-cell level that AML cells harboring FLT3-ITD (i.e., MV4-11 cells) required more Mcl-1 siRNA to achieve cell death than those with WT (i.e., KG1a cells).
Results
Controlled delivery of Mcl-1 siRNA to AML cells
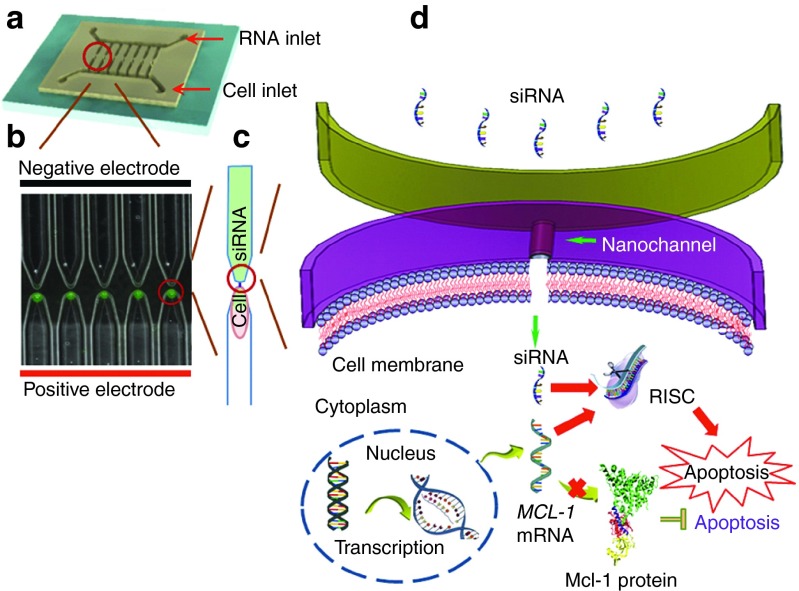
Microchannel-nanochannel-microchannel array based NEP biochips were fabricated by a soft lithography-based DNA combing and imprinting method.25 After secondary machining with femtosecond laser, the biochip was sealed and packaged as an integrated device incorporated with microfluidic channels inside for cell and gene/biomolecular loading.25 bulk electroporation was widely used as a conventional method but it could not provide dosage control. In NEP, precise drug/gene delivery is achieved by a focused electric field through a nanochannel juxtaposed to a single cell. The focused electric field nanoporates the cell and provides a high electrophoretic mobility to move charged molecules into the cell during poration. The delivered dosage can be controlled by the electric voltage, pulse length, and number of pulses. The nanochannel also serves as a diffusion barrier to prevent any further delivery after poration, and this achieves precise dosage control at the single-cell level.
There are 300 microchannels on each side of the array, with a nanochannel between each pair of microchannels. Individual cells were loaded to one side of the microchannels array, while Mcl-1 siRNA was loaded to the other side of the microchannels array (Figure 1a). The Mcl-1 siRNA delivered into cells by electroporation could complementarily bind to MCL1 mRNA and thus block its translation. As a result, a reduced level of MCL1 mRNA would contribute to the decrease of the encoded antiapoptotic protein (Figure 1b–d). Although many conventional methods can deliver Mcl-1 siRNA into cells and downregulate MCL1 mRNA, the dosage is difficult to control, which makes it impossible to investigate its mechanism and develop dosage-dependent therapy. Our NEP technology, on the other hand, demonstrated excellent dosage control and can be used for studying the dynamics of cell apoptosis process.
Figure 1.
Precise delivery of Mcl-1 siRNA to acute myeloid leukemia cells by nanochannel electroporation (NEP) device for downregulation of MCL1 mRNA and inducing apoptosis. (a) NEP array with single cell in each channel. (b) Schematic diagram of Mcl-1 siRNA function in cell.
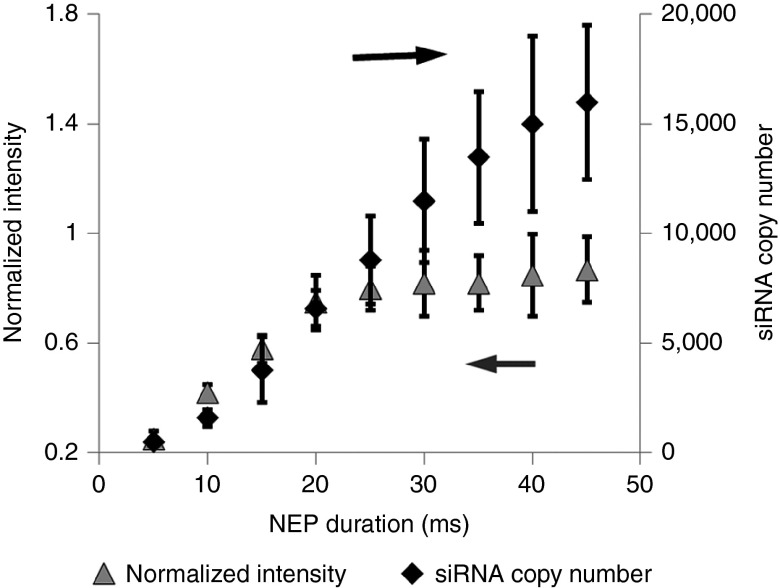
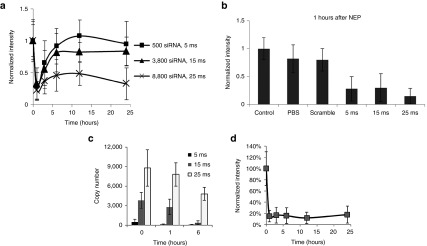
The delivered doses of Mcl-1 siRNA were quantified by both single-cell quantitative reverse transcription PCR (qRT-PCR) and fluorescent intensity, emitted from FAM-labeled Mcl-1 siRNA at different NEP pulse durations in KG1a cell (Figure 2). Both detection methods revealed a nearly linear trend for the pulse duration less than 25 ms when the delivered Mcl-1 siRNA copy number was less than 9,000. When the pulse duration of NEP was increased to 25 ms or higher, the fluorescence intensity showed a nonlinear increase because of fluorescence signal saturation within a single cell, while qRT-PCR detection still showed a linear increase. Although qRT-PCR is more sensitive at higher dosages, 25 ms pulse duration is enough to cause Mcl-1 siRNA-induced cell death for both cell lines and patient blasts, thus the simpler MB-based fluorescence measurement by NEP is a viable method for intracellular RNA detection. The approximate copy number can be determined by single-cell qRT-PCR. The distributions of the Mcl-1 mRNA copy number in KG1a and MV4-11 are shown in Supplementary Figure S1. Among individual cells, the distributions of the MCL1 mRNA levels are centered in each cell line and the copy numbers are significantly different between two cell lines (P < 0.001). There are around 300 ± 50 copies of MCL1 mRNA in each KG1a cell and around 500 ± 50 copies in each MV4-11 cell.
Figure 2.
Delivery of Mcl-1 siRNA calibrated by single-cell qRT-PCR and fluorescence intensity detection.
Threshold of Mcl-1 siRNA-induced apoptotic cell death
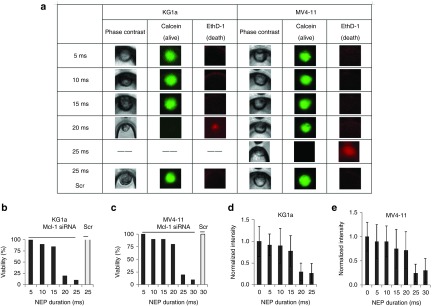
Different doses of Mcl-1 siRNA were delivered by NEP to KG1a and MV4-11 cells. After 24 hours, we measured the residual MCL1 mRNA by delivering MBs into some Mcl-1 siRNA-transfected cells using NEP. Cell viability was also examined for the transfected cells by using the Calcein-EthD-1 kit. Figure 3a shows that most KG1a cells remained alive at 15 ms NEP pulse duration time or ~3,800 Mcl-1 siRNA copies. Cell death was observed at 20 ms pulse duration time or >6,000 Mcl-1 siRNA copies. On the other hand, in MV4-11 cells, cell death was not observed till the NEP pulse duration time was increased to 25 ms or ~8,800 copies of Mcl-1 siRNA. Scramble siRNA was also delivered at 25 ms duration time for KG1a cells and 30 ms duration time for MV4-11 cells. Scramble-treated cells remained alive in both cell lines, indicating that the observed cell death was caused by the delivered Mcl-1 siRNA, not the electroporation process. Figure 3b and Figure 3c show the cell death rate at different dosages of Mcl-1 siRNA. Cell viability means the percentage of living cells among the total cells. For each cell viability test, we transfected cells several times with multiple cells each time. Nearly 80% KG1a cells (n = 10) were killed 24 hours after NEP of 20 ms (~6,000 copies) Mcl-1 siRNA delivery to each cell, while only 20% of MV4-11 cells (n = 10) were killed at the same Mcl-1 siRNA dosage. More Mcl-1 siRNA is needed to induce the apoptosis of FLT3-ITD AML cells than that in the WT cells.
Figure 3.
Comparison of critical threshold of Mcl-1 siRNA and downregulation of mRNA. (a) Live and dead kit detection. (b,c) Cell viability of KG1a and MV4-11 with Mcl-1 siRNA and scramble (Scr) siRNA (n = 5~10 for each cell line). (d,e) mRNA in KG1a and MV4-11 (n = 5~10 for each cell line), 24 hours after siRNA delivery.
With the increase of Mcl-1 siRNA dosage by prolonging the NEP pulse length, the MCL1 mRNA levels decreased in both cell lines measured by MB-based fluorescence intensity. As shown in Figure 3d and Figure 3e, ~80% MCL1 mRNA decrease (P < 0.05, compared with mRNA level in scramble-treated cells) was observed at 20 ms (~6,000 copies) Mcl-1 siRNA delivery in KG1a cells 24 hours after transfection, while the MCL1 mRNA level decreased to the same level at 25 ms (>8,000 copies) Mcl-1 siRNA delivery in MV4-11 cells (P < 0.01).
Validation in patient blasts
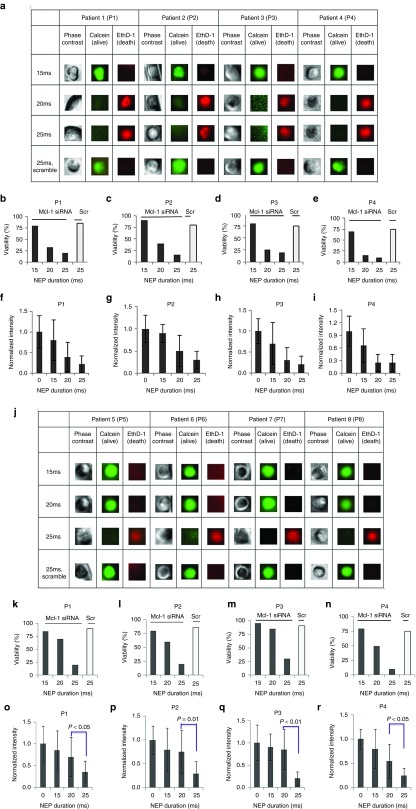
Next, we validated our observation in primary blasts from eight patients (four in WT FLT3 group and four in FLT3-ITD group). Different doses of Mcl-1 siRNA were delivered into patient blast cells (n = 15~20 for each patient sample) by NEP, then the cell viability and MCL1 mRNA downregulation were assessed. Consistent with the cell line results, approximately 75% of the cells from patients 1–4 with WT FLT3 are dead 24 hours after Mcl-1 siRNA delivery by NEP with 20 ms pulse duration (Figure 4a–e), whereas blast cells with FLT3-ITD from patient 5–8 required a higher dose of siRNA (Figure 4j–n). When 25 ms NEP was applied to the blasts from patient 5–8, the delivery of approximately 9,000 copies of Mcl-1 siRNA can induce more than 50% cell death. The corresponding MCL1 mRNA levels were also measured at different siRNA doses. Lower siRNA doses (e.g., 20 ms NEP pulse) were needed to reduce >50% MCL1 mRNA levels in the patient sample 1–4 (Figure 4f–i), while higher siRNA doses (e.g., 25 ms NEP pulse) were required to reduce >50% MCL1 mRNA in the patient sample 5–8 (Figure 4o–r).
Figure 4.
Comparison of critical threshold of Mcl-1 siRNA and downregulation of mRNA in patient blasts. Patient 1 to 4 had WT FLT3 and patient 5 to 8 harbored FLT3- internal tandem duplications (ITD). (a) Live and dead kit detection of wild-type acute myeloid leukemia (AML) patient blasts 1–4. (b–e) Cell viability of blast cells from patient 1–4 with Mcl-1 siRNA and scramble (Scr) siRNA (n = 10~15 for each patient sample). (f–i). MCL1 mRNA level in blast cells from patient 1–4 (n = 5~10 for each patient sample). (j). Live and dead kit detection of harbored FLT3-ITD AML patient blasts 5–8. (k–n) Cell viability of blast cells from patient 5–8 with Mcl-1 siRNA and scramble (Scr) siRNA (n = 10~15 for each patient sample). (o–r) MCL1 mRNA level in blast cells from patient 5–8 (n = 5~10 for each patient sample). Twenty-four hours after siRNA delivery.
Time dependence of intracellular MCL1 mRNA and Mcl-1 siRNA changes after siRNA delivery
To further investigate the activity of Mcl-1 siRNA, we monitored theMCL1 mRNA level change over time in KG1a cells at different Mcl-1 siRNA dosages. Three different Mcl-1 siRNA doses, ~500, ~3,800 copies, and ~8,800 copies (corresponding to 5, 15, and 25 ms NEP pulse duration) were used. Figure 5a shows a rapid and significant MCL1mRNA decrease at 1 hour after Mcl-1 siRNA delivery at all dosages. This means that Mcl-1 siRNA delivered at a level similar to that of the MCL1 mRNA in the cell (i.e., ~500 versus ~300 copies) was sufficient to silence MCL1 mRNA. In contrast, little changes of the MCL1 mRNA level were observed in scramble siRNA and phosphate-buffered saline (PBS)-treated controls (Figure 5b).
Figure 5.
Single-cell detection of time-dependent changes of MCL1 mRNA and Mcl-1 siRNA level at different siRNA dosages. (a) Time-dependent changes of MCL1 mRNA downregulation and self-protection recovery. (b) Comparison of control (without nanochannel electroporation (NEP)), phosphate-buffered saline, scramble siRNA and Mcl-1 siRNA with different dosages on mRNA downregulation. (c) Time-lapse change of Mcl-1 siRNA level at different siRNA dosages. (d) MCL1 mRNA level change with single NEP of low dosage (5 ms) after pretreatment of Act D.
The siRNA-dependent MCL1 mRNA decrease, however, did not lead to cell death. At a low Mcl-1 siRNA dosage (~500 copies), we observed a rapid MCL1 mRNA recovery. At 3 hours after siRNA transfection, the MCL1 mRNA level returned to >60% of its original level and nearly back to the normal level at 6 hours. Even with a higher siRNA dosage (~3,800 copies), we still observed significant MCL1 mRNA recovery after the initial fast decrease. Even at a very high siRNA dosage (~8,800), the MCL1 mRNA recovery was weak but still observable. Interestingly, the MCL1 mRNA level decreased again at this Mcl-1 siRNA dosage when the cells reached apoptotic death (between 12 and 24 hours as determined by the Calcein-EthD-1 kit, data not shown). To better understand the influence of Mcl-1 siRNA on MCL1 mRNA downregulation, we also measured the intracellular Mcl-1 siRNA residue at all three dosages by qRT-PCR 1 and 6 hours after Mcl-1 siRNA delivery. Figure 5 shows that the siRNA residue dropped to less than 100 copies at 1 hour and was undetectable at 6 hours when the delivered dosage was low, i.e., around 500 copies (Figure 5c). While the delivered Mcl-1 siRNA dosage was ~3,800 copies, there were >2,000 copies at 1 hour and <200 copies at 6 hours (Figure 5c). For the high-dosage delivery (~8,800 copies), there were plenty of Mcl-1 siRNA residues at both 1 and 6 hours (Figure 5c). The decrease of Mcl-1 siRNA residue at 0, 1, and 6 hours was very similar to that at the ~3,800 and ~8,800 dose levels. In addition, we pretreated KG1a cells with a commonly used transcription inhibitor, actinomycin D (Act D) at 20 nmol/l for 4 hours, and then conducted the single NEP with low-dose (5 ms) treatment. Act D, per se, at 20 nmol/l, did not cause significant downregulation of MCL1 mRNA level at 4 and 15 hours, which indicated that downregulation of MCL1 mRNA after NEP was caused by Mcl-1 siRNA at 1, 3, 6, and 12 hours. Act D treatment by itself downregulated MCL1 mRNA at 27 hours, which indicated the activity of the drug. We observed no recovery of MCL1 mRNA with the low dose of Mcl-1 siRNA delivery after pretreatment of Act D (Figure 5d).
Study Mcl-1 siRNA therapy strategy
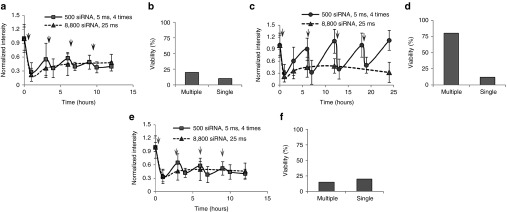
The dynamics of Mcl-1 siRNA and mRNA levels in single-cell studies suggested two siRNA therapy strategies, one with high single dose and the other one with multiple low doses. Here, we compared the efficacy of each strategy in KG1a cell. We chose 25 ms NEP single pulse with 8,800 copies of siRNA as the first strategy. Because NEP pulse of 5 ms with 500 copies of Mcl-1 siRNA can effectively down regulate MCL-1 mRNA after 1 hour (Figure 5b), multiple siRNA injections of 5 ms NEP pulse were used as the second strategy. Moreover, Figure 5a shows that MCL1 mRNA began to restore 3 hours after delivery. We preferred a longer time interval to minimize the dosage used and cell damage. However, too long interval would lead to strong MCL1 mRNA recovery in cells. We tested different time periods and found that 3 hours was a good choice. This also agrees with Figure 5 that significant mRNA recovery would occur after 3 hours. Thus, we chose 3 hours as the interval between two injections in the second strategy for continuous downregulation of MCL1 mRNA. Figure 6a shows that the MCL1 mRNA level remained approximately 50% reduced in both cases although a slightly mRNA recovery was observed after each injection in the multiple delivery strategy. Cell death was observed after 12 hours in both cases (Figure 6b). However, in the single high-dose strategy, we injected approximately 8,800 copies of siRNA, while in the multiple low-dose strategy we only injected a total of approximately 2,000 copies. Based on our previous result, 15 ms single NEP pulse which delivered 3,800 copies of Mcl-1 siRNA was not enough to cause KG1a cell death (Figure 3). Here four injections with 500 copies of Mcl-1 siRNA each time (total 2,000 copies of siRNA) can cause the cell death. Thus, the multiple low-dose strategy has higher efficacy than the single high-dose strategy.
Figure 6.
Study on oligonucleotide-based therapy strategy. Arrows represent the nanochannel electroporation (NEP) delivery in multiple delivery strategy. (a) MCL1 mRNA level change with either optimized multiple NEP strategy of low dosage (500 copies) and short interval (3 hours) treatment or single NEP of high-dosage (8,800 copies) treatment. (b) Cell viability with either multiple NEP of low dosage (500 copies) and short interval (3 hours) treatment or single NEP of high-dosage (8,800 copies) treatment. (c) MCL1 mRNA level change with either multiple NEP strategy of low dosage (500 copies) and long interval (6 hours) treatment or single NEP of high-dosage (8,800 copies) treatment. (d) Cell viability with either multiple NEP of low dosage (500 copies) and long interval (6 hours) treatment or single NEP of high-dosage (8,800 copies) treatment. (e) Validation of multiple NEP with long dosage and short interval in patient blasts. (f) Cell viability of patient blasts with respective treatment.
Furthermore, we hypothesized that the key to the successful siRNA therapy was to continuously inhibit the level of MCL1 mRNA to a certain level, e.g., >50% decrease. We conducted another experiment with the same NEP pulse durations and the same number of delivery times. The only variation was the prolonged pulse interval, as 6 hours. Figure 6c showed that the longer intervals provide enough time for MCL1 mRNA to fully recover. As expected, the multiple low doses with the longer interval strategy could not cause cell death (Figure 6d). The same trend was observed in patient blasts, that only the multiple low doses with a short time interval could be as effective as a single high dose with 22% less total required siRNA (Figure 6e,f).
Discussion
In this study, we first demonstrated an integrated single-cell transfection and analysis system by combining a newly developed NEP technology, MB-based fluorescence measurements, and single-cell qRT-PCR. NEP allows precise transfection of single cell or a small number of cells (e.g., <100 cells) with minimal cell damage, not achievable by any other existing methods. It will be helpful to stem cell, cancer stem cell, or circulation tumor cell research in the future since they all have a very limited number of cells. Microinjection is the only current method capable of single-cell transfection. However, it often causes severe cellular damage for small cells. Our platform does not require any pretreatment of the cell and NEP can be performed with a regular culture media. Thus, our platform is more benign to fragile cells such as primary AML blasts. It can also deliver molecular probes such as MBs into the transfected cells without any cell fixation or labeling. However, MB-based fluorescence measurements are qualitative. Single-cell qRT-PCR is an established quantitative method, but the assay is more expensive and time-consuming. By calibrating MB-based fluorescence measurements using single-cell qRT-PCR, NEP can be a powerful analytical tool for cell biology and drug delivery studies.
We used this system to investigate the effect of Mcl-1 siRNA on MCL1 mRNA silencing and inducing apoptotic cell death for a wild-type FLT3 AML cell line, KG1a and a FLT3-ITD AML cell line, MV4-11. Our system was able to identify the threshold Mcl-1 siRNA levels for both cell types. The results showed that although the MCL1 mRNA copy number in MV4-11 was only 200 more than that in KG1a, more than 2,000 extra Mcl-1 siRNAs were required to kill MV4-11 cells. We also validated this observation in primary patient blasts. This partially explains why patients with FLT3-ITD have stronger chemotherapy resistance. It also indicates that Mcl-1 downregulation by siRNA may enhance chemotherapy in FLT3-ITD patients.
Following the time-dependent changes of both MCL1 mRNA and Mcl-1 siRNA in single cells, our system was able to reveal the intracytoplasmic dynamics of MCL1 mRNA downregulation and self-protection recovery. Delivery of Mcl-1 siRNA, even at a low dosage, was able to quickly silence most of the target mRNA, however, self-protection recovery of cells allowed the restoration of the target mRNA. It requires a great deal of Mcl-1 siRNA residue or repeated and timely Mcl-1 siRNA delivery to keep the intracellular MCL1 mRNA lower than a certain level for a certain time to eventually induce cell apoptosis. Successful siRNA based therapies require long-term inhibition of the expression of certain proapoptotic genes to overcome cell's self-protection recovery.
Based on above-mentioned observation, we studied two siRNA therapy strategies: multiple low doses with short delivery interval versus single high dose. The former strategy can cause cell death as effective as the latter one. However the total siRNA required by multiple low-dose strategy is much lower, which not only lowers the clinical cost but also reduced the side effect of large amount of free siRNA. The reason why the single high-dose strategy is not as good as the multiple low-dose strategy may be because the delivered extra free siRNA are degraded in the cell.
The information revealed by this study provides insight into cancer cell biology and can help the design of siRNA based therapeutic strategy. Next step would be to treat patient samples with lipoplex-formulated siRNA with a selection of concentrations and exposure times empirically and measure the outcome of patient samples such as apoptosis and at the same time to measure the delivered concentration in cells. The intracellular level of delivered siRNA by lipoplex and outcomes will be compared to our NEP results and then the concentration and exposure time will be optimized based on our NEP results—enough dosage for a minimal time period.
Our current 2D NEP method can transfect several hundreds of cells to reveal mechanisms of cancer cell apoptosis. For other NEP application in cancer biology research, which requires large amount of cells, a high-throughput 3D NEP design/fabrication/testing is underway in our laboratory to transfect millions of cells simultaneously for potential cell-engineered clinical therapy.
Materials and Methods
NEP device fabrication. The micro-ridge patterned stamps were made up of SU-8 photoresist (MicroChem, Westborough, MA) spin-coated on silicon wafers. Stamps were cast with Sylgard 184 poly(dimethyl siloxane) (PDMS, Sigma-Aldrich, St. Louis, MO) that was mixed in a resin to initiator ratio of 10 to 1. The nanowire combing was carried out with 0.5 wt% calf thymus DNA (75 kbp, USB Co., Santa Clara, CA) in 5% wt% NaCl solution to produce an array of stretched DNA nanowires on the micro-ridge patterned PDMS stamp.
The stamp with DNA nanowires was then coated with gold by E-Beam Evaporator (Denton DV-502A, Denton Vacuum LLC, Moorestown, NJ). Gold is used because it is easy to etch away after the imprinting process and can be used to control the nanowire/nanochannel diameter. Prepolymer (99 wt% 1,4-butanediol diacrylate and 1 wt% Irgacure 651 initiator, Sigma-Aldrich, St. Louis, MO) was used as an imprinting resin and cured using ultraviolet light (wavelength, 365 nm; intensity, 30 mW 16 cm) for 3 minutes under nitrogen. The stamp was then peeled off the slide, leaving behind the array of microchannels connected by gold-coated DNA nanowires. To remove gold-coated DNA nanowires, the slide was soaked in gold etchant (GE8111, Transene Company, Danvers, MA) for 48 hours and then thoroughly rinsed with DI water, leaving behind embedded nanochannels connecting the microchannels. The chip was treated with plasma (10% oxygen, 300 mw, 1 minute) and soaked in Piranha solution (70% H2SO4/30% H2O2, 50:50) for 3 hours to make it hydrophilic. The chip was rinsed with DI water three times and soaked in 0.1% bovine serum albumin containing DI water for 8 hours to reduce the adhesion between the loaded cells and the substrate.
Cell culture. KG1a cells were cultured in 25 T flasks containing 5 ml of Roswell Park Memorial Institute culture medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS16000, Invitrogen). MV4-11 cells were cultured in 25 T flasks containing 5 ml of Roswell Park Memorial Institute culture medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. Cells were seeded into T flasks at a concentration of 3 × 105 viable cells per ml, incubated at 37 °C in a humidified atmosphere containing 5% CO2, and subcultured every 2 days. AML patient blasts were obtained from the Ohio State University Leukemia Tissue Bank. The primary blasts were grown in Roswell Park Memorial Institute 1640 media supplemented with 20% of fetal bovine serum, in the presence of FLT3, Stem Cell Factor, IL-3, and IL-6 (StemSpan CC100, Stemcell Technologies, Canada). All patients provided written informed consent in accordance with the Declaration of Helsinki under an Institutional Review Board approved protocol for discovery studies according to OSU institutional guidelines for tissue collection and the use of the tissue in research.
NEP delivery of Mcl-1 siRNA. Suspension cells were centrifuged at 1,000×g for 3 minutes to remove the culture medium and washed with DPBS buffer for two times to remove residual medium components. The final deposit was resuspended in PBS for 5 minutes. 200–300 cells in 5 µl suspension were placed into the inlet reservoir on the biochip through a microfluidic channel by a pipette. The device was then sealed on top with soft silicone gel to prevent any possible leakage from the inlet, and then the cells were manipulated using the spinning method (1,000×g, 3 minutes) to move them into individual microchannels and close to the nanochannel inlets. Mcl-1 siRNA was placed in the other inlet reservoir at a concentration of 100 nmol/l. In this study, we focused on precise delivery at the single-cell level to figure out the threshold and dynamics of induced apoptosis. Considering that the variation of concentration might result in different copy numbers of siRNA delivered into cells, the same siRNA concentration and NEP voltage were used in the whole study. An electronic pulser from a Bio-RAD Gene Pulser Xcell electroporation system was used to provide the required voltage pulse sequences (250 V, different pulse durations). Palladium wires (0.25 mm in diameter, Invitrogen/Molecular Probes) were used as electrodes and were connected to an electronic pulser. After poration, unused Mcl-1 siRNA was removed and the reservoir was washed with PBS for ten times.
Bulk electroporation delivery of Mcl-1 siRNA. Cells were centrifuged at 1,000×g for 3 minutes to remove the culture medium and resuspended in PBS for 5 minutes. Around 2 × 105 cells in 100 μl electroporation buffer was prepared and then mixed with 10% Mcl-1 siRNA at the final concentration of 100 nmol/l. The mixture was sucked into the electroporation tip and the electroporation pipette was placed into the stage for electroporation with a 20 ms pulse under 1,400 V. The mixture was then transferred to a six-well cell culture plate and incubated at 37 °C in a humidified atmosphere containing 5% CO2.
Cell viability assay. LIVE/DEAD Viability/Cytotoxicity Kit (Invitrogen) was applied to check cell viability following the protocol.
MB detection for MCL1 mRNA. MCL-1 Molecular Beacon: 5′-FAM-CCT AGC TTG GCT TTG TGT CCT TGG CGG CTA GG-BHQ2a-Q-3′.
MCL-1-specific siRNA was purchased from Ambion (sense: 5′-GGUCCAUGU UUUCAAAGAUTT-3′; anti-sense: 5′AUCUUUGAAAACAUGGA CCAT-3′).
Commercial scrambled negative siRNA was purchased from Invitrogen in USA. PCR primers are commercial products purchased from Taqman in USA, and the product ID is 4440878.
The fluorescence microscope was purchased from Nikon. It is a Nikon Eclipse TiE inverted epi-fluorescent microscope with a Photometrics Evolve EMCCD camera. NIS-Elements AR software was used for acquisition and analysis. The 60 and 100× are Plan Apo oil objectives with 1.4 NA, while the other objectives are dry Plan Fluor objectives.
For results shown in Figures 3 and 4, fresh media was added into the reservoir and the microfluidic channel after NEP with Mcl-1 siRNA, and the whole device was incubated at 37 °C for 24 hours. The device was then taken out for the secondary NEP with Mcl-1 MB (Sigma). For results shown in Figures 5 and 6, Mcl-1 MB was delivered by NEP at 500 nmol/l after 1, 3, 6, 12, and 24 hours. Fifteen minutes after the injection of MB, the device was placed under a microscope for fluorescence detection. Actinomycin D (Act D) was applied to block transcription. Act D was added to cell culture at a concentration of 20 nmol/l and incubated for 4 hours at 37 °C in a humidified atmosphere containing 5% CO2. NEP was then carried out to check the inhibition effect on transcription. The intensity of fluorescence signal was normalized to scrambled negative control siRNA.
Cell lysate for single-cell PCR. Cells in the biochips were trypsinized for 5 minutes after NEP and then transferred to cell culture dish. Individual cells were picked with a micro capillary under a dissection microscope and then washed with 0.1% bovine serum albumin in PBS twice. Subsequently, cells were individually picked and introduced into the RT reaction solution (without RT and dNTP) and treated at 95 °C for 5 minutes.
Single-cell reverse transcription PCR. Total cell lysate was used as a template in a 7.5 μl reaction. RT buffer, RNase inhibitor, and dNTP were added for RT reaction. RT reaction was carried out according to the manufacture's suggestions. Mcl-1 primers and Mcl-1 siRNA/MCL1mRNA primers were purchased from ABI. Briefly, the total RNA was first reverse transcribed into cDNA using the TaqMan MicroRNA reverse transcription kit (Applied Biosystems).
Single-cell level real-time PCR. The q-PCR amplification of cDNA was then performed using TaqMan MicroRNA assay (Applied Biosystems). All reactions were duplicated to obtain the Mcl-1 siRNA expression levels because of the small sample size. Duplication was found to be accurate enough. TaqMan universal PCR master mix of ABI was used according to manufacturer's suggestion in a 10 μl system. The conditions used were as follows: 95 °C for 10 minutes, followed by 40 cycles at 95 °C for 15 seconds, and 60 °C for 1 minute. All the reactions were run on ABI One-Step plus Detection System. The Ct value of the Mcl-1 siRNA expression was determined and the copy number was calculated according to the standard curve. To measure endogenous MCL1 expression at the mRNA level, the total cell lysate was transcribed into cDNA using the first-strand cDNA synthesis kit (Invitrogen). The cDNA was then amplified by qRT-PCR (Applied Biosystems). Ct values were determined and the copy number was calculated according to the standard curve.
Standard curve. The dynamic range and sensitivity of the Mcl-1 siRNA quantification scheme were first evaluated using a synthetic Mcl-1 siRNA. Mcl-1 siRNA was quantified based on the A260 value and diluted over seven orders of magnitude. The siRNA TaqMan siRNA assay showed excellent linearity between the log of target input and Ct value, demonstrating that the assay has a dynamic range of at least 7 logs.
SUPPLEMENTARY MATERIAL Figure S1. Endogenous level of MCL1 mRNA in MV4-11 cells and KG1a cells.
Acknowledgments
This work was supported by NSF under grant NSEC EEC-0914790, and NIH under grant R01 CA102031 and R01 CA135332. It wa partly supported by Leukemia SPORE (Specialized Program of Research Excellence) Grant from P50 CA140158. The authors thank Donna Bucci, the OSU Leukemia Tissue Bank, and OSU Comprehensive Cancer Center for assistance with the AML samples. K.G. fabricated NEP devices, designed and performed experiments, analyzed and interpreted data, and wrote the manuscript; X.H. performed transcript block, analyzed and interpreted data, and revised the manuscript; C.C. performed apoptosis assay; L.J.L. and G.M. proof-read the manuscript. The authors declare no competing financial interests.
Supplementary Material
Endogenous level of MCL1 mRNA in MV4-11 cells and KG1a cells.
References
- Puente, XS, Pinyol, M, Quesada, V, Conde, L, Ordóñez, GR, Villamor, N et al. (2011). Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeld, AK, Marcucci, G, Liyanarachchi, S, Döhner, K, Schwind, S, Maharry, K et al. (2012). Heritable polymorphism predisposes to high BAALC expression in acute myeloid leukemia. Proc Natl Acad Sci USA 109: 6668–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsoni, A, Pagano, L, Latagliata, R, Casini, M, Cerri, R, Crugnola, M et al. (2004). Survival of elderly patients with acute myeloid leukemia. Haematologica 89: 296–302. [PubMed] [Google Scholar]
- Jiang, Q, Crews, LA, Barrett, CL, Chun, HJ, Court, AC, Isquith, JM et al. (2013). ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci USA 110: 1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsagel, PL and Kuehl, WM (2011). Comprehensive identification of somatic mutations in chronic lymphocytic leukemia. Cancer Cell 20: 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory, S, Huang, DC and Adams, JM (2003). The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene 22: 8590–8607. [DOI] [PubMed] [Google Scholar]
- Gascoyne, RD, Adomat, SA, Krajewski, S, Krajewska, M, Horsman, DE, Tolcher, AW et al. (1997). Prognostic significance of Bcl-2 protein expression and Bcl-2 gene rearrangement in diffuse aggressive non-Hodgkin's lymphoma. Blood 90: 244–251. [PubMed] [Google Scholar]
- Yoshimoto, G, Miyamoto, T, Jabbarzadeh-Tabrizi, S, Iino, T, Rocnik, JL, Kikushige, Y et al. (2009). FLT3-ITD up-regulates MCL-1 to promote survival of stem cells in acute myeloid leukemia via FLT3-ITD-specific STAT5 activation. Blood 114: 5034–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser, SP, Lee, EF, Trounson, E, Bouillet, P, Wei, A, Fairlie, WD et al. (2012). Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev 26: 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenbuecher, F, Markova, B, Kasper, S, Carius, B, Stauder, T, Böhmer, FD et al. (2009). A novel molecular mechanism of primary resistance to FLT3-kinase inhibitors in AML. Blood 113: 4063–4073. [DOI] [PubMed] [Google Scholar]
- Kasper, S, Breitenbuecher, F, Heidel, F, Hoffarth, S, Markova, B, Schuler, M et al. (2012). Targeting MCL-1 sensitizes FLT3-ITD-positive leukemias to cytotoxic therapies. Blood Cancer J 2: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirewalt, DL, Kopecky, KJ, Meshinchi, S, Appelbaum, FR, Slovak, ML, Willman, CL et al. (2001). FLT3, RAS, and TP53 mutations in elderly patients with acute myeloid leukemia. Blood 97: 3589–3595. [DOI] [PubMed] [Google Scholar]
- Whitman, SP, Archer, KJ, Feng, L, Baldus, C, Becknell, B, Carlson, BD et al. (2001). Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a cancer and leukemia group B study. Cancer Res 61: 7233–7239. [PubMed] [Google Scholar]
- Zamore, PD, Tuschl, T, Sharp, PA and Bartel, DP (2000). RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33. [DOI] [PubMed] [Google Scholar]
- Drinnenberg, IA, Weinberg, DE, Xie, KT, Mower, JP, Wolfe, KH, Fink, GR et al. (2009). RNAi in budding yeast. Science 326: 544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, R (2006). Plants, RNAi, and the Nobel Prize. Science 314: 1242–1243. [DOI] [PubMed] [Google Scholar]
- Lei, EP and Corces, VG (2006). A long-distance relationship between RNAi and Polycomb. Cell 124: 886–888. [DOI] [PubMed] [Google Scholar]
- Matzke, M and Matzke, AJ (2003). RNAi extends its reach. Science 301: 1060–1061. [DOI] [PubMed] [Google Scholar]
- Fearon, ER and Cadigan, KM (2005). Cell biology. Wnt signaling glows with RNAi. Science 308: 801–803. [DOI] [PubMed] [Google Scholar]
- Allshire, R (2002). Molecular biology. RNAi and heterochromatin–a hushed-up affair. Science 297: 1818–1819. [DOI] [PubMed] [Google Scholar]
- Beebe, SJ, Fox, PM, Rec, LJ, Willis, EL and Schoenbach, KH (2003). Nanosecond, high-intensity pulsed electric fields induce apoptosis in human cells. FASEB J 17: 1493–1495. [DOI] [PubMed] [Google Scholar]
- Buerstedde, JM and Takeda, S (1991). Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 67: 179–188. [DOI] [PubMed] [Google Scholar]
- van de Stolpe, A, Pantel, K, Sleijfer, S, Terstappen, LW and den Toonder, JMJ (2011). Circulating tumor cell isolation and diagnostics: toward routine clinical use. Cancer Res 71: 5955–5960. [DOI] [PubMed] [Google Scholar]
- Dick, JE (2009). Looking ahead in cancer stem cell research. Nat Biotechnol 27: 44–46. [DOI] [PubMed] [Google Scholar]
- Gao, K, Li, L, He, L, Hinkle, K, Wu, Y, Ma, J et al. (2014). Design of a microchannel-nanochannel-microchannel array based nanoelectroporation system for precise gene transfection. Small 10: 1015–1023. [DOI] [PubMed] [Google Scholar]
- Boukany, PE, Morss, A, Liao, WC, Henslee, B, Jung, H, Zhang, X et al. (2011). Nanochannel electroporation delivers precise amounts of biomolecules into living cells. Nat Nanotechnol 6: 747–754. [DOI] [PubMed] [Google Scholar]
- White, AK, VanInsberghe, M, Petriv, OI, Hamidi, M, Sikorski, D, Marra, MA et al. (2011). High-throughput microfluidic single-cell RT-qPCR. Proc Natl Acad Sci USA 108: 13999–14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, GT, Seo, YJ and Kim, BH (2004). A highly discriminating quencher-free molecular beacon for probing DNA. J Am Chem Soc 126: 6528–6529. [DOI] [PubMed] [Google Scholar]
- Zheng, R, Levis, M, Piloto, O, Brown, P, Baldwin, BR, Gorin, NC et al. (2004). FLT3 ligand causes autocrine signaling in acute myeloid leukemia cells. Blood 103: 267–274. [DOI] [PubMed] [Google Scholar]
- Yeh, HY, Yates, MV, Mulchandani, A and Chen, W (2008). Visualizing the dynamics of viral replication in living cells via Tat peptide delivery of nuclease-resistant molecular beacons. Proc Natl Acad Sci USA 105: 17522–17525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, DY, Raj, A, Marras, SA, Kramer, FR and Tyagi, S (2005). Mechanism of mRNA transport in the nucleus. Proc Natl Acad Sci USA 102: 17008–17013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Endogenous level of MCL1 mRNA in MV4-11 cells and KG1a cells.