Abstract
Streptococcus infantarius subsp. infantarius (Sii) and Streptococcus gallolyticus subsp. macedonicus are members of the Streptococcus bovis/Streptococcus equinus complex (SBSEC) associated with human infections. SBSEC-related endocarditis was furthermore associated with rural residency in Southern Europe. SBSEC members are increasingly isolated as predominant species from fermented dairy products in Europe, Asia and Africa. African variants of Sii displayed dairy adaptations to lactose metabolism paralleling those of Streptococcus thermophilus including genome decay. In this study, the aim was to assess the prevalence of Sii and possibly other SBSEC members in dairy products of East and West Africa in order to identify their habitat, estimate their importance in dairy fermentation processes and determine geographic areas affected by this potential health risk. Presumptive SBSEC members were isolated on semi-selective M17 and SM agar media. Subsequent genotypic identification of isolates was based on rep-PCR fingerprinting and SBSEC-specific16S rRNA gene PCR assay. Detailed identification was achieved through application of novel primers enhancing the binding stringency in partial groES/groEL gene amplification and subsequent DNA sequencing. The presence of S. thermophilus-like lacS and lacZ genes in the SBSEC isolates was determined to elucidate the prevalence of this dairy adaptation. Isolates (n = 754) were obtained from 72 raw and 95 fermented milk samples from Côte d'Ivoire and Kenya on semi-selective agar media. Colonies of Sii were not detected from raw milk despite high microbial titers of approximately 106 CFU/mL on M17 agar medium. However, after spontaneous milk fermentation Sii was genotypically identified in 94.1% of Kenyan samples and 60.8% of Kenyan isolates. Sii prevalence in Côte d'Ivoire displayed seasonal variations in samples from 32.3% (June) to 40.0% (Dec/Jan) and isolates from 20.5% (June) to 27.7% (Dec/Jan) present at titers of 106–108 CFU/mL. lacS and lacZ genes were detected in all Kenyan and 25.8% (June) to 65.4% (Dec/Jan) of Ivorian Sii isolates. Regional differences in prevalence of Sii and dairy adaptations were observed, but no clear effect of dairy animal, fermentation procedure and climate was revealed. Conclusively, the high prevalence of Sii in Kenya, Côte d'Ivoire in addition to Somalia, Sudan and Mali strongly indicates a pivotal role of Sii in traditional African dairy fermentations potentially paralleling that of typical western dairy species S. thermophilus. Putative health risks associated with the consumption of high amounts of live Sii and potential different degrees of evolutionary adaptation or ecological colonization require further epidemiologic and genomic investigations, particularly in Africa.
Keywords: Streptococcus infantarius subsp. infantarius, Streptococcus bovis/Streptococcus equinus, complex, Streptococcus macedonicus, Streptococcus thermophilus, Dairy fermentation, Lactose metabolism
1. Introduction
Streptococcus infantarius subsp. infantarius (Sii) and Streptococcus gallolyticus subsp. macedonicus (originally designated as Streptococcus macedonicus) are increasingly found in fermented food products worldwide, in particular after spontaneous fermentations. Both species are members of the Streptococcus bovis/Streptococcus equinus complex (SBSEC) which is associated with several human infections including bacteremia, endocarditis and colonic cancer (Boleij et al., 2011; Herrera et al., 2009; Schlegel et al., 2000, 2003).
Sii was recently shown to be highly prevalent and predominant among the lactic acid bacteria (LAB) in African spontaneously fermented dairy products originating from cow, goat and camel milk in Mali, Sudan, Kenya, Somalia and Tanzania (Abdelgadir et al., 2008; Isono et al., 1994; Jans et al., 2012a; Wullschleger et al., 2013). S. gallolyticus subsp. macedonicus was first isolated from Greek Kasseri cheese and later also detected in cheese from France, Italy and Slovakia (Callon et al., 2004; Chebeňová-Turcovská et al., 2011; Franciosi et al., 2009; Lombardi et al., 2004; Pacini et al., 2006; Tsakalidou et al., 1998). Further studies report the prevalence of members of the SBSEC in traditionally fermented dairy and plant products from Asia (Bangladesh) and North America (Mexico) (Díaz-Ruiz et al., 2003; Rashid et al., 2007; Renye et al., 2011).
Interestingly, genome analysis of dairy strains Sii CJ18 and S. gallolyticus subsp. macedonicus ACA-DC 198 revealed genetic decay paralleling that of Streptococcus thermophilus, however in a less advanced state for Sii CJ18 (Jans et al., 2012b, 2013; Papadimitriou et al., 2012). In contrast to the Sii type strain classified within an elevated risk group, African isolates of Sii displayed phenotypic and genotypic adaptation of lactose metabolism. African strains harbor instead of a lactose phosphotransferase system (PTS) a LacS/LacZ mediated lactose uptake system similar to S. thermophilus (Jans et al., 2013). Strains carrying this adaptation were defined as African Sii variants (Jans et al., 2012c). However, the taxonomic relationship with pathogenic members of the SBSEC and epidemiological implications with endocarditis among rural communities in Europe (Giannitsioti et al., 2007) demands further analysis of their prevalence in food products and of their putative health risks. Accurate subspecies identification is crucial for the estimation of putative health risks as they are increasingly linked to specific species and subspecies within the SBSEC. Recommended identification with sufficient discriminatory power is therefore based on the heat shock protein encoding genes groES and groEL (Chen et al., 2008).
In this study, we aim to provide an enhanced comparison of the prevalence and habitat of African variants of Sii and other members of the SBSEC in African raw and fermented dairy products of different animal and geographical origin. This study represents the first large scale investigation to estimate the importance of Sii in the traditional fermentation processes and assesses the distribution of this potential pathogen in East vs. West Africa. A highly discriminative identification approach was used comprising rep-fingerprinting, pre-screening via a 16S rRNA gene-specific PCR assay and the development of novel primers of enhanced stringency for groES/groEL sequencing. In addition, we assessed the prevalence of lacS/lacZ genes as major dairy adaptations. New isolates from East Africa (Kenya) and West Africa (Côte d'Ivoire) obtained during this study were combined with data previously obtained from Kenya, Mali, Sudan and Somalia for a comprehensive overview of SBSEC prevalence in dairy products across Africa.
2. Material and methods
2.1. Culture conditions, bacterial strains, chemicals and enzymes
Streptococcus spp. reference strains were grown aerobically at 37 °C for 24 h on M17 agar media (Biolife, Milan, Italy). The enumeration and isolation of bacteria from dairy samples were generally performed aerobically at 30 °C for 24 h on M17 agar media (Biolife) and at 43 °C for 48 h on S. macedonicus (SM) agar media (Pacini et al., 2006). SM agar media was produced with the following modifications (Wullschleger, 2009): peptone from casein (15 g/L, Merck, Darmstadt, Germany) was used instead of 7.5 g/L peptone from casein and 7.5 g/L peptone from gelatin; 0.5 g/L potassium ascorbate instead of 0.5 g/L ascorbic acid; 19 g/L glycerol phosphate instead of 19 g/L disodium glycerol phosphate. Unless noted otherwise, all chemicals and enzymes were obtained from Sigma-Aldrich (Buchs, Switzerland).
Reference and type strains used in this study were obtained from the Culture Collection of the University of Gothenburg (CCUG, Gothenburg, Sweden) and the Deutsche Stammsammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany): Sii CCUG 43820T, Streptococcus lutetiensis (=S. infantarius subsp. coli) CCUG 43822, S. gallolyticus subsp. gallolyticus DSM16831T, S. gallolyticus subsp. macedonicus DSM15879T, S. bovis DSM20480T and S. thermophilus DSM20259.
2.2. African SBSEC strain collection
African Sii and S. gallolyticus subsp. macedonicus strains (culture collection of the Laboratory of Food Biotechnology, ETH Zurich) previously isolated from Mali, Kenya and Somalia (Jans et al., 2012a; Wullschleger, 2009; Wullschleger et al., 2013) were used for comparative analysis of strains. A comprehensive database comprising rep-PCR fingerprints of over 500 African LAB isolates including members of the SBSEC was previously established in GelCompar II (Applied-Maths, Sint-Martens-Latem, Belgium) and used in this study as a comparison tool (Jans, 2011; Jans et al., 2012a).
2.3. Dairy product samples
Cow (n = 14), camel (n = 25) and goat (n = 15) raw milk samples were collected at herd and market levels in the cities Nairobi, Isiolo, Garissa and Marsabit (Kenya) in December 2009 through July 2010 (Table 1). Additional samples comprised partly fermented raw milk samples (after 24 h) of goat (n = 4) and camel (n = 14) origin. Fermented sour milk suusac (camel milk, n = 17) and mala (cow milk, n = 1) were collected in Garissa and Nairobi in the same time period. Additional sour milk samples (cow, n = 76) were collected in Côte d'Ivoire in December 2010 through January 2011 and in June 2011 (Table 2). Côte d'Ivoire samples were obtained in Abobo (n = 25), Azito (n = 3), Beago (n = 3), Bingerville (n = 5), Lievre rouge (n = 12), N'dotré (n = 1), Port-Bouet (n = 23) and Songon (n = 4).
Table 1.
Bacterial counts in colony forming units (CFU) and prevalence of S. infantarius subsp. infantarius (Sii) and S. gallolyticus subsp. macedonicus (Sgm) determined from unfermented Kenyan raw milk samples (n = 72) of camel, cow and goat origin in this and in a previous study.
| Sample type | Samples/isolates | Agar media log10 CFU/mL (average ± SDa) |
pH (average ± SDa) | Prevalence Sii (and SBSEC members) |
Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M17 | SM | MRS | KFS | Samples % |
Isolates % |
||||||
| Sii | Sgm | Sii | Sgm | ||||||||
| Raw milk from Kenya | |||||||||||
| Camel raw milk | |||||||||||
| Herd level | 14/40 | 1.6 ± 2.0* | 2.6 ± 1.2* | n.d. | n.d. | 6.1 ± 0.2(2)* | - | - | - | - | This study |
| Herd level | 17/59 | 2.7 ± 0.8C | n.d. | 2.2 ± 1.5B | 0.7 ± 1.1* | 6.5 ± 0.1(17)A | - | - | - | - | Jans et al. (2012a) |
| Intermediate level | 5/26 | 4.5 ± 1.1BC | n.d. | 4.7 ± 1.1A | 2.1 ± 1.9A | 6.4 ± 0.2(5)A | - | - | - | - | Jans et al. (2012a) |
| Market level | 11/55 | 5.6 ± 1.1A | 3.0 ± 2.3A | n.d. | n.d. | 6.4 ± 0.1(3)AB | - | - | - | - | This study |
| Market level | 4/50 | 6.8 ± 0.5A | n.d. | 6.6 ± 0.8A | 3.7 ± 1.0A | 6.2 ± 0.0(4)BC | - | - | - | - | Jans et al. (2012a) |
| Milk 24 h | 14/69 | 6.5 ± 1.2* | 6.3 ± 1.4* | n.d. | n.d. | 6.5 ± 0.1(12)* | - | - | - | - | This study |
| Cow raw milk | |||||||||||
| Herd level | 6/56 | 3.8 ± 1.2BC | 3.9 ± 1.1A | n.d. | n.d. | n.d. | - | - | - | - | This study |
| Market level | 8/50 | 3.5 ± 1.8BC | 2.5 ± 1.6* | n.d. | n.d. | 6.5 ± 0.2(3)A | - | - | - | - | This study |
| Goat raw milk | |||||||||||
| Herd level | 6/36 | 3.2 ± 0.8BC | 2.8 ± 1.0A | n.d. | n.d. | 6.2 ± 0.2(6)C | - | - | - | - | This study |
| Market level | 9/73 | 3.6 ± 1.4* | 3.1 ± 1.7A | n.d. | n.d. | 6.1 ± 0.3(2)* | 11.1 | - | 2.7 | - | This study |
| Milk 24 h | 4/10 | 4.1 ± 0.7B | 2.9 ± 2.0* | n.d. | n.d. | 6.6(1)* | - | - | - | - | This study |
Sii: S. infantarius subsp. infantarius; Sgm: S. gallolyticus subsp. macedonicus; a) SD: standard deviation; superscript figure in brackets = number of pH measurements; n.d.: not determined; “–” below detection limit.Statistics comment:*) samples not normally distributed according to Shapiro–Wilk test (alpha = 0.05) were excluded from t-test and analyzed in a pair-wise Kruskal–Wallis test (alpha = 0.05). Values per column M17, SM, MRS, KFS and pH not connected by the same capital letter (A, B, C) are significantly different (t test, alpha = 0.05).
Table 2.
Bacterial counts in colony forming units (CFU), prevalence of S. infantarius subsp. infantarius (Sii) and S. gallolyticus subsp. macedonicus (Sgm) and prevalence of lacS/lacZ genes in dairy adapted Sii determined from fermented dairy samples (n = 95) in comparison with those of other countries.
| Sample type | Animal origin | Samples/isolates | Agar media log10 CFU/mL (average ± SDa) |
pH (average ± SDa) | Prevalence S. infantarius subsp. infantarius (and SBSEC members) |
% Sii with lacS/lacZ genes | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M17 | SM | MRS | KFS | Samples % |
Isolates % |
||||||||
| Sii | Sgm | Sii | Sgm | ||||||||||
| Kenya | |||||||||||||
| Suusac | Camel | 17/88 | 6.1 ± 0.5C | 6.4 ± 0.9* | n.d. | n.d. | 4.3 ± 0.1(15)* | 94.1 | - | 78.4 | - | 100 | This study |
| Suusac | Camel | 15/439 | 8.6 ± 0.4A | 7.3 ± 0.9c | 8.0 ± 0.5A | 8.0 ± 0.8* | 5.1 ± 1.0(15)* | 93.3 | 6.7 | 62.8 | 0.4 | 100 | Jans (2011); Jans et al. (2012a); Wullschleger (2009) |
| Sour milk mala | Cow | 1/5 | 6.2* | 5.8* | n.d. | n.d. | 5.5(1)* | 100.0 | - | 20.0 | - | 100 | This study |
| Somalia | |||||||||||||
| Suusac | Camel | 11/192 | 8.5 ± 0.1* | 5.8 ± 0.7c* | 8.4 ± 0.5A | 5.6 ± 2.5* | 4.5 ± 0.2(11)* | 72.7 | 45.5 | 19.8 | 4.2 | 100 | Jans (2011); Jans et al. (2012a); Wullschleger (2009) |
| Sour milk | Goat | 1/27 | 8.3 ± 0.1* | 6.6 ± 0.0* | 8.3 ± 0.0* | 5.9 ± 0.5* | 5.1(1)* | 100.0 | - | 11.1 | - | This study and Wullschleger (2009) | |
| Côte d’Ivoire | |||||||||||||
| Sour milk (June) | Cow | 31/151 | 9.0 ± 1.0* | n.d.b | n.d. | n.d. | 4.5 ± 0.2(31)* | 32.3 | - | 20.5 | - | 25.8 | This study |
| Sour milk (Dec/Jan) | Cow | 45/94 | 7.9 ± 0.7B | n.d.b | n.d. | n.d. | 5.5 ± 0.1(45)* | 40.0d | 6.7 d | 27.7 | 5.3 | 65.4 | This study |
| Mali | |||||||||||||
| Fènè | Cow | 19/57 | n.d. | 6.5 ± 1.8* | n.d. | n.d. | 4.8 ± 1.2(19)* | 36.8 | 31.6 | 19.3 | 21.1 | 55.5 | Wullschleger (2009) |
| Sudan | |||||||||||||
| Gariss | Camel | 9/180 | 8.0 ± 0.4B | n.d. | 8.2 ± 0.3A | n.d. | 4.0 ± 0.2(9) | 100.0 | - | 68.3 | - | n.d. | Abdelgadir et al. (2008) |
Sii: S. infantarius subsp. infantarius; Sgm: S. gallolyticus subsp. macedonicus; a) SD: standard deviation; superscript figure in brackets = number of pH measurements; b) SM agar media not used as explained in Section 3.2; c) determined from 2 (Kenya) and 5 (Somalia) selected samples (Wullschleger, 2009); d) based on the following calculation: 17 samples with Sii only, 2 samples with Sgm only, 1 sample with Sii + Sgm yielding 18 (40.0%) with Sii and 3 (6.7%) with Sgm and a total of 20 (44.4%) positive for SBSEC; n.d.: not determined; “–” below detection limit. Statistics comment: *) samples not normally distributed according to Shapiro–Wilk test (alpha = 0.05) were excluded from t-test and analyzed in a pair-wise Kruskal–Wallis test (alpha = 0.05). Values per column M17, SM, MRS, KFS and pH not connected by the same capital (A, B, C) letter are significantly different (t test, alpha = 0.05).
The sampling procedure and storage of samples in the field using dry ice were performed as previously described (Jans et al., 2012a).
2.4. Enumeration and isolation of bacteria
Dairy samples were serially diluted (1:10) and plated onto M17 (Biolife) and SM agar media designed for the semi-selective isolation of coccoid LAB and SBSEC members, respectively. M17 agar media were incubated aerobically at 30 °C for 24 h and SM agar media at 43 °C for 48 h. Cell counts were expressed in colony forming units (CFU) per mL using the arithmetic weighted mean. The isolation of bacteria and presumptive members of the SBSEC from dairy products was performed as previously described (Jans et al., 2012a, 2012c). The selection criteria were to randomly pick 3 isolates per colony morphology from different sections of the agar media with 10 < X < 300 CFU. Single colonies were picked, streaked onto the corresponding agar media and incubated. Isolates were stored at − 80 °C in the corresponding media broth containing 30% (v/v) glycerol.
2.5. Genotypic identification of isolates
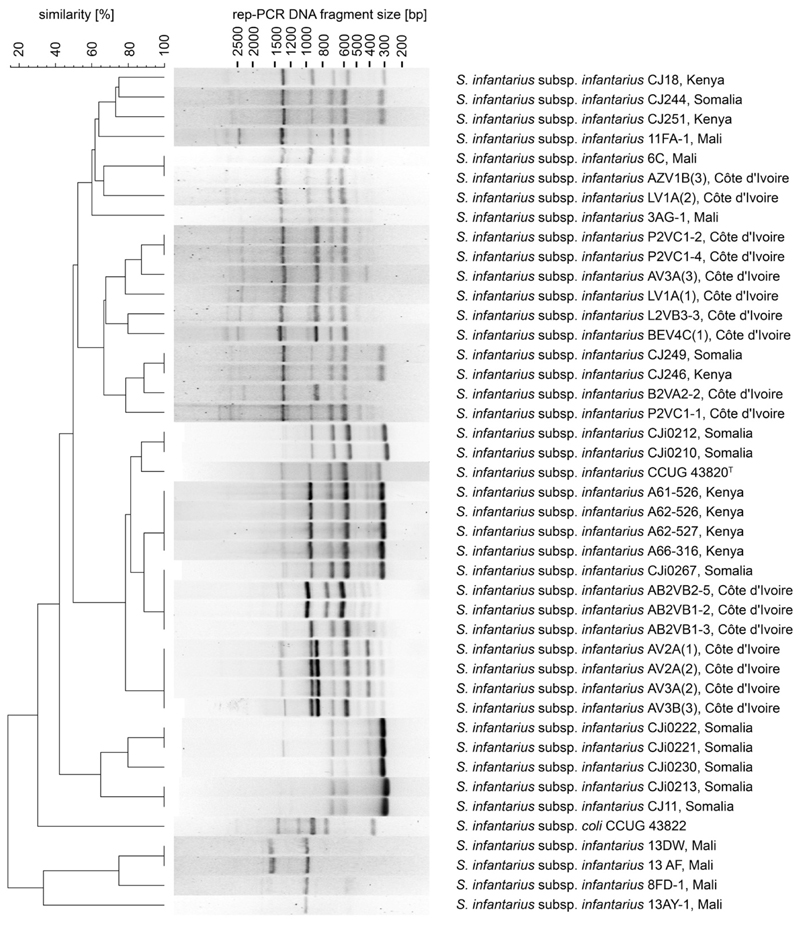
DNA for PCR and restriction fragment length polymorphism (RFLP) assays was isolated from bacterial single colonies after a short cell lysis procedure (Goldenberger et al., 1995). Subsequent genotypic identification of bacterial isolates was performed as previously described using a modified rep-PCR assay with (GTG)5 primer for genomic fingerprinting (Gevers et al., 2001; Jans et al., 2012a; Wullschleger, 2009). The fingerprints were clustered using GelCompar II software (Applied-Maths, Sint-Martens-Latem, Belgium). Calculations were based on the Jaccard similarity coefficient using an UPGMA dendrogram type, 1.30% position tolerance and 2.00% optimization. Sii was previously reported to feature a characteristic subspecies-specific rep-PCR fingerprint (Abdelgadir et al., 2008; Jans et al., 2012a, 2012c), which was used to cluster the isolates for presumptive Sii.
Taxonomically closely related species of the SBSEC, such as Sii and S. infantarius subsp. coli (=S. lutetiensis), required the application of a series of rep-PCR fingerprinting, 16S rRNA gene RFLP and groES/groEL sequencing for accurate identification. Preliminary clustering of isolates via rep-PCR fingerprinting and pre-screening using a 16S rRNA gene specific PCR/RFLP assay (Gevers et al., 2001; Jans et al., 2012a, 2012d) reduced necessary groES/groEL sequencing reactions while allowing accurate identification to subspecies level. SBSEC species classification was validated by a modified PCR/RFLP assay targeting the groES and groEL genes (Chen et al., 2008) whose partial DNA amplicons were sequenced for reliable subspecies identification. For that, the base composition of highly degenerated primers ES5-29F and EL1265R designed by Chen et al. (2008) was optimized in order to enhance binding stringency in PCR amplifications. This was performed through comparative analysis of conserved DNA sequences of groES and groEL sequences of members of the SBSEC available under GenBank accession numbers ABJK02000017, NZ_GL397173, FR824043, NC_013798, AP012054, EU140553, NC_016749, NZ_DS572689, CP003295, EU140554 and EU140555. In silico sequence analysis of the amplified groES/groEL segment clearly indicated the discrimination power within the amplified DNA fragment (Chen et al., 2008). Novel primers were named ES5-29F-inf (5′-TGA AAC CAT TAG GTG ACC GTG TGG T-3′) and EL1265R-inf (5′-CAA GTT CAA GTT CAG CGA CTT TWG-3′) targeting the same binding sites as the original primers. PCR assays using the ES5-29F-inf/EL1265R-inf primers were performed with an initial cycle of 95 °C for 3 min, followed by 40 cycles of 95 °C for 30 s, 54 °C for 30 s and 72 °C for 2 min. Final replication was performed at 72 °C for 10 min.
PCR products for sequencing and RFLP were purified using the GFX DNA purification kit (GE Healthcare Europe, Glattbrugg, Switzerland) according to the instructions. Restriction endonucleases AclI, MseI and XbaI (New England Biolabs, Ipswich, MA, USA) were used according to the conditions specific for groES/groEL and 16S rRNA gene RLFP assays (Chen et al., 2008; Jans et al., 2012d). Sanger sequencing reactions were performed at GATC-Biotech (Koblenz, Germany) using primers ES5-29F-inf and EL1265R. DNA sequences of genes groES and groEL were assembled in BioEdit (Hall, 1999). Sequences were aligned in MEGA4.0 (Tamura et al., 2007) using the ClustalW algorithm and then trimmed to equal lengths to calculate a sequence identity matrix in comparison with reference strains from GenBank and other strains sequenced in-house. Construction of phylogenetic trees was performed in MEGA4.0 using the neighbor-joining method and a bootstrap test with 1000 repetitions followed by the computation of evolutionary distances using the Maximum Composite Likelihood method (Felsenstein, 1985; Saitou and Nei, 1987; Tamura et al., 2004, 2007). PCR-amplicons from S. gallolyticus subsp. macedonicus strains previously isolated from fermented dairy products in Mali and Somalia (Wullschleger, 2009) were sequenced and included in the comparison.
The prevalence of lactose metabolism genes lacZ and lacS among SBSEC isolates was assessed as an indication for dairy adaptations. Both genes were targeted in PCR assays using S. thermophilus- and African Sii-specific primers lacS-8 (5′-GCG TGA CGT GCT TCA GTC-3′), lacS18.1 (5′- GAT TGA ATA CAG TTG TTG GTT TG-3′), lacZ-6.2 (5′-TTC CTC AAG AAT CAA ATG CTG-3′) and lacZ-17rev (5′-CCA CAA GAC CAA ATG ATA ACA C-3′) (Jans et al., 2012c). PCR assays for lacS and lacZ were both performed with an initial cycle of 95 °C for 2 min, followed by 35 cycles of 95 °C for 30 s, 58 °C for 30 s and 72 °C for 1 min. Final replication was performed at 72 °C for 7 min.
All PCR reactions were performed with 2× concentrated master mix (Thermo Scientific, St. Leon-Rot, Germany), 1 μM of each primer, 1 μL of DNA extract from the short lysis procedure and sterile double-distilled H2O to a final volume of 25 μL. All primers were obtained from Microsynth (Balgach, Switzerland).
Amplified DNA fragments were visualized under UV light after agarose gel (Sysmex Digitana AG, Horgen, Switzerland) electrophoresis using 1% for general analysis, 1.5% for rep-PCR and 2% for RFLP assays and ethidium bromide staining (2.5 mg/L).
2.6. Statistical analysis
Statistical analysis of cell counts and sample properties was performed using JMP 10.0 for Windows (SAS Institute Inc., Cary, NC, USA) and SPSS 20.0 (IBM Corp., Armonk, NY, USA). Data were analyzed for normal distribution using a Shapiro–Wilk test (alpha = 0.05). A one-way paired t test (alpha = 0.05) was used for all calculations with normally distributed samples. Transformation of not-normally distributed samples did not enable inclusion in a t-test. Not-normally distributed samples were analyzed in a non-parametric pair-wise Kruskal–Wallis test (alpha = 0.05).
2.7. Genbank accession numbers
DNA sequences obtained during this study were deposited in GenBank under accession number range KC113270–KC113281 (Table A.1).
3. Results
3.1. Enumeration of bacteria
African dairy product samples were analyzed for the presence of SBSEC members to determine their prevalence and distribution in East and West Africa. Mean cell counts from raw milk samples along supply chains on M17 and SM agar media averaged from 1.6 ± 2.0 to 5.6 ± 1.1 log10 CFU/mL and 2.6 ± 1.2 to 3.9 ± 1.1 log10 CFU/mL, respectively. Large variations between samples were indicated by high standard deviations and non-normally distributed data. Counts significantly (Kruskal–Wallis, p < 0.05) increased for raw camel milk from herd to market level from 1.6 ± 2.0 to 5.6 ± 1.1 log10 CFU/mL on M17 agar media (Table 1). Market level camel milk was not significantly different in pH and cell count compared to our previous study (Jans et al., 2012a), but showed larger variations. Cell counts in goat and cow milk samples did not significantly change between herd and market level. However, they were both significantly lower than those of camel market milk. pH values of 6.1 ± 0.2 to 6.5 ± 0.2 indicated varying degrees of acidification of the products compared to standard pH of camel (pH 6.2–6.5), goat (pH 6.5–6.8) and cow (pH 6.6–6.7) milk (Farah and Fischer, 2004; Jans et al., 2012a; Park et al., 2007).
Fermented milk samples suusac (Kenya) and sour milk (Côte d'Ivoire) featured mean cell counts on M17 agar media between 6.1 ± 0.5 log10 CFU/mL and 9.0 ± 1.0 log10 CFU/mL, respectively (Table 2). Significantly different cell counts on M17 (Kruskal–Wallis, p < 0.05) were observed between Kenyan suusac samples of 6.1 ± 0.5 log10 CFU/mL and Ivorian December/January-samples of 7.9 ± 0.7 log10 CFU/mL and Ivorian June samples of 9.0 ± 1.0 log10 CFU/mL. pH was reduced to a value of 4.3 ± 0.1 for suusac which was significantly different from all other products except fènè. pH of Ivorian sour milk samples displayed significant (Kruskal–Wallis, p < 0.05) seasonal variations from 4.5 ± 0.2 to 5.5 ± 0.1 (Table 2). Predominant bacteria were isolated on M17 and SM agar media from raw and fermented dairy products for subsequent identification.
3.2. Identification of presumptive members of the SBSEC and prevalence in dairy products
A total of 754 bacterial isolates were obtained from dairy samples for identification and determination of the prevalence of Sii and other members of the SBSEC. All 754 isolates were first clustered by rep-PCR fingerprinting and compared to the established African LAB/SBSEC fingerprint database to yield presumptive Sii.
Isolates (n = 389) from Kenyan raw milk samples displayed no rep-PCR fingerprint profile typical for Sii. To reduce the likelihood of false-negative identification of members of the SBSEC due to uncategorized rep-fingerprints, a total of 192 randomly selected raw milk isolates obtained from both agar media were further subjected to the SBSEC-specific PCR assay targeting the 16S rRNA gene (Jans et al., 2012d). None of the 192 isolates tested of major rep fingerprint clusters were identified as members of the SBSEC, thereby confirming rep-PCR clustering. Only two presumptive Sii isolates were found in one partly fermented raw goat milk market sample from Garissa (Table 1).
Fermented milk samples yielded a total of 365 isolates comprising 93 Kenyan, 27 Somali and 245 Ivorian isolates. All presumptive Sii isolates including type strain CCUG 43820T displayed a subspecies-specific rep-PCR fingerprint backbone consisting of DNA fragments of approximate sizes 585–595 bp, 700–720 bp, 920–950 bp and 1320–1350 bp (Fig. 1). Minor variations in comparison with rep-PCR fingerprints the African LAB/SBSEC fingerprint database include an additional fragment at 310–320 bp or a double band at 920–950 bp and additional bands at 2400 bp and 2700–2800 bp. DNA fragment sizes were calculated in GelCompar II. The subsequent SBSEC-specific PCR assay identified 86.4% (SM agar media) and 70.5% (M17 agar media) of all Kenyan isolates as member of the SBSEC. A total of 69 out of 88 (78.4%) isolates from fermented Kenyan camel milk, fermented cow milk (20.0%) and fermented Somali goat milk (11.1%) were identified as Sii (Table 2). Mean prevalence of Sii (isolates n = 120) in fermented milk products from Kenya and Somalia was 60.8%.
Fig. 1.
Dendrogram of rep-PCR fingerprints from representative African Sii isolates originating from dairy products of Kenya, Somalia, Mali and Côte d'Ivoire in comparison with reference and type strains. Fingerprints were clustered using GelCompar II software (Applied-Maths).
SBSEC prevalence among suusac samples (camel milk) was high in Kenya and less pronounced in fermented Ivorian cow milk samples. Out of 17 suusac samples, Sii or other SBSEC members were detected in 16 samples (94.1%, detection limit on SM agar media 103 CFU/mL). In contrast, 10 out of 31 fermented Ivorian samples (cow milk) collected in June (32.3%) and 20 out of 45 of those collected in December/January (44.4%) yielded Sii (17 samples), S. gallolyticus subsp. macedonicus (2 samples) or both species (1 sample) (Table 2). Possible preliminary indications for a seasonal dependency of prevalence were detected between Ivorian December isolates (27.7%) compared to those collected in June (20.5%). Only five isolates from Côte d'Ivoire products collected in December were identified as S. gallolyticus subsp. macedonicus.
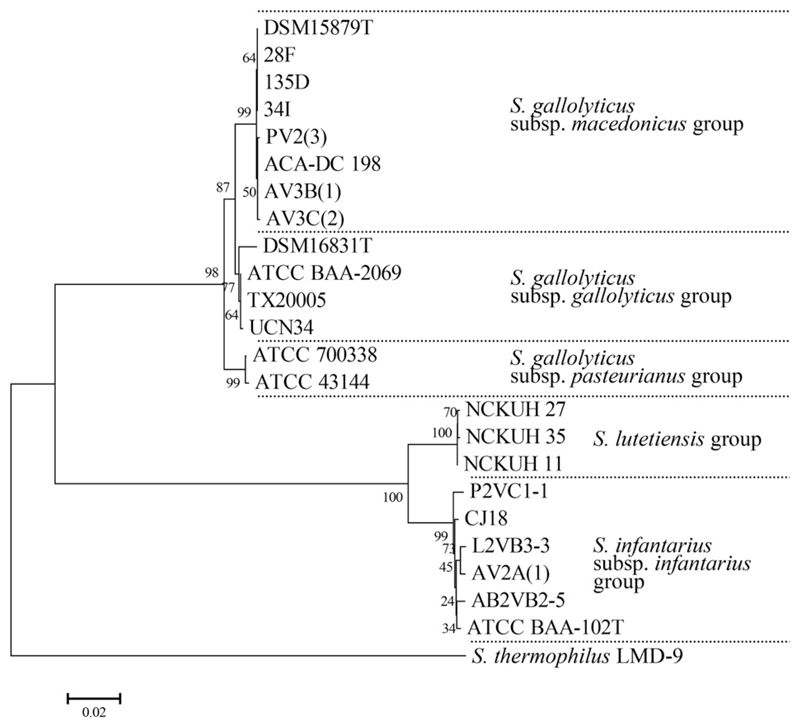
Subspecies identification of all members of the SBSEC was performed via partial groES and groEL sequencing. Based on rep-PCR fingerprints and 16S rRNA gene PCR-RFLP data, a random selection from different sample regions was performed to yield six isolates of the S. gallolyticus group and four of the S. infantarius group for subspecies identification using novel primers with enhanced binding stringency (Section 2.5). All S. gallolyticus isolates revealed highest DNA sequence identities of 99.8% and 100% to S. macedonicus ACA-DC 198 whereas all S. infantarius isolates showed 99.3–99.6% identity to Sii CJ18 (Table A.1). The phylogenetic tree confirmed the identification and classification of SBSEC isolates within the S. gallolyticus subsp. macedonicus or Sii subspecies indicating the evolutionary conservation of the groES/groEL genes to subspecies level and high discrimination power (Fig. 2). Based on rep-PCR fingerprint clustering, S. gallolyticus subsp. macedonicus or Sii identification was assigned to all isolates featured within the corresponding cluster of fingerprints allowing the assessment of prevalence across all samples described above.
Fig. 2.
Phylogenetic neighbor-joining tree of aligned and trimmed partial groES and groEL gene sequences (1402–1441 bp) of S. gallolyticus subsp. macedonicus and Sii strains in comparison with reference strains and S. thermophilus LMD-9 as out-group. The optimal tree with the sum of branch length = 0.47843285 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances indicated by the horizontal bar below the figure are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). There were a total of 1369 positions in the final dataset.
3.3. Prevalence of lacS and lacZ genes among Sii and SBSEC isolates
The prevalence of lacS and lacZ genes with S. thermophilus-specific primers was tested as an indication for dairy adaptation as previously reported for African variants of Sii in Kenya, Somalia and Mali (Jans et al., 2012b, 2012c). All Kenyan Sii isolates harbored both lacS and lacZ genes (Table 2). All tested isolates harbored both genes and none were detected which featured only lacS or lacZ. In Côte d'Ivoire, the prevalence of both lacS and lacZ genes was lower and season dependent with 25.8% and 65.4% for samples collected in June and December/January, respectively. Neither lacS nor lacZ genes were detected in S. gallolyticus subsp. macedonicus and non-S. infantarius SBSEC isolates from Côte d'Ivoire and Kenya, suggesting a specific prevalence of this dairy adaptation only for Sii.
4. Discussion
Fermented dairy and cereal foods play an important role as weaning and staple food with high nutritional value, increased microbial safety and storage properties (Motarjemi, 2002). However, the ingestion of high quantities of Sii through fermented dairy products especially by children representing 30% (38.6 millions) of the total population of the countries studied (United Nations Department of Economic and Social Affairs Population Division, 2011) has to be critically assessed for any putative health risks associated with this bacterial species.
Sii isolation from milk products was agar media dependent. SM agar media seemed to be more selective for members of the SBSEC compared to M17 agar media whereas M17 agar media provided a more general overview of coccoid LAB present (Jans et al., 2012a) and thus a more comprehensive picture of the fermentative LAB microflora. Therefore, M17 medium allows a better estimation on the predominance of Sii and other SBSEC members over typical dairy fermentation cocci such as Lactococcus spp. or S. thermophilus.
The prevalence of Sii was previously described in small sample batches of fermented dairy products from Mali, Sudan, Somalia and Kenya (Abdelgadir et al., 2008; Jans et al., 2012a; Wullschleger et al., 2013). However, their potential presence in unfermented raw milk of those regions as well as their prevalence in other African regions such as West Africa (Côte d'Ivoire) was unknown. Consolidated data from our study and other studies suggest that unfermented raw milk does not harbor significant numbers of Sii. Instead, raw milk microflora was previously described to predominately contain Streptococcus agalactiae, Lactococcus spp. and Enterococcus spp. on M17 agar media (Jans et al., 2012a).
Fermented dairy products featured high prevalence of Sii with high average titers between 6 and 8 log10 CFU/mL (Table 2). Our data is in agreement with previous studies showing that traditionally fermented dairy products in Côte d'Ivoire, Mali, Sudan, Somalia and Kenya displayed a high prevalence of Sii in 32–100% of all samples, with 19–78% of all isolates identified as Sii (Table 2), and a tendency of higher prevalence in East vs. West Africa (Abdelgadir et al., 2008; Jans et al., 2012a; Wullschleger, 2009; Wullschleger et al., 2013). Therefore, traditionally fermented dairy products seem to be a major reservoir of dairy adapted African variants of Sii with no other natural reservoir yet.
Despite the absence of Sii, unfermented raw milk products in regions handled under minimal process and storage conditions featured enhanced microbial growth. Therefore, they may exhibit significant health risks likely associated with the consumption of such unfermented raw market milk which was shown to harbor Escherichia coli, S. agalactiae and staphylococci (Jans et al., 2012a; Kaindi et al., 2012; Kouamé-Sina et al., 2012; Njage et al., 2012, 2013). Mean cell counts observed in raw milk of 1.6–6.7 log10 (Table 1) were comparable to those reported for camel milk market chains ranging from 2.6 to 6.7 log10 CFU/mL (Jans et al., 2012a), total bacterial counts of cow milk in Mali ranging from 2.9 to 7.3 log10 CFU/mL (Bonfoh et al., 2003) and total coliforms of cow milk from farm to market in Côte d'Ivoire from 3.9 to 5.9 log10 CFU/mL (Kouamé-Sina et al., 2010). Spontaneous growth of the indigenous microflora occurred during transport and storage under elevated temperatures and in contrast to traditional milk fermentation processes, pH values did not decrease below pH 5 to provide microbial protection (Holzapfel et al., 1995). All samples including fermented and unfermented displayed large heterogeneity as indicated by large variations in pH, cell counts and often non-normal distributions. Strong pH fluctuations in fermented products indicate low process control and reduced product safety due to higher pH values.
Seasonal variations of cell counts and product properties were previously reported for the Malian sour milk fènè with a positive correlation between temperature and cell counts (Wullschleger et al., 2013). Although based only on a small number of sampling points, samples from Côte d'Ivoire suggested the opposite temperature correlation. Samples collected in June featured significantly higher cell counts and significantly lower pH than those collected in December. In Côte d'Ivoire, the climate is on average warm and dry in December through January (25.6–27.7 °C, 9.6–23.2 mm rain) whereas the condition in June is on average slightly colder in combination with heavy precipitation (24.9–25.9 °C, 164.8–227.1 mm rain). These temperature differences in Côte d'Ivoire are less pronounced as in Mali (21.9–25.7 in November/December vs. 31.7–33.2 °C in June) whereas precipitation is generally higher in Côte d'Ivoire (World Bank, 2012). These humid conditions with high precipitation in June in Côte d'Ivoire might lead to increased contamination of milk (Henry et al., 1990) and possibly less favorable growth conditions for Sii.
Sii displayed different prevalence in samples of East Africa (Kenya, Somalia and Sudan: 72.7–100.0%) and West Africa (Côte d'Ivoire and Mali: 32.3–40.0%) (Abdelgadir et al., 2008; Jans et al., 2012a; Wullschleger et al., 2013). High prevalence recorded both in the arid sampling regions in Kenya, Somalia and Mali as well as the more tropical regions of Côte d'Ivoire suggests wide tolerance of Sii to different climatic environments.
An animal specific association of Sii is not evident. Besides camel milk products, Sii was isolated from fermented cow and goat milk products in Mali, Kenya and Somalia (Jans, 2011; Jans et al., 2012a; Wullschleger et al., 2013). Nevertheless, a preference towards camel milk or a specific fermentation process and climate cannot be excluded.
Dairy adaptations based on the prevalence of lacS/lacZ genes were less frequent in West African Sii isolates compared to isolates from East Africa. Whether this lower prevalence of lacS/lacZ harboring Sii in Côte d'Ivoire is actually due to the lower prevalence of these genes or a possible mutation within the primer binding sites is uncertain at this stage and will require further genome sequencing. Similar findings on the lower prevalence of lacS and lacZ genes were previously determined in a small sample size from Mali (Jans et al., 2012c). This suggests different levels of evolutionary dairy adaptations between the Kenyan lineage and that of Côte d'Ivoire and therefore different levels of competitiveness. Hence, the thought of a potential ongoing ecological colonization by the most adapted strains should be further elucidated.
Other members of the SBSEC such as S. gallolyticus subsp. macedonicus were commonly isolated from Southern European cheeses. Few isolates of this subspecies were also obtained from African dairy products. Interestingly, they seem to be less prevalent in Sub-Saharan African fermented dairy products whereas Sii was so far not isolated from European dairy products. This suggests the existence of a previously unknown fermentation microflora in Africa containing high titers of dairy adapted variants of Sii in strong contrast to the intensively characterized microflora of European dairy products.
In conclusion, this study establishes a comprehensive prevalence overview of Sii and members of the SBSEC in dairy products from East to West Africa previously limited to East Africa or semi-arid and arid regions. For accurate subspecies identification of Sii and SBSEC members, groES/groEL gene sequencing proved to be the preferred identification method offering higher discrimination power than RFLP or 16S rRNA gene sequencing. Sii displays a high prevalence especially in fermented dairy products of Kenya, Côte d'Ivoire, Somalia, Sudan and Mali strongly indicating these products as a major habitat. The decreasing prevalence towards West Africa might be related to regional differences of dairy production, dairy animals (cow vs. goat vs. camel), and degree of evolutionary adaptation or ecological colonization. Similarly, the lower prevalence of lacS/lacZ genes among Ivorian isolates compared to Kenya might be related to the same factors and suggests also different strain lineages in these countries. Due to the unclear pathogenicity of Sii and the risk associated with many other members of the SBSEC, potential health risks for consumers of fermented milk products have to be considered. Further research needs to assess epidemiology and potential health risks. The presence of members of the SBSEC in traditional fermented products in Europe, Asia and North America supports the importance of elucidating their role in food fermentations.
Acknowledgments
This study was supported by the UBS Optimus Foundation (Switzerland) in the frame of an Innovation Phase Project. The funding party had no role in the design, conduction, analysis of experiments and preparation of the manuscript. The authors acknowledge additional support from NCCR North–South (Switzerland) funded by the Swiss National Science Foundation and the Swiss Agency for Development and Cooperation. The authors also acknowledge support received by the consortium Afrique One “Ecosystem and Population Health: Expanding Frontiers in Health” funded by the Wellcome Trust (WT087535MA). The authors thank Jennifer Integlia for assistance in laboratory analyses.
Abbreviations
- SBSEC
Streptococcus bovis/Streptococcus equinus complex
- Sii
Streptococcus infantarius subsp. infantarius
Appendix A
References
- Abdelgadir W, Nielsen DS, Hamad S, Jakobsen M. A traditional Sudanese fermented camel's milk product, Gariss, as a habitat of Streptococcus infantarius subsp. infantarius. Int J Food Microbiol. 2008;127:215–219. doi: 10.1016/j.ijfoodmicro.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Boleij A, Muytjens CMJ, Bukhari SI, Cayet N, Glaser P, Hermans PWM, Swinkels DW, Bolhuis A, Tjalsma H. Novel clues on the specific association of Streptococcus gallolyticus subsp. gallolyticus with colorectal cancer. J Infect Dis. 2011;203:1101–1109. doi: 10.1093/infdis/jiq169. [DOI] [PubMed] [Google Scholar]
- Bonfoh B, Wasem A, Traoré AN, Fané A, Spillmann H, Simbé CF, Alfaroukh IO, Nicolet J, Farah Z, Zinsstag J. Microbiological quality of cows' milk taken at different intervals from the udder to the selling point in Bamako (Mali) Food Control. 2003;14:495–500. [Google Scholar]
- Callon C, Millet L, Montel MC. Diversity of lactic acid bacteria isolated from AOC Salers cheese. J Dairy Res. 2004;71:231–244. doi: 10.1017/s0022029904000159. [DOI] [PubMed] [Google Scholar]
- Chebeňová-Turcovská V, Ženišová K, Kuchta T, Pangallo D, Brežná B. Culture-independent detection of microorganisms in traditional Slovakian bryndza cheese. Int J Food Microbiol. 2011;150:73–78. doi: 10.1016/j.ijfoodmicro.2011.07.020. [DOI] [PubMed] [Google Scholar]
- Chen H-J, Tsai J-C, Chang T-C, Hung W-C, Tseng S-P, Hsueh P-R, Teng L-J. PCR-RFLP assay for species and subspecies differentiation of the Streptococcus bovis group based on groESL sequences. J Med Microbiol. 2008;57:432–438. doi: 10.1099/jmm.0.47628-0. [DOI] [PubMed] [Google Scholar]
- Díaz-Ruiz G, Guyot JP, Ruiz-Teran F, Morlon-Guyot J, Wacher C. Microbial and physiological characterization of weakly amylolytic but fast-growing lactic acid bacteria: a functional role in supporting microbial diversity in pozol, a Mexican fermented maize beverage. Appl Environ Microbiol. 2003;69:4367–4374. doi: 10.1128/AEM.69.8.4367-4374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah Z, Fischer A. Milk and Meat From the Camel — Handbook on Products and Processing. vdf Hochschulverlag AG Zürich. 2004 [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Franciosi E, Settanni L, Cavazza A, Poznanski E. Biodiversity and technological potential of wild lactic acid bacteria from raw cows' milk. Int Dairy J. 2009;19:3–11. [Google Scholar]
- Gevers D, Huys G, Swings J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol Lett. 2001;205:31–36. doi: 10.1111/j.1574-6968.2001.tb10921.x. [DOI] [PubMed] [Google Scholar]
- Giannitsioti E, Chirouze C, Bouvet A, Béguinot I, Delahaye F, Mainardi JL, Celard M, Mihaila-Amrouche L, Moing VL, Hoen B Association pour l' Etude et la Prévention de l' Endocardite Infectieuse (AEPEI) Study Group. Characteristics and regional variations of group D streptococcal endocarditis in France. Clin Microbiol Infect. 2007;13:770–776. doi: 10.1111/j.1469-0691.2007.01753.x. [DOI] [PubMed] [Google Scholar]
- Goldenberger D, Perschil I, Ritzler M, Altwegg M. A simple universal DNA extraction procedure using SDS and proteinase K is compatible with direct PCR amplification. PCR Methods Appl. 1995;4:368–370. doi: 10.1101/gr.4.6.368. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Henry FJ, Patwary Y, Huttly SRA, Aziz KMA. Bacterial contamination of weaning foods and drinking water in rural Bangladesh. Epidemiol Infect. 1990;104:79–85. doi: 10.1017/s0950268800054558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera P, Min Kwon Y, Ricke SC. Ecology and pathogenicity of gastrointestinal Streptococcus bovis. Anaerobe. 2009;15:44–54. doi: 10.1016/j.anaerobe.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Holzapfel WH, Geisen R, Schillinger U. Biological preservation of foods with reference to protective cultures, bacteriocins and food-grade enzymes. Int J Food Microbiol. 1995;24:343–362. doi: 10.1016/0168-1605(94)00036-6. [DOI] [PubMed] [Google Scholar]
- Isono Y, Shingu I, Shimizu S. Identification and characteristics of lactic acid bacteria isolated from Masai fermented milk in northern Tanzania. Biosci Biotechnol Biochem. 1994;58:660–664. [Google Scholar]
- Jans C. Biodiversity of Lactic Acid Bacteria in Raw Camel Milk Products of East Africa Including Genomic and Functional Characterization of Predominant Lactose-adapted Streptococcus infantarius subsp. infantarius. PhD Thesis No. 19835; ETH Zurich, Zurich, Switzerland: 2011. [Google Scholar]
- Jans C, Bugnard J, Njage PMK, Lacroix C, Meile L. Lactic acid bacteria diversity of African raw and fermented camel milk products reveals a highly competitive, potentially health-threatening predominant microflora. LWT Food Sci Technol. 2012a;47:371–379. [Google Scholar]
- Jans C, Follador R, Lacroix C, Meile L, Stevens MJA. Complete genome sequence of the African dairy isolate Streptococcus infantarius subsp. infantarius strain CJ18. J Bacteriol. 2012b;194:2105–2106. doi: 10.1128/JB.00160-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans C, Gerber A, Bugnard J, Njage PMK, Lacroix C, Meile L. Novel Streptococcus infantarius subsp. infantarius variants harboring lactose metabolism genes homologous to Streptococcus thermophilus. Food Microbiol. 2012c;31:33–42. doi: 10.1016/j.fm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Jans C, Lacroix C, Meile L. A novel multiplex PCR/RFLP assay for the identification of Streptococcus bovis/Streptococcus equinus complex members from dairy microbial communities based on the 16S rRNA gene. FEMS Microbiol Lett. 2012d;326:144–150. doi: 10.1111/j.1574-6968.2011.02443.x. [DOI] [PubMed] [Google Scholar]
- Jans C, Follador R, Hochstrasser M, Lacroix C, Meile L, Stevens MJA. Comparative genome analysis of Streptococcus infantarius subsp. infantarius CJ18, an African fermented camel milk isolate with adaptations to dairy environment. BMC Genomics. 2013;14:200. doi: 10.1186/1471-2164-14-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaindi DWM, Schelling E, Wangoh J, Imungi J, Farah Z, Meile L. Risk factors for symptoms of gastrointestinal illness in rural town Isiolo, Kenya. Zoonoses Public Health. 2012;59:118–125. doi: 10.1111/j.1863-2378.2011.01425.x. [DOI] [PubMed] [Google Scholar]
- Kouamé-Sina SM, Bassa A, Dadié A, Makita K, Grace D, Dje M, Bonfoh B. Analyse des risques microbiens du lait cru local à Abidjan (Côte d'Ivoire) Rev Afr Santé Prod Anim (RASPA) (S) 2010:35–42. [Google Scholar]
- Kouamé-Sina SM, Makita K, Costard S, Grace D, Dadié A, Dje M, Bonfoh B. Hazard identification and exposure assessment for bacterial risk assessment of informally marketed milk in Abidjan, Côte d'Ivoire. Food Nutr Bull. 2012;33:223–234. doi: 10.1177/156482651203300402. [DOI] [PubMed] [Google Scholar]
- Lombardi A, Gatti M, Rizzotti L, Torriani S, Andrighetto C, Giraffa G. Characterization of Streptococcus macedonicus strains isolated from artisanal Italian raw milk cheeses. Int Dairy J. 2004;14:967–976. [Google Scholar]
- Motarjemi Y. Impact of small scale fermentation technology on food safety in developing countries. Int J Food Microbiol. 2002;75:213–229. doi: 10.1016/s0168-1605(01)00709-7. [DOI] [PubMed] [Google Scholar]
- Njage PMK, Jans C, Wangoh J, Lacroix C, Meile L. Detection, isolation and molecular characterisation of shigatoxigenic O157 and non-O157 Escherichia coli in raw and fermented camel milk. Afr J Microbiol Res. 2012;6:6031–6038. [Google Scholar]
- Njage PMK, Dolci S, Jans C, Wangoh J, Lacroix C, Meile L. Biodiversity and enterotoxigenic potential of staphylococci isolated from raw and spontaneously fermented camel milk. Br Microbiol Res J. 2013;3:128–138. [Google Scholar]
- Pacini F, Cariolato D, Andrighetto C, Lombardi A. Occurrence of Streptococcus macedonicus in Italian cheeses. FEMS Microbiol Lett. 2006;261:69–73. doi: 10.1111/j.1574-6968.2006.00330.x. [DOI] [PubMed] [Google Scholar]
- Papadimitriou K, Ferreira S, Papandreou NC, Mavrogonatou E, Supply P, Pot B, Tsakalidou E. Complete genome sequence of the dairy isolate Streptococcus macedonicus ACA-DC 198. J Bacteriol. 2012;194:1838. doi: 10.1128/JB.06804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YW, Juárez M, Ramos M, Haenlein GFW. Physico-chemical characteristics of goat and sheep milk. Small Ruminant Res. 2007;68:88–113. [Google Scholar]
- Rashid MH, Togo K, Ueda M, Miyamoto T. Identification and characterization of dominant lactic acid bacteria isolated from traditional fermented milk Dahi in Bangladesh. World J Microbiol Biotechnol. 2007;23:125–133. [Google Scholar]
- Renye JA, Jr, Somkuti GA, Van Hekken DL, Guerrero Prieto VM. Short communication: characterization of microflora in Mexican Chihuahua cheese. J Dairy Sci. 2011;94:3311–3315. doi: 10.3168/jds.2011-4177. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schlegel L, Grimont F, Collins MD, Régnault B, Grimont PAD, Bouvet A. Streptococcus infantarius sp. nov., Streptococcus infantarius subsp. infantarius subsp. nov. and Streptococcus infantarius subsp. coli subsp. nov., isolated from humans and food. Int J Syst Evol Microbiol. 2000;50:1425–1434. doi: 10.1099/00207713-50-4-1425. [DOI] [PubMed] [Google Scholar]
- Schlegel L, Grimont F, Ageron E, Grimont PAD, Bouvet A. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int J Syst Evol Microbiol. 2003;53:631–645. doi: 10.1099/ijs.0.02361-0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tsakalidou E, Zoidou E, Pot B, Wassill L, Ludwig W, Devriese LA, Kalantzopoulos G, Schleifer KH, Kersters K. Identification of streptococci from Greek Kasseri cheese and description of Streptococcus macedonicus sp. nov. Int J Syst Bacteriol. 1998;48:519–527. doi: 10.1099/00207713-48-2-519. [DOI] [PubMed] [Google Scholar]
- United Nations Department of Economic and Social Affairs Population Division. World Population Prospects: the 2010 Revision. United Nations; New York, NY USA: 2011. [Google Scholar]
- World Bank. [Accessed: April-19-2013];World Bank Climate Change Knowledge Portal [Online] 2012 Available at http://data.worldbank.org/country/
- Wullschleger S. Biodiversity and microbial safety of artisanal Malian sour milk fènè and development of adapted starter cultures for controlled production. PhD thesis No. 18287; ETH Zurich, Zurich, Switzerland: 2009. [Google Scholar]
- Wullschleger S, Lacroix C, Bonfoh B, Sissoko-Thiam A, Hugenschmidt S, Romanens E, Baumgartner S, Traoré I, Yaffee M, Jans C, Meile L. Analysis of lactic acid bacteria communities and their seasonal variations in a spontaneously fermented dairy product (Malian fènè) by applying a cultivation/genotype-based binary model. Int Dairy J. 2013;29:28–35. [Google Scholar]