Abstract
Ultrasensitive impedimetric lectin biosensors recognising different glycan entities on serum glycoproteins were constructed. Lectins were immobilised on novel mixed self-assembled monolayer containing 11-mercaptoundecanoic acid for covalent immobilisation of lectins and betaine terminated thiol to resist non-specific interactions. Construction of biosensors based on Concanavalin A (Con A), Sambucus nigra agglutinin type I (SNA) and Ricinus communis agglutinin (RCA) on polycrystalline gold electrodes was optimised and characterised with a battery of tools including electrochemical impedance spectroscopy, various electrochemical techniques, QCM, FTIR spectroscopy, AFM, XPS and compared with a protein/lectin microarray. The lectin biosensors were able to detect glycoproteins from 1 fM (Con A), 10 fM (RCA) or 100 fM (SNA) with a linear range spanning 6 (SNA), 7 (RCA) or 8 (Con A) orders of magnitude. Furthermore, a detection limit for the Con A biosensor down to 1 aM was achieved in a sandwich configuration. A non-specific binding of proteins for the Con A biosensor was only 6.1% (probed with an oxidised invertase) of the signal towards its analyte invertase and a negligible non-specific interaction of the Con A biosensor was observed in diluted human sera (1000x), as well. The performance of the lectin biosensors was finally tested by glycoprofiling of human serum samples from healthy individuals and those having rheumatoid arthritis, which resulted in distinct glycan pattern between these two groups.
Keywords: Lectin biosensors, electrochemical impedance spectroscopy (EIS), self-assembled monolayer (SAM), glycoproteins, ultrasensitive detection, sulfobetaine, human serum
Introduction
Since the introduction of DNA microarrays in 19951, the technology has been heavily applied in analysis of genome-wide expression in order to get information about possible functions of novel or poorly characterised genes2 and for diagnostic purposes, as well3. Even though DNA microarray technology has shed light on many physiological states by analysis of expression of gene clusters, there is usually very low correlation between RNA and protein abundance detected in single-cell organisms4 as well as in higher ones, including humans5. Since quantitative analysis of proteins is central to proteomics with a focus on design of novel drugs, diagnosis of diseases and their therapeutic applications, protein microarrays were successfully launched to address these issues.6
Analysis of finely tuned post-translational modifications (PTMs) of proteins7, considering addition of a small functionality such as single phosphorylation can change activity of a protein up to 108 times8, is an additional challenge for current analytical technology. Glycosylation is another and highly abundant form of PTM of proteins and it is estimated that 70% of proteins in humans together with 80% of membrane-bound proteins are glycosylated.9 Glycan mediated recognition plays an important role in cell physiology (fertilisation, immune response, differentiation of cells, cell-matrix interaction, cell-cell adhesion etc.)10 Glycans, as highly abundant ligands on the surface of cells, are naturally involved in pathological processes triggered by adhesion of viruses, bacteria and parasites to host cells, in neurological disorder and in tumour growth and metastasis.3,11 Thus, better understanding of glycan mediated pathogenesis can establish a “policy” to develop more efficient strategies for disease treatment with few recent studies as good examples e.g. “neutralisation” of various forms of viruses12 or more efficient vaccines against various diseases13. Changes of protein glycosylation can be effectively applied in early stage diagnostics of several diseases, including different forms of cancer with known glycan-based biomarkers.14 Moreover, many previously established and even commercially successful strategies used to treat diseases are currently being revisited in light of glycan recognition in order to lower side effects, enhance serum half-life or to decrease cellular toxicity.3,15 Recently, the first glyco-engineered antibody was approved to the market, what was called by the authors “a triumph for glyco-engineering”.16
Glycomics focuses on revealing finely tuned reading mechanisms in the cell orchestra based on graded affinity, avidity and multivalency of glycans (i.e. sugar chains covalently attached to proteins and lipids).17 Glycans are ideal information coding tools since they can form enormous numbers of possible unique sequences from basic building units. The theoretical number of all possible hexamers for glycans is 8 orders of magnitude larger than the theoretical number of peptides and 11 orders of magnitude larger than the theoretical number of DNA sequences.18 It is estimated that the size of the cellular glycome can be up to 500,000 glycan modified biomolecules (proteins and lipids) formed from 7,000 unique glycan sequences.19 This variation can explain human complexity in light of a paradoxically small genome. This glycan complexity together with similar physico-chemical properties of glycans is the main reason why progress in the field of glycomics has been behind advances in genomics and proteomics.20
Traditional glycoprofiling protocols rely on glycan release from a biomolecule with subsequent quantification by an array of techniques including capillary electrophoresis, liquid chromatography and mass spectrometry.21 There is an alternative way for glycoprofiling by application of lectins (natural glycan recognizing proteins18,22) in combination with various transducing protocols.11b,23 The most powerful glycoprofiling tool relies on lectins arrayed on solid surfaces for direct analysis of glycoproteins, glycolipids, membranes and even glycans on the surface of intact cells.24 Even though lectin microarrays offer high throughput assay protocols with a minute consumption of samples and reagents, there are some drawbacks such as the need to fluorescently label the sample or the lectin, which negatively affects the performance of detection11a,b, relatively high detection limits and quite narrow working concentration ranges.
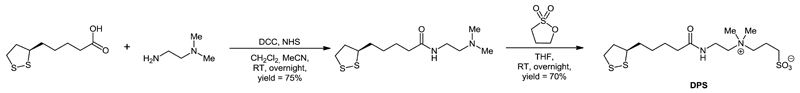
The use of nanotechnology, sophisticated patterning protocols and advanced detection platforms can help overcome the drawbacks of lectin microarray technology allowing it to work in a label-free mode of operation, with high sensitivity, low detection limits, a wide concentration window and in some cases, real time analysis of a binding event is possible.10b,25 In our recent work we focused on development of ultrasensitive impedimetric lectin biosensors with detection limits down to the single-molecule level based on controlled architecture at the nanoscale25e,26, but such biosensors were prone to non-specific interactions. The main aim of this manuscript was to develop a patterning protocol based on a mixed SAM layer containing one derivative available for covalent immobilisation of lectins and a sulfobetaine derivative (DPS, Scheme 1) forming an interfacial layer (Fig. S1) effectively blocking non-specific interactions.27 This strategy allowed us to detect changes in a glycoprofile in complex human samples with a detection limit down to fM level. The biosensors, based on three different lectins, were calibrated using 4 different glycoproteins (Fig. S2) and finally its reliability was proved in complex samples, suggesting this concept can be integrated into an array format of analysis.
Scheme 1.
Synthesis of DPS.
Experimental section
Chemicals
11-mercaptoundecanoic acid (MUA), potassium hexacyanoferrate(III), potassium hexacyanoferrate(II) trihydrate, sodium chroride, potassium chloride, 1,3-propane sultone, N-hydroxysuccinimide (NHS), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), sodium sulphate, N,N-dimethylethylene diamine, dicyclohexylcarbdiimide (DCC), sodium periodate, ethylene glycol, ethanolamine, acetonitrile, dichloromethane, tetrahydrofuran (THF), glycine, hydrochloric acid, Tween 20, fetuin (FET, 8.7% of N-acetylneuraminic acid), asialofetuin (ASF, ≤ 0.5% of N-acetylneuraminic acid), invertase from baker´s yeasts (INV), transferrin (TRF), Ricinus communis agglutinin (handle with special care since it is a toxin) and concanavalin A were purchased from Sigma Aldrich (USA). (R)-Lipoic acid was purchased from TCI Europe. SNA I lectin from Sambucus nigra was purchased from Gentaur (Belgium). Ethanol for UV/VIS spectroscopy (ultra pure) was purchased from Slavus (Slovakia). Zeba™ spin desalting columns (40k MWCO) for protein purification were purchased from Thermo Scientific (UK). All buffer components were dissolved in deionised water (DW).
Preparation of a biorecognition surface
On an electrode modified by a mixed SAM, as described in Supporting information, the biorecognition elements (different lectins) were immobilised using standard amine coupling with the carboxylic groups of MUA activated by a 1 to 1 mixture of 0.2 M EDC and 0.05 M NHS. The incubation of the electrode in a mixture of EDC/NHS took 15 min and the surface was washed by DW. Lectins were covalently immobilised on the activated SAM layer in a 10 μM stock solution (40 µl) in a 10 mM phosphate buffer solution (PBS) pH 7.0 containing 0.05% Tween 20 by incubation for 1 h in the dark and at a room temperature. After the immobilisation was completed, the surface was gently rinsed by DW and incubated with 10 mM M HCl for 3 min, rinsed by DW, incubated with 10 mM glycine-HCl (pH 2.5) for 3 min and finally again rinsed by DW. This procedure was introduced for removal of non-covalently bound lectin molecules from the electrode.28 Incubation of the biosensor with the analyte was performed by incubation with a stock solution of a glycoprotein or a sample (40 µl) for 20 min.
Assay procedure
All electrochemical impedance spectroscopy (EIS) measurements were performed in an electrolyte containing 5 mM potassium hexacyanoferrate(III), 5 mM potassium hexacyanoferrate(II) and 0.1 M KCl. The analysis was run at 50 different frequencies (ranging from 0.1 Hz up to 100 kHz) under Nova Software 1.9 (Ecochemie, Netherlands). The acquired data were evaluated by the same software using a Nyquist plot with an equivalent circuit R(C[RW]), employed. The charge transfer resistance (RCT) parameter was used as the measure for the calibration of the biosensor and for real sample measurements. Each analyte/sample was measured at least in triplicate with an independent biosensor device and results are shown with a standard deviation (± SD) calculated in Excel. Human serum samples were diluted in 10 mM PBS buffer, pH 7.4. All stock solutions (lectins, standard glycoproteins and human sera) were stored at -20 °C in aliquots.
Results and discussion
Characterisation of SAM modified gold surface
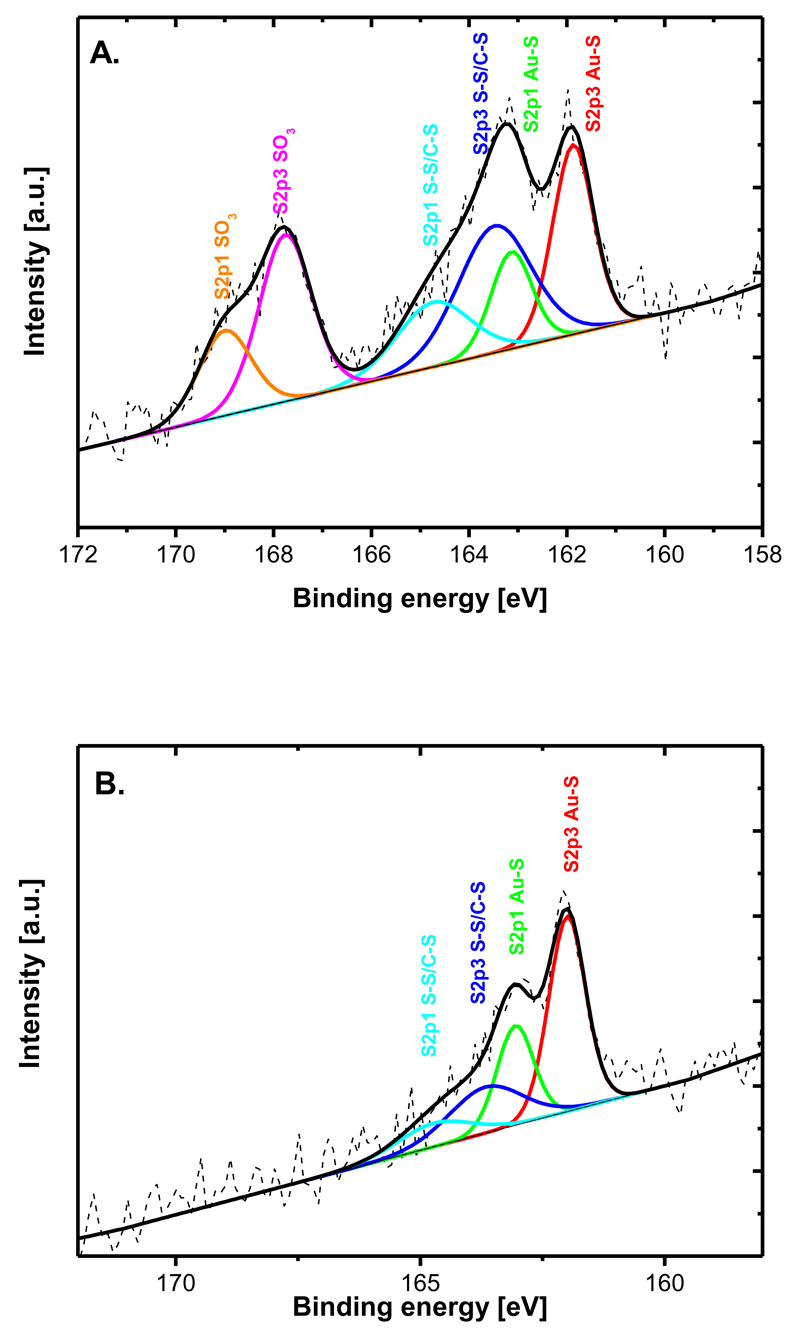
In order to get information about the surface coverage of DPS within a single and mixed SAM layers on a planar gold surface, characterisation by XPS and an electrochemical reduction of the SAM were applied. XPS data were fitted in a way to fit the S2p3 spectrum with two doublets at 161.9 eV and 163.5 eV, respectively, according to a previous study.29 XPS showed a clear difference in the presence of various functionalities. When pure SAM layers of MUA and DPS were compared (Fig. 1), peaks attributed to SO32- and S-S groups were observed only in the spectra of DPS (Fig. 1A). More detailed analysis of a mixed SAM composed of MUA and DPS by XPS revealed increased amounts of DPS (signal from SO32- group was plotted) as the amount of DPS in a mixture with MUA was decreasing with the highest surface amount reached at a ratio of DPS:MUA of 1 to 1 (Fig. S3).
Fig. 1.
XPS of two different SAM layers showing present functional groups. A) pure SAM from a sulfobetaine derivative and B) pure SAM from MUA.
The results from reductive desorption of thiols can be used to get quantitative information about the surface coverage of thiols.30 A pure DPS SAM layer revealed a surface density of 1.97 molecules nm-2, a value that is in excellent agreement with values from 1.8 to 2.1 molecules nm-2 obtained previously for thioctic acid31. Moreover, quantification of DPS in mixed SAM layers by reductive desorption was possible since the onset potential of reductive desorption of DPS and MUA was different. Analysis of a mixed SAM sample by reductive desorption provided a value of 2.66 molecules nm-2 of DPS in a SAM prepared from a 1 to 1 mixture of MUA and DPS (Fig. S3). Higher surface coverage of DPS within a mixed SAM compared to a pure SAM can be explained by a lower repulsion between SO3- groups of neighbouring DPS molecules present in the mixed SAM. Recently it was observed that the density of a mixed SAM composed of 2-aminoethanethiol and 2-mercaptoethane sulfonic acid is higher compared to corresponding pure monolayers, what was ascribed to the strong molecular interactions between these two components.32 There are other reports describing a strong interaction between a SO3- terminated thiol and a diluting one terminated in –NH2 or –OH functional group influencing contact angle or zeta potential of mixed SAMs with changes in the monolayer composition.33
QCM experiments
A surface coverage of 11.3 pmol cm-2 for Con A lectin immobilised on the surface prepared from a 1 to 1 mixture of MUA and DPS was obtained from a QCM experiment (Fig. S4). Con A is composed of four identical subunits (Fig. S5). When a hard sphere model34 was applied for calculation of a theoretical surface coverage of a Con A tetramer, a value of 4.8 pmol cm-2 was calculated. This calculated result does not correspond to the value obtained from a QCM experiment. Since Con A can dissociate into subunits under certain physico-chemical conditions35, we tried to calculate what would be a theoretical surface coverage for a monomer of Con A, revealing a value of 12.1 pmol cm-2, applying the same hard sphere model. These calculations suggest Con A dissociates into subunits after incubation with a heavily negatively charged surface occupying almost a full monolayer.
Since INV has 18 glycosylation sites with glycans terminated in mannose units23a we ran QCM measurements to see if it is possible to form a sandwich configuration. In this experiment, the Con A biosensor was incubated with INV and then Con A was injected over the biorecognition layer to complete a sandwich configuration (Fig. S6). The QCM experiment showed that INV was bound to the Con A biosensor with a surface density of 0.67 pmol cm-2 (Fig. S7), revealing a molar ratio INV/Con A of 0.055. This is in a good agreement with values of molar ratio from 0.02 to 0.17 obtained on various peptide aptamer surfaces.36 The second Con A layer on a layer of INV was formed with a surface density of 2.75 pmol cm-2 (Fig. S7), suggesting that every INV molecule on average interacted with 4 molecules of Con A present in the outer layer. There are two possible outputs of this observation. The first one is the observed decrease in the detection limit of the Con A biosensor, which is further explored in section The Con A biosensor. The second more important application would be the formation of a sandwich configuration with a possibility to vary lectins involved in the formation of a 2nd lectin layer. Such an approach can provide additional information about possible changes in the glycan profile at different glycosylation sites, which can be a useful tool in more detailed glycoprofiling of various proteins applicable in diagnostics.
AFM experiments
AFM images showed the surface is densely populated by Con A molecules (Fig. 2). A detailed analysis of Con A spots revealed the presence of different forms of Con A with the heights of h=(3.6 ± 0.4) nm with a full width at half maximum FWHM=(18 ± 1) nm, h=(5.4 ± 0.7) nm with FWHM=(26 ± 4) and h=(7.6 ± 0.4) nm with FWHM=(26 ± 4). These results suggest besides the presence of monomers of Con A, most likely other forms of Con A such as dimers and tetramers (intact Con A molecules), might be present, as well. Indication that the spot of Con A with h=7.6 nm is an intact Con A tetramer is supported by the thickness of Con A layer of 7.7 nm previously published.37 For example, a Con A monomer38 has the size 42 x 40 x 39 Å, a dimer of Con A39 has the size of 61 x 86 x 91 Å and a tetramer39 of Con A has the size of 67 x 113 x 122 Å. The lateral size of all Con A spots in AFM images is larger due to the tip convolution effect.40 Moreover, a high density of Con A monomers on the surface is in good agreement with a similar conclusion made from the QCM results about preferential adsorption of Con A on the surface in a monomer form. This is supported by calculations made from a hard sphere model, when high fraction dimers and tetramers of Con A on the surface cannot make the surface coverage as read from the QCM experiment. A detailed analysis of AFM images revealed an increase of the roughness factor Rq from 0.8 nm for bare gold to 1.0 nm for gold modified by the SAM prepared from a 1 to 1 mixture of MUA and DPS (Fig. S8). Moreover, the surface roughness increased significantly to a value of 1.8 nm after immobilisation of Con A lectin (Fig. 2).
Fig. 2.
AFM image of the surface after immobilisation of Con A.
The Con A biosensor
The oxidised form of INV (oxINV) was prepared using sodium periodate in order to “destroy” glycan determinants on a protein backbone. This oxidised protein was used as a negative control during characterisations of the Con A biosensor. The FTIR spectra of oxINV showed a decrease in the peaks corresponding to -OH groups (3250-3350 nm) and a small peak appeared at 1738 nm, attributed to the presence of free aldehyde groups (data not shown) after oxidation.
The performance of the Con A biosensor was tested on five different surfaces prepared from a mixture of MUA:DPS having different molar ratio in a liquid phase (1:0, 3:1, 1:1, 1:3 and 0:1). When, the Con A biosensor was built on a pure MUA layer (1:0) incubation with INV and oxINV did not provide a well resolved semicircle in a Nyquist plot and thus RCT could not be obtained, as already shown26a. The Con A biosensor prepared on a SAM from a 3:1 mixture did not discriminate between INV and oxINV, suggesting that the response is only due to non-specific interaction e.g. electrostatic interactions with predominant –COOH groups (Fig. S9A). The Con A biosensor prepared on the SAM from a 1:1 and 1:3 mixture showed the ability of the Con A biosensor to selectively detect INV (Fig. 3 and Fig. S9B), but the Con A biosensor built-up on a surface deposited from 1:1 MUA:DPS mixture provided better performance (Fig. 3). The Con A biosensor prepared on a pure DPS SAM layer was completely resistant to any protein binding (Fig. S9C). Thus, for further experiments the Con A biosensor prepared on a mixed SAM layer from 1:1 mixture MUA:DPS was used, showing excellent selectivity for INV in comparison to oxINV detection (Fig. S10).
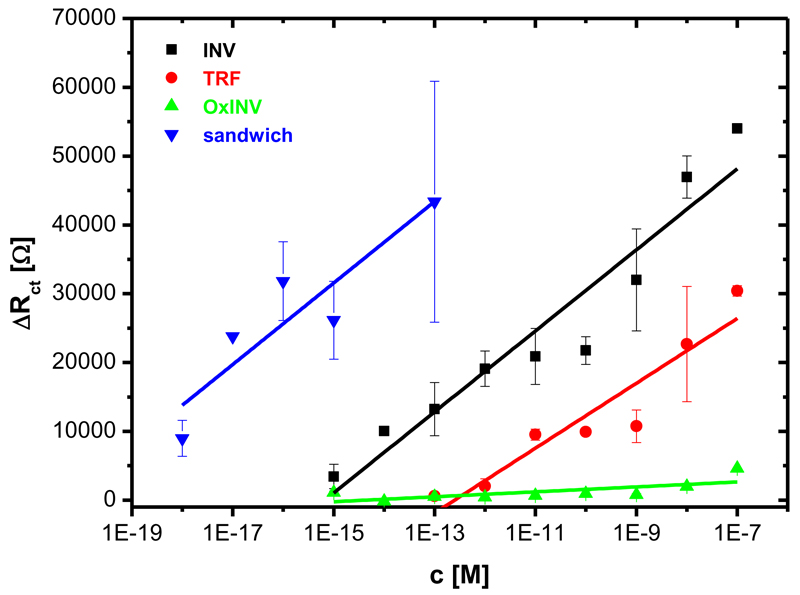
Fig. 3.
Calibration of the Con A impedimetric biosensor with various glycoproteins such as invertase (INV), transferrin (TRF) and oxidised invertase (oxINV) applied as a control. Moreover, a calibration for the sandwich biosensor configuration with additional outer lectin layer applied (see Fig. S6) is shown, as well. Each analyte was measured at least in triplicate with an independent biosensor device and results are shown with a standard deviation (± SD) calculated in Excel.
Besides strong interaction Con A exhibits with oligomannose glycans (Scheme 1) such as on INV (KD in the range 0.1-0.4 μM)23a, Con A also interacts weakly with complex glycans containing few mannose units and terminated in galactose or N-acetylneuraminic acid (Scheme 1) such as on TRF (KD in the range 1-3 μM)23a. Thus, the Con A biosensor was calibrated with 3 different glycoproteins INV, oxINV and TRF, showing only 6.1% of the sensitivity for oxINV in comparison to INV (Fig. 3). Moreover, the Con A biosensor showed 68% of the sensitivity for TRF in comparison to INV and a detection limit for TRF that was significantly shifted to higher values (≈ 1 pM), when compared to the detection limit for INV (≈ 1 fM). Linear range for the Con A biosensor towards INV was calculated from concentration at which the signal for INV was above the signal for oxINV, considered to be blank. Quite a wide linear range spanning 4 orders of magnitude for biosensors based on EIS was observed41, but our lectin biosensor offering linear response over 8 orders of magnitude has a feature essential for analysis of glycoproteins in complex samples where the amount of glycoproteins can vary over a large concentration window.
As already indicated from the QCM measurements, formation of a sandwich configuration has the potential to further decrease the detection limit of the Con A biosensor down to ≈ 1 aM, what is clearly shown in Fig. 3. The detection limit of our biosensor is much lower than for any lectin biosensor prepared to date10b,35, except for our recent lectin biosensor based on a 3-D nanointerface.26b The best detection limit for the lectin biosensors prepared by other groups was 20 fM41b or 150 fM41a. Moreover, the biosensor offers a linear response in the concentration window of at least 5 orders of magnitude.
The SNA and RCA lectin biosensors
The composition of an interfacial SAM layer being optimal for the Con A biosensor (1:1 MUA:DPS) was chosen for construction of the SNA and RCA biosensors, since all lectins have approximately the same size (112-140 kDa). Linear range for the SNA and RCA biosensors were calculated from concentration at which the signal for the analyte was above the background signal, shown as a thin line (Fig. 4 and Fig. 5).
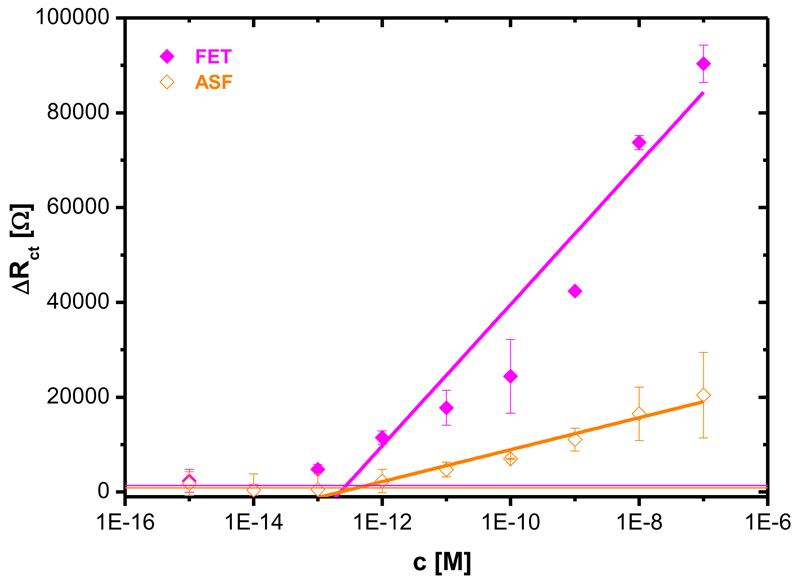
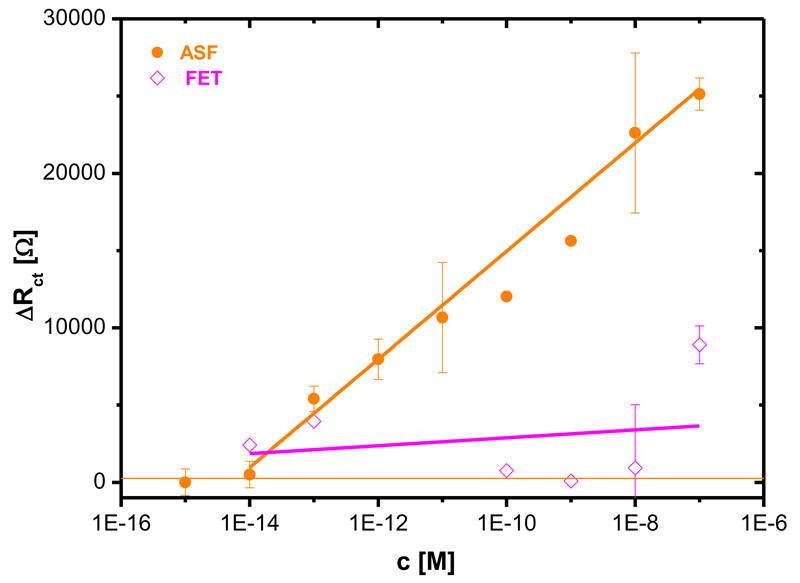
Fig. 4.
Calibration of the SNA biosensor with two glycoproteins (FET: fetuin with 8.7% of N-acetylneuraminic acid, ASF: asialofetuin with 0.5% of N-acetylneuraminic acid). Each analyte was measured at least in triplicate with an independent biosensor device and results are shown with a standard deviation (± SD) calculated in Excel.
Fig. 5.
Calibration of the RCA biosensor with two glycoproteins (FET: fetuin with 8.7% of N-acetylneuraminic acid, ASF: asialofetuin with 0.5% of N-acetylneuraminic acid and galactose exposed). Each analyte was measured at least in triplicate with an independent biosensor device and results are shown with a standard deviation (± SD) calculated in Excel.
The SNA biosensor recognising N-acetylneuraminic acid was calibrated with two analytes FET (8.7% of N-acetylneuraminic acid) and ASF (≤ 0.5% of N-acetylneuraminic acid). The SNA biosensor started to interact with FET at a concentration of 100 fM, while ASF was recognised by the SNA biosensor at a concentration of 1 pM (Fig. 4). The sensitivity ratio of the SNA biosensor for these two analytes derived from the linear part of their concentration dependence, of 4.8 is in agreement with a previously obtained value of 7.6.26b
The RCA biosensor detecting terminal galactose on glycans was calibrated with FET and ASF, as well. ASF, which is a better analyte for the RCA biosensor compared to FET, was detected at concentration of 10 fM with a much higher sensitivity (13x) compared to a quite scattered response of the RCA biosensor for FET (Fig. 5). Quite scattered signal for low-affinity analytes was observed by others, as well, when ELISA-like41b and EIS analytical protocols41 with lectins were applied. This might be caused by a subtle change in the washing protocol in between measurements.
Lectin microarray
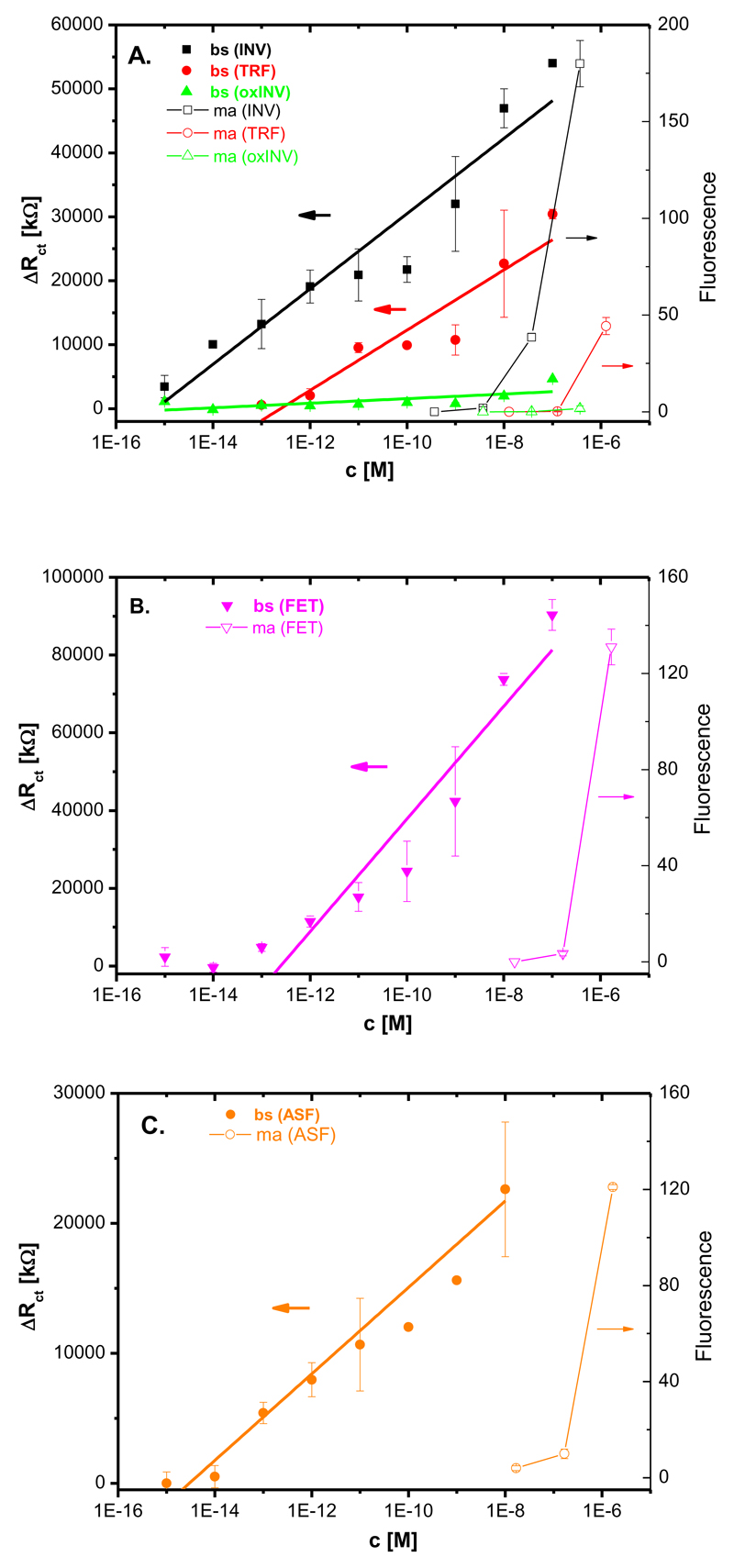
The lectin biosensors were validated against state-of-the-art glycoprofiling tool, a lectin microarray with all lectins used for comparison (Fig. S11). Con A was incubated with three different glycoproteins (INV, oxINV and TRF) spotted on the chip and three different concentrations (0.1; 0.01 and 0.001 mg ml-1) were quantified. SNA was incubated with FET and ASF (data not shown due to low signal) spotted and RCA was incubated with FET and ASF spotted at the same concentrations and results were quantified. Analysis of the lectin microarray was applied for construction of calibration curves, which were compared to calibration curves of all lectin biosensors (Fig. 6), indicating that all lectin biosensors offered few orders of magnitude lower detection limits for their analytes and a much wider working concentration range compared to the lectin microarray.
Fig. 6.
A direct comparison of working concentration range for the lectin biosensors (bs) and lectin microarray (ma) applying A) Con A lectin, B) SNA lectin and C) RCA lectin. Each analyte was measured at least in triplicate with an independent biosensor device and results are shown with a standard deviation (± SD) calculated in Excel.
Analysis of human samples
Furthermore, the application potential of the lectin biosensors was tested for the analysis of human samples from healthy individuals and from those suffering rheumatoid arthritis. At first, an optimal dilution of human serum was needed to be found in order to detect glycoproteins with high sensitivity, but at the same time lowering the effect of non-specific interactions from such complex samples. A dilution of the human serum samples 1:1000 was selected since at this dilution, non-specific response of the sample on the reference surface without any lectin immobilised was negligible, while response with the Con A biosensor was well above non-specific signal (Fig. S12).
The performance of the Con A and SNA biosensors was further validated by standard addition of 1 pM INV and 1 pM FET to human serum diluted 1:1000, respectively. The results showed a recovery of 91% for the Con A biosensor with added INV and of 72% for the SNA biosensor with added FET. A relatively low recovery index for FET might indicate quite quick (1 h of incubation) formation of complexes of FET with other proteins present in human serum via N-acetylneuraminic acid (attached to FET), which is quite often involved in a wide range of interactions within a cell or with other cells; or removal of terminal N-acetylneuraminic acid from FET by sialidases.10a,42 Thus, the lectin biosensor can be applied for kinetic analysis of the fate of various glycoproteins in complex samples such human serum, which might have physiological consequences.
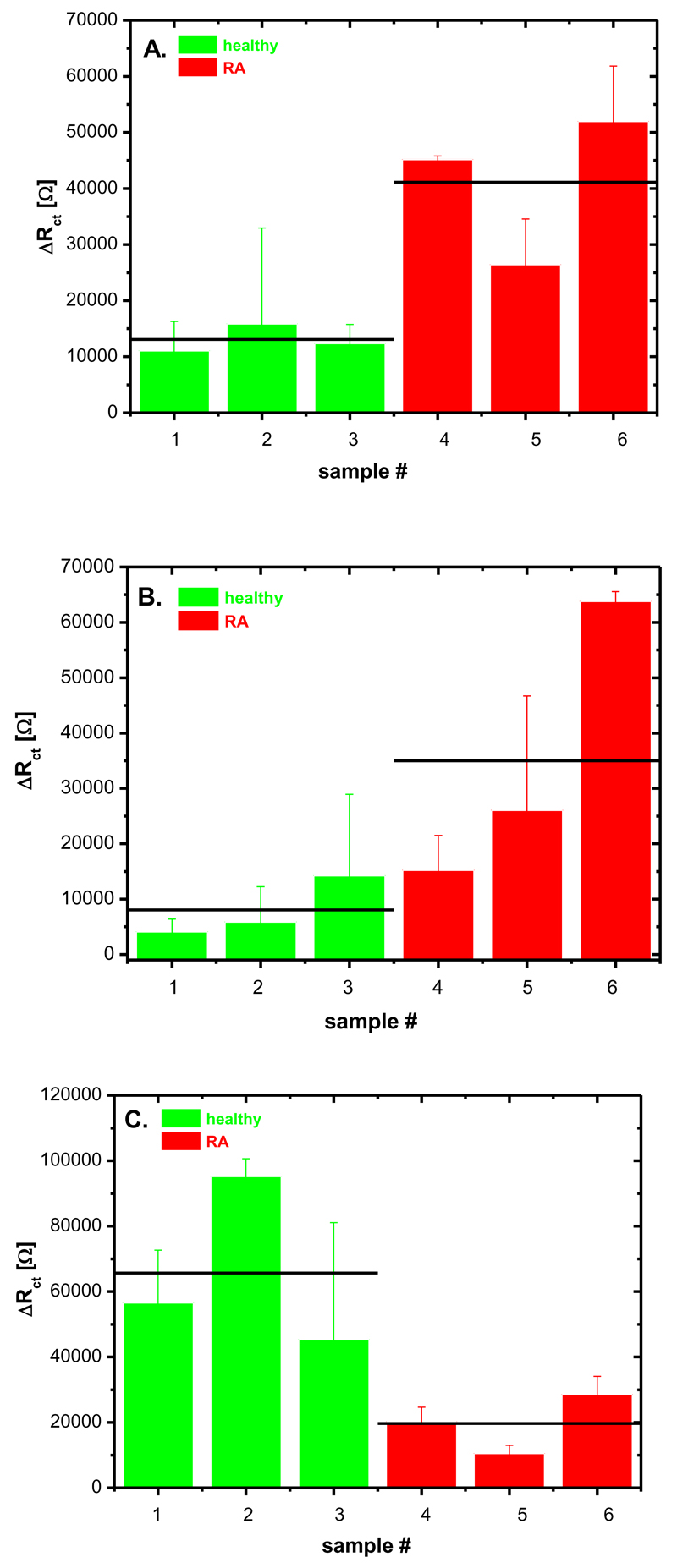
Finally, all three lectin biosensors were applied in the analysis of three human serum samples from healthy individuals and three human serum samples from people affected by rheumatoid arthritis (RA). Circulating antibodies (IgG) in the blood carry complex type glycans of a biantennary structure as shown in the graphical abstract. These biomolecules are hardly ever completed with both N-acetylneuraminic acids present and in patients with RA galactose or even N-acetylglucosamine can be exposed.10a The severity of the RA disease tends to correlate with the extent of the glycosylation change.10a Results obtained by the application of the lectin biosensors corroborated prior results that serum samples from healthy individuals had weaker signal on Con A biosensor compared to samples from patients with RA indicating that mannose units are not that exposed on IgG in serum from healthy individuals (Fig. 7A). The signal on the RCA biosensor showed a similar pattern as in the case of the Con A biosensor, suggesting healthy individuals do not have exposed galactose residues available for RCA lectin binding (Fig. 7B). The SNA biosensor exhibited higher signals for samples from healthy individuals compared to the RA samples, suggesting in healthy individuals, N-acetylneuraminic acid is still present (Fig. 7C). Further experiments are needed to see if lectin biosensors can be applied for the analysis of the severity of RA disease from glycoprofiling of human serum. In order to achieve this goal, the signal from the lectin biosensors has to be correlated with standard clinical methods, an effort, which is currently under way.
Fig. 7.
Glycoprofile of three human serum samples from healthy individuals and three samples from patients with rheumatoid arthritis (RA) analysed by the A) Con A biosensor, B) RCA biosensor and C) SNA biosensor. Each sample was measured at least in triplicate with an independent biosensor device and results are shown with a standard deviation (± SD) calculated in Excel.
Conclusions
A mixed SAM composed of a newly synthesized thioctic acid derivative DPS and functional thiol MUA provided an interface resisting non-specific interactions, while allowing covalent immobilisation of lectins. The lectin biosensors prepared by immobilisation of three different lectins on the gold electrode surface provided high sensitivity of detection of glycoproteins with a detection limit down to the fM level with a wide linear range of operation. The study suggests lectin biosensors outperform lectin microarrays in terms of sensitivity and utilisable working concentration range with a great potential of the lectin biosensors for searching for new disease biomarkers, which can be present in biological samples at extremely low concentrations. Moreover, reliability of biosensing was tested by standard addition with recovery of 91% for INV on the Con A biosensor and 72% for FET on the SNA biosensor. Surprisingly low recovery index for FET on the SNA biosensor might indicate an interaction with serum components via N-acetylneuraminic acid or removal of N-acetylneuraminic acid from the FET by the action of sialidase. Thus, the lectin biosensors can be applied for monitoring of kinetic parameters of interaction of a particular glycoprotein with this sample, which have a diagnostic value. Finally, comparison of a glycoprofile of serum samples from healthy individuals and those having RA showed distinct glycoprofile differences with a potential to detect severity of disease progression in the future.
Supplementary Material
Supporting Information. The file provides information about electrode pre-treatment and SAM formation; human samples; synthesis of sulfobetaine; oxidation of the glycan; experimental procedures (lectin arrays, QCM, XPS, FTIR and AFM); results from QCM experiments; structure of Con A lectin; structure of glycans on standard glycoproteins; image of a sandwich biosensor configuration; performance of the Con A biosensor built on various SAM interfaces; representative Nyquist plot for the Con A biosensor; raw data from lectin microarray and figure showing optimisation of dilution of human sera. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgment
The financial support from the Slovak scientific grant agency VEGA 2/0127/10 and from the Slovak research and development agency APVV 0282-11 is acknowledged. The research leading to these results has received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement n. 311532.
Abbreviations
- ASF
asialofetuin
- Con A
concanavalin A
- DPS
(R)-3-((2-(5-(1,2-dithiolan-3-yl)pentanamido)ethyl)dimethylammonio)propane-1-sulfonate
- DW
deionised water
- EIS
electrochemical impedance spectroscopy
- FET
fetuin
- FTIR
Fourier transform infrared spectroscopy
- GAL
galactose
- INV
invertase
- MAN
mannose
- MUA
11-mercaptoundecanoic acid
- OxINV
oxidised invertase
- QCM
quartz crystal microbalance
- RCA
Ricinus communis agglutinin
- RT
room temperature
- SAM
self-assembled monolayer
- SNA I
Sambucus nigra agglutinin
- TRF
transferrin
- XPS
X-ray photoelectron spectroscopy.
References
- 1.Schena M, Shalon D, Davis RW, Brown PO. Science. 1995;270:467. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Eisen MB, Spellman PT, Brown PO, Botstein D. Proceedings of the National Academy of Sciences. 1998;95:14863. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, et al. Nature. 2000;403:503. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Gygi SP, Rochon Y, Franza BR, Aebersold R. Molecular and Cellular Biology. 1999;19:1720. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gry M, Rimini R, Stromberg S, Asplund A, Ponten F, Uhlen M, Nilsson P. BMC Genomics. 2009;10:365. doi: 10.1186/1471-2164-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) MacBeath G, Schreiber SL. Science. 2000;289:1760. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]; (b) Zhu H, Snyder M. Current Opinion in Chemical Biology. 2003;7:55. doi: 10.1016/s1367-5931(02)00005-4. [DOI] [PubMed] [Google Scholar]; (c) Lee J-R, Magee DM, Gaster RS, LaBaer J, Wang SX. Expert Review of Proteomics. 2013;10:65. doi: 10.1586/epr.12.67. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Rusmini F, Zhong Z, Feijen J. Biomacromolecules. 2007;8:1775. doi: 10.1021/bm061197b. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Science. 2001;293:1074. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Nature. 1995;376:313. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 9.Arnaud J, Audfray A, Imberty A. Chemical Society Reviews. 2013 doi: 10.1039/c2cs35435g. [DOI] [PubMed] [Google Scholar]
- 10.(a) Varki A. Essentials of glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2009. [PubMed] [Google Scholar]; (b) Bertók T, Katrlík J, Gemeiner P, Tkac J. Microchimica Acta. 2013;180:1. doi: 10.1007/s00604-012-0876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Gemeiner P, Mislovicová D, Tkác J, Svitel J, Pätoprst1 V, Hrabárová E, Kogan G, Kozár T. Biotechnology Advances. 2009;27:1. doi: 10.1016/j.biotechadv.2008.07.003. [DOI] [PubMed] [Google Scholar]; (b) Katrlík J, Švitel J, Gemeiner P, Kožár T, Tkac J. Medicinal Research Reviews. 2010;30:394. doi: 10.1002/med.20195. [DOI] [PubMed] [Google Scholar]; (c) Hirabayashi J. Nat Chem Biol. 2009;5:198. doi: 10.1038/nchembio0409-198. [DOI] [PubMed] [Google Scholar]; (d) Krishnamoorthy L, Bess JW, Preston AB, Nagashima K, Mahal LK. Nat Chem Biol. 2009;5:244. doi: 10.1038/nchembio.151. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Schauer R, Kamerling JP. ChemBioChem. 2011;12:2246. doi: 10.1002/cbic.201100421. [DOI] [PubMed] [Google Scholar]; (f) Song X, Lasanajak Y, Xia B, Heimburg-Molinaro J, Rhea JM, Ju H, Zhao C, Molinaro RJ, Cummings RD, Smith DF. Nat Meth. 2011;8:85. doi: 10.1038/nmeth.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. Science. 2011;334:255. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Burton DR, Poignard P, Stanfield RL, Wilson IA. Science. 2012;337:183. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Doores KJ, Fulton Z, Hong V, Patel MK, Scanlan CN, Wormald MR, Finn MG, Burton DR, Wilson IA, Davis BG. Proceedings of the National Academy of Sciences. 2010;107:17107. doi: 10.1073/pnas.1002717107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Pejchal R, Doores KJ, Walker LM, Khayat R, Huang P-S, Wang S-K, Stanfield RL, Julien J-P, Ramos A, Crispin M, Depetris R, et al. Science. 2011;334:1097. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Anthony RM, Kobayashi T, Wermeling F, Ravetch JV. Nature. 2011;475:110. doi: 10.1038/nature10134. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Science. 2008;320:373. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, et al. Nature. 2012;492:118. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Li F, Ravetch JV. Science. 2011;333:1030. doi: 10.1126/science.1206954. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Kim J-H, Resende R, Wennekes T, Chen H-M, Bance N, Buchini S, Watts AG, Pilling P, Streltsov VA, Petric M, Liggins R, et al. Science. 2013;340:71. doi: 10.1126/science.1232552. [DOI] [PubMed] [Google Scholar]
- 14.(a) Kim EH, Misek DE. International Journal of Proteomics. 2011;2011 doi: 10.1155/2011/343582. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ferens-Sieczkowska M, Kowalska B, Kratz EM. Biomarkers. 2013;18:10. doi: 10.3109/1354750X.2012.719035. [DOI] [PubMed] [Google Scholar]; (c) Chandler KB, Goldman R. Molecular & Cellular Proteomics. 2013 doi: 10.1074/mcp.R112.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Gilgunn S, Conroy PJ, Saldova R, Rudd PM, O'Kennedy RJ. Nat Rev Urol. 2013;10:99. doi: 10.1038/nrurol.2012.258. [DOI] [PubMed] [Google Scholar]
- 15.(a) Beck A, Sanglier-Cianférani S, Van Dorsselaer A. Analytical Chemistry. 2012;84:4637. doi: 10.1021/ac3002885. [DOI] [PubMed] [Google Scholar]; (b) Schmaltz RM, Hanson SR, Wong C-H. Chemical Reviews. 2011;111:4259. doi: 10.1021/cr200113w. [DOI] [PubMed] [Google Scholar]; (c) van Bueren JJL, Rispens T, Verploegen S, van der Palen-Merkus T, Stapel S, Workman LJ, James H, van Berkel PHC, van de Winkel JGJ, Platts-Mills TAE, Parren PWHI. Nat Biotech. 2011;29:574. doi: 10.1038/nbt.1912. [DOI] [PubMed] [Google Scholar]
- 16.Beck A, Reichert JM. mAbs. 2012;4:419. doi: 10.4161/mabs.20996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Nat Meth. 2005;2:817. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]
- 18.Gabius H-J, André S, Jiménez-Barbero J, Romero A, Solís D. Trends in biochemical sciences. 2011;36:298. doi: 10.1016/j.tibs.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Cummings RD. Molecular BioSystems. 2009;5:1087. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 20.Bertozzi CR, Kiessling LL. Science. 2001;291:2357. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 21.(a) Furukawa J-i, Fujitani N, Shinohara Y. Biomolecules. 2013;3:198. doi: 10.3390/biom3010198. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rakus JF, Mahal LK. Annual Review of Analytical Chemistry. 2011;4:367. doi: 10.1146/annurev-anchem-061010-113951. [DOI] [PubMed] [Google Scholar]; (c) Smith DF, Cummings RD. Molecular & Cellular Proteomics. 2013 doi: 10.1074/mcp.R112.027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.(a) Oliveira C, Teixeira JA, Domingues L. Critical Reviews in Biotechnology. 2013;33:66. doi: 10.3109/07388551.2012.670614. [DOI] [PubMed] [Google Scholar]; (b) Murphy P, André S, Gabius H-J. Molecules. 2013;18:4026. doi: 10.3390/molecules18044026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(a) Mislovičová D, Katrlík J, Paulovičová E, Gemeiner P, Tkac J. Colloids and Surfaces B: Biointerfaces. 2012;94:163. doi: 10.1016/j.colsurfb.2012.01.036. [DOI] [PubMed] [Google Scholar]; (b) Mislovičová D, Gemeiner P, Kozarova A, Kožár T. Biologia. 2009;64:1. [Google Scholar]
- 24.(a) Hirabayashi J, Kuno A, Tateno H. ELECTROPHORESIS. 2011;32:1118. doi: 10.1002/elps.201000650. [DOI] [PubMed] [Google Scholar]; (b) Hirabayashi J, Yamada M, Kuno A, Tateno H. Chemical Society Reviews. 2013 doi: 10.1039/c3cs35419a. [DOI] [PubMed] [Google Scholar]; (c) Krishnamoorthy L, Mahal LK. ACS Chemical Biology. 2009;4:715. doi: 10.1021/cb900103n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(a) Reuel NF, Mu B, Zhang J, Hinckley A, Strano MS. Chemical Society Reviews. 2012;41:5744. doi: 10.1039/c2cs35142k. [DOI] [PubMed] [Google Scholar]; (b) Q GJ, Stephen C, Marian K, Lokesh J. Biochemical Society Transactions. 2010;38:1333. [Google Scholar]; (c) Cunningham S, G JQ, Kane M, Joshi L. Analyst. 2010;135:2471. doi: 10.1039/c0an00276c. [DOI] [PubMed] [Google Scholar]; (d) Sanchez-Pomales G, Zangmeister RA. International Journal of Electrochemistry. 2011;2011 [Google Scholar]; (e) Reuel NF, Ahn J-H, Kim J-H, Zhang J, Boghossian AA, Mahal LK, Strano MS. Journal of the American Chemical Society. 2011;133:17923. doi: 10.1021/ja2074938. [DOI] [PubMed] [Google Scholar]; (f) Tkac J, Davis JJ. In: Engineering the Bioelectronic Interface: Applications to Analyte Biosensing and Protein Detection. Davis JJ, editor. Royal Society of Chemistry; Cambridge: 2009. [Google Scholar]
- 26.(a) Bertok T, Gemeiner P, Mikula M, Gemeiner P, Tkac J. Microchimica Acta. 2013;180:151. doi: 10.1007/-s00604-012-0902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bertok T, Sediva A, Katrlik J, Gemeiner P, Mikula M, Nosko M, Tkac J. Talanta. 2013;108:11. doi: 10.1016/j.talanta.2013.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.(a) Shen C-H, Lin J-C. Colloids and Surfaces B: Biointerfaces. 2013;101:376. doi: 10.1016/j.colsurfb.2012.07.035. [DOI] [PubMed] [Google Scholar]; (b) Kim D, Chae MK, Joo HJ, Jeong I-h, Cho J-H, Lee C. Langmuir. 2012;28:9634. doi: 10.1021/la300043m. [DOI] [PubMed] [Google Scholar]; (c) Kim G, Yoo CE, Kim M, Kang HJ, Park D, Lee M, Huh N. Bioconjugate Chemistry. 2012;23:2114. doi: 10.1021/bc300339b. [DOI] [PubMed] [Google Scholar]; (d) Chen S, Liu L, Jiang S. Langmuir. 2006;22:2418. doi: 10.1021/la052851w. [DOI] [PubMed] [Google Scholar]; (e) Ostuni E, Chapman RG, Liang MN, Meluleni G, Pier G, Ingber DE, Whitesides GM. Langmuir. 2001;17:6336. [Google Scholar]
- 28.Davis JJ, Tkac J, Laurenson S, Ferrigno PK. Analytical chemistry. 2007;79:1089. doi: 10.1021/ac061863z. [DOI] [PubMed] [Google Scholar]
- 29.Castner DG, Hinds K, Grainger DW. Langmuir. 1996;12:5083. [Google Scholar]
- 30.Tkac J, Davis JJ. Journal of Electroanalytical Chemistry. 2008;621:117. [Google Scholar]
- 31.(a) Volkert AA, Subramaniam V, Ivanov MR, Goodman AM, Haes AJ. ACS Nano. 2011;5:4570. doi: 10.1021/nn200276a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dong Y, Abaci S, Shannon C, Bozack MJ. Langmuir. 2003;19:8922. [Google Scholar]
- 32.Ooi Y, Hobara D, Yamamoto M, Kakiuchi T. Langmuir. 2005;21:11185. doi: 10.1021/la051160x. [DOI] [PubMed] [Google Scholar]
- 33.(a) Shen C-H, Lin J-C. Langmuir. 2011;27:7091. doi: 10.1021/la200906b. [DOI] [PubMed] [Google Scholar]; (b) Shen CH, Lin JC. In: Lim C, Goh JH, editors. 13th International Conference on Biomedical Engineering; Springer Berlin Heidelberg; 2009. [Google Scholar]
- 34.Lahiri J, Isaacs L, Tien J, Whitesides GM. Analytical Chemistry. 1999;71:777. doi: 10.1021/ac980959t. [DOI] [PubMed] [Google Scholar]
- 35.Cairo CW, Gestwicki JE, Kanai M, Kiessling LL. Journal of the American Chemical Society. 2002;124:1615. doi: 10.1021/ja016727k. [DOI] [PubMed] [Google Scholar]
- 36.Davis JJ, Tkac J, Humphreys R, Buxton AT, Lee TA, Ko Ferrigno P. Analytical chemistry. 2009;81:3314. doi: 10.1021/ac802513n. [DOI] [PubMed] [Google Scholar]
- 37.Yonzon CR, Jeoung E, Zou S, Schatz GC, Mrksich M, Van Duyne RP. Journal of the American Chemical Society. 2004;126:12669. doi: 10.1021/ja047118q. [DOI] [PubMed] [Google Scholar]
- 38.Becker JW, Reeke GN, Wang JL, Cunningham BA, Edelman GM. Journal of Biological Chemistry. 1975;250:1513. [PubMed] [Google Scholar]
- 39.Bouckaert J, Poortmans F, Wyns L, Loris R. Journal of Biological Chemistry. 1996;271:16144. doi: 10.1074/jbc.271.27.16144. [DOI] [PubMed] [Google Scholar]
- 40.(a) Yu L, Huang M, Wang PG, Zeng X. Analytical Chemistry. 2007;79:8979. doi: 10.1021/ac071453q. [DOI] [PubMed] [Google Scholar]; (b) Zhang Y, Luo S, Tang Y, Yu L, Hou K-Y, Cheng J-P, Zeng X, Wang PG. Analytical Chemistry. 2006;78:2001. doi: 10.1021/ac051919+. [DOI] [PubMed] [Google Scholar]
- 41.(a) La Belle JT, Gerlach JQ, Svarovsky S, Joshi L. Analytical Chemistry. 2007;79:6959. doi: 10.1021/ac070651e. [DOI] [PubMed] [Google Scholar]; (b) Nagaraj V J, Aithal S, Eaton S, Bothara M, Wiktor P, Prasad S. Nanomedicine. 2010;5:369. doi: 10.2217/nnm.10.11. [DOI] [PubMed] [Google Scholar]
- 42.Cohen M, Varki A. Omics: a journal of integrative biology. 2010;14:455. doi: 10.1089/omi.2009.0148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.