Abstract
A DNA fragment containing the promoter of the mouse metallothionein-I gene fused to the structural gene of rat growth hormone was microinjected into the pronuclei of fertilized mouse eggs. Of 21 mice that developed from these eggs, seven carried the fusion gene and six of these grew significantly larger than their littermates. Several of these transgenic mice had extraordinarily high levels of the fusion mRNA in their liver and growth hormone in their serum. This approach has implications for studying the biological effects of growth hormone, as a way to accelerate animal growth, as a model for gigantism, as a means of correcting genetic disease, and as a method of farming valuable gene products.
The introduction of foreign genes into mammalian embryos could form the basis of a powerful approach for studying gene regulation and the genetic basis of development. Recent studies have clearly established the feasibility of introducing foreign DNA into the mammalian genome by microinjection of DNA molecules of interest into pronuclei of fertilized eggs, followed by insertion of the eggs into the reproductive tracts of foster mothers1–7. The data suggest that the so-called transgenic animals that develop from this procedure integrate the foreign DNA into one of the host chromosomes at an early stage of embryo development. As a result, the foreign DNA is generally transmitted through the germ line3,8,9. In some cases, expression of these foreign genes has also been detected; low levels of rabbit β-globin and viral thymidine kinase (TK) activity were measured in transgenic mice after introducing the respective genes6,7. It is possible to obtain regulated expression of viral TK by fusing the structural gene to the mouse metallothionein-I (MT-I) gene promoter and inducing expression with heavy metals2,9.
The possibility of introducing genes encoding secreted regulatory peptides into animals via this technology has several potential applications. The growth hormone (GH) gene whose protein product has profound developmental effects is particularly convenient for testing this possibility. Based on our success in achieving regulatable expression of herpes TK using the MT-I promoter9, we fused this promoter to the rat GH structural gene which has been shown to direct GH synthesis in mouse cells10. We show here that when this gene construct (MGH) is introduced into eggs it is expressed efficiently, giving rise to greater than normal levels of growth hormone in transgenic mice. The most obvious consequence of this high level of growth hormone expression is gigantism. This success raises several issues of genetic and practical importance.
Generation of animals carrying MGH fusion genes
A fragment of the cloned rat GH gene, from which the 5' regulatory region had been deleted, was fused to the MT-I promoter region, generating the plasmid pMGH (see Fig. 1a). In this construction, the fusion occurs in exon 1, retaining the initiation codon of the GH gene and the 5' regulatory region of the MT-I gene. Thus, the fusion gene is predicted to direct transcription of a mRNA containing 68 bases contributed by MT-I, followed by 1 base contributed by an XhoI linker, then the rat GH mRNA sequence starting at nucleotide 7. This construction preserves the AUG initiation codon for GH synthesis located at position 124–126 (Fig. 1c), the four intervening sequences and the poly(A) site of the rat GH gene (Fig. 1b), and 3 kilobases (kb) of downstream chromosomal sequences.
Fig. 1.
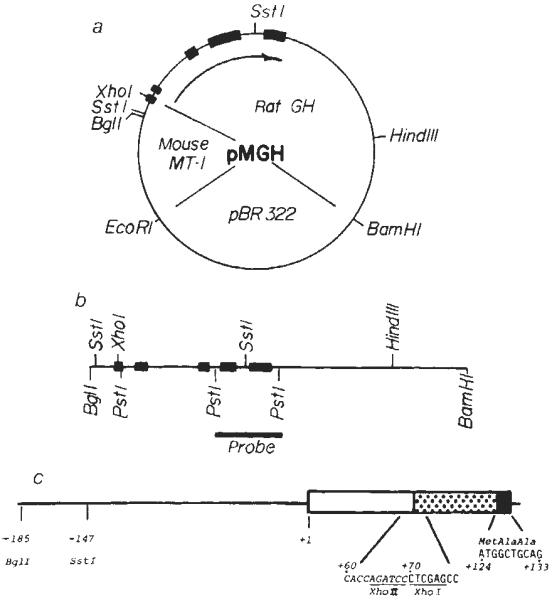
Construction of the fusion plasmid pMGH. a, The unique BglII site of the MT-I genomic clone28 was converted to a XhoI site by digesting with BglII, followed by filling in the sticky ends with Klenow fragment of DNA polymerase in the presence of all four dNTPs; then XhoI linkers (CCTCGAGG) were ligated to the blunt ends and bacterial strain RR1 was transformed with this DNA. The PvuII site of pBR322 was converted to a BamHI site by a similar procedure, then the 4.1-kb XhoI–BamHI fragment containing the MT-I promoter and pBR was ligated to a 4.8-kb XhoI–BamHI fragment containing the rat GH structural gene (see refs 10, 30) to give pMGH (8.9 kb). b, The fragment used for injection was a 5.0-kb BglI–BamHI fragment which was isolated from an agarose gel by the NaClO4 method29. For genomic Southern blots, a 1.0-kb PstI fragment spanning exons 4 and 5 was isolated and nick-translated9 and used as a hybridization probe. c, The predicted structure of exon 1 of the fusion gene. The line and open box represent MT-1 untranslated sequences, the stippled box represents GH untranslated sequences and the solid box represents the beginning of the GH coding region (see refs 28 and 30 for complete sequence information).
A 5.0-kb fragment extending from the BglI site of MT-I (−185) to BamHI (see Fig. 1b) was restricted from pMGH, separated from the other fragments on an agarose gel and used for injection into eggs. This linear fragment with heterologous ends was chosen because our recent experience suggests that such fragments integrate into host DNA more efficiently than supercoiled plsmids. Although this fragment contains only 253 base pairs (bp) of MT-I sequence, we have shown previously that this includes sequences essential for heavy metal regulation of the MT-I promoter11,12. The male pronuclei of fertilized eggs were microinjected with 2 pl containing ~600 copies of this fragment and 170 eggs were inserted into the reproductive tracts of foster mothers; 21 animals developed from these eggs2.
When they were weaned, total nucleic acids were extracted from a piece of tail and used for DNA dot hybridization to determine which animals carried MGH sequences. Seven of the animals gave hybridization signals above background (Fig. 2a) and their DNA was analysed further by restriction with SstI and Southern blotting. This analysis show ed that all seven had an intact 1.7-kb SstI fragment predicted from the restriction map shown in Fig. 1. All the animals having a prominent 1.7-kb hybridizing band also revealed a 3.3-kb band. This band is predicted from a circularized version of the 5.0-kb BglI–BamHI fragment. Two of the mice having multiple copies (MGH-10 and -19) gave a 5.0-kb hybridizing fragment when digested with HindIII, an enzyme that cuts only once within the injected fragment. Surprisingly, BamHI also generates a 5.0-kb hybridizing fragment in all the DNAs analysed (MGH-10, -14, -16 and -19), suggesting that this restriction site was restored during circularization, whereas BglI and EcoRI (which does not cut within the fragment) gave larger fragments (data not shown). We conclude that all the MGH-positive animals have at least one intact BglI–BamHI insert and four of the mice have a tandem head-to-tail duplication of this 5.0-kb fragment; however, the details of how and when this fragment circularized and integrated are not discernible. One of the mice, MGH-10, transmitted the MGH genes to half (10 of 19) of its offspring, suggesting that these genes are stably integrated into one of its chromosomes.
Fig. 2.
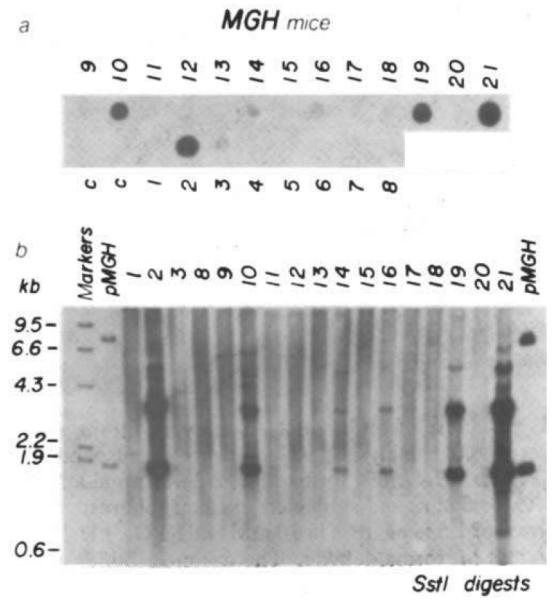
Analysis of DNA from transgenic mice for MGH sequences. a, DNA dot hybridization9 was used to detect mice with MGH sequences and quantitate their abundance. A nick-translated PstI probe (see Fig. 1b) was used (6 h exposure). Numbers correspond to the MGH mice examined; c, controls, b, DNA (5 μg) from MGH mice was restricted with SstI electrophoresed on a 1% agarose gel, transferred to nitrocellulose and hybridized with the nick-translated PstI probe shown in Fig. 1b. pMGH (13 pg, left and 130 pg, right) was included as a hybridization standard. Markers are HindIII-cut λ DNA end-labelled with 32P.
Rapid growth of mice carrying MGH genes
At 33 days post-parturition, all of the mice were weaned and maintained on a solid diet supplemented with water containing 5,000 p.p.m. ZnSO4 (76 mM); their growth rate was recorded periodically. The dose of Zn was chosen on the basis of experiments which indicated that 5,000 p.p.m. induced almost maximal levels of MT-I mRNA in the liver without imparing the breeding potential of mice maintained on this diet for several months. Figure 3 shows the growth of the seven mice carrying MGH sequences. The littermates without MGH sequences served as convenient controls; the weights for each sex were averaged separately. Six of the mice with MGH sequences showed substantially more weight gain (up to 1.8-fold) than the controls. With the exception of MGH-16, there was a correlation between weight gain and MGH gene dosage (Table 1). The animal carrying the most copies of MGH sequences, MGH-21, died after 7 weeks of rapid growth; other mice have, however, grown larger. Most of the animals were already larger than normal before the Zn diet was instituted. Furthermore, one of the animals (MGH-19) was removed from the Zn diet on day 56 and it continued to grow at an accelerated rate. Thus, the effect of Zn on growth rate is uncertain. Perhaps mice carrying several copies of the MGH gene produce excess GH constitutively without a requirement for heavy metal induction. Thus, the question of Zn-dependent growth will be most easily answered by analysis of offspring of mice such as MGH-16 that contain only a few copies of the MGH gene.
Fig. 3.
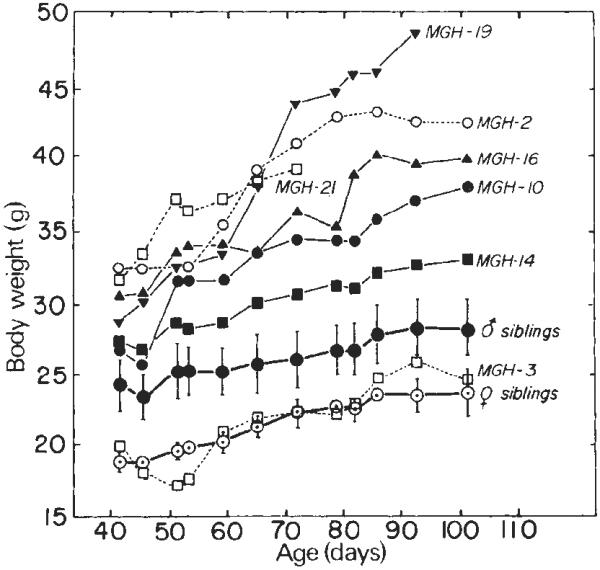
Growth of MGH mice. Microinjected eggs (SJL × C57) were transferred to oviducts of foster mothers on 14 April; 21 animals were born 3 weeks later. At 33 days old they were weaned and the drinking water was supplemented with 76 mM ZnSO4. The body weights of the males are shown as solid symbols; the mean weight (± s.d.) of 11 siblings not containing MGH sequences is also shown. The female weights are represented by open symbols; means (± s.d.) of three siblings are indicated also. MGH-21 died on day 72. A partial hepatectomy was performed on MGH-19 on day 56 and it was taken off Zn thereafter; it was killed on day 100. All mice were taken off Zn after day 83.
Table 1.
MGH genes, mRNA, GH levels, and growth of transgenic mice
| No. of MGH genes | No. of MGH mRNA molecules | Growth hormone | Growth* | |||
|---|---|---|---|---|---|---|
| Mouse | Sex | per cell | per cell | (μg ml−1) | (g) | Ratio |
| MGH-2 | ♀ | 20 | 800 | 57.0 | 41.2 | 1.87 |
| MGH-3 | ♀ | 1 | <50 | 0.87 | 22.5 | 1.02 |
| MGH-10 | ♂ | 8 | <50 | 0.28 | 34.4 | 1.32 |
| MGH-14 | ♂ | 2 | <50 | 0.31 | 30.6 | 1.17 |
| MGH-16 | ♂ | 2 | <50 | 17.9 | 36.4 | 1.40 |
| MGH-19 | ♂ | 10 | 1,500 | 32.0 | 44.0 | 1.69 |
| MGH-21 | ♀ | 35 | 3,000 | 112.0 | 39.3 | 1.78 |
| Female littermates | 0 | 0 | 0.16 | 22.0 | ||
| (n = 3) | ±0.1 | ±0.8 | ||||
| Male littermates | 0 | 0 | 0.15 | 26.0 | ||
| (n = 11) | ±0.08 | ±2 | ||||
The number of MGH genes per cell was estimated by DNA dot hybridization and scintillation counting (Fig. 2a). MGH mRNA molecules per cell were estimated by densitometric scanning of the autoradiogram from RNA slot hybridization (Fig. 4) and assuming 2.5 × 105 mRNA molecules per liver cell. GH was measured by RIA as described elsewhere10.
Animal weights when 74 days old. The ratio of body weight compared with littermates of the same sex is indicated (see Fig. 3).
Expression of the MGH gene in the liver
Based on previous studies with mice carrying metallothionein–TK fusion genes, maximal expression of MGH was expected in the liver, intestine and kidney. Evaluation of the tissue specificity of MGH gene expression was initiated by analysis of its activity in liver. A partial hepatectomy on day 56 allowed isolation and quantification of RNA for MGH-specific sequences. Figure 4b shows the results of RNA slot hybridization. The level of MGH mRNA expression in the liver correlated with the growth of the transgenic mice. The largest animals, MGH-2, -19 and -21, contained large amounts of MGH mRNA in the liver, whereas MGH-10, -14 and -16, which showed slower growth rates, contained ~50-fold less MGH RNA, not visible on the exposure shown in Fig. 4. Analysis of samples exposed to base hydrolysis before spotting the nucleic acids on to nitrocellulose (Fig. 4a) or treated with RNases A plus T1 after hybridization (not shown) established that RNA rather than potentially contaminating DNA was hybridizing to the probe. From these data, we estimate that 30 μg of liver RNA from MGH-2, -19 and -21 contains the same amount of GH mRNA as 13, 25 and 50 ng of poly(A)+ rat pituitary RNA, respectively. The RNA standard used contains about 10% GH mRNA on the basis of Rot analysis; therefore, we estimate that there are ~800–3,000 MGH mRNA molecules per liver cell in these transgenic mice (Table 1).
Fig. 4.
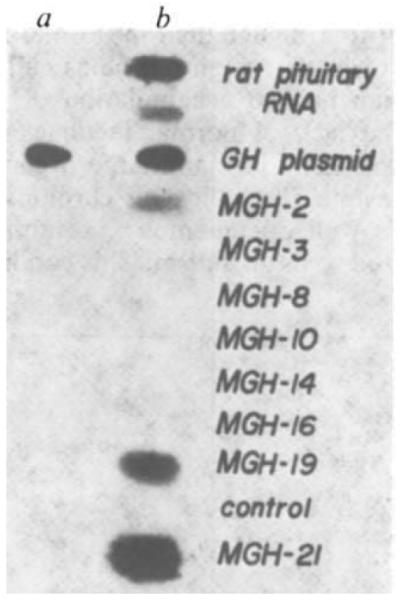
Slot-blot analysis of liver RNAs from transgenic mice. Mouse liver RNA was purified by homogenizing a liver slice in 1 × SET (1% SDS, 5 mM EDTA, 10 mM Tris pH 7.5), 200 μg ml−1 proteinase K (Boehringer) followed by phenol/chloroform extraction, ethanol precipitation, DNase I treatment (Worthington; DPFF), a second phenol/chloroform extraction, then re-precipitation with ethanol and finally resuspension for A260 determination. Liver RNA (30 μg) was diluted to 100 μl in 2X SSC. Samples were brought to 0.5 M NaOH or 0.5 M NaCl and incubated at 65 °C for 30 min. 140 μl of 20 × SSC and 160 μl of 12.3 M formaldehyde were added, incubated at 65 °C for another 15 min and applied to a nitrocellulose sheet resoaked in 20 × SSC through a slot-blot device31. The nitrocellulose was baked at 80 °C for 2 h, prehybridized for 5 h and then hybridized with nick-translated 12P-rGH cDNA32 (5 × 106 c.p.m. in 4 ml at 42 °C overnight), washed in 0.1 × SSC at 42 °C, and exposed to X-ray film at −70 °C for 24 h. a, Samples were treated with 0.5 M NaOH and neutralized with HCl. b, Samples were incubated with an equivalent amount of NaCl. 40 and 20 ng of rat pituitary poly(A)+ RNA in the presence of 30 μg of control liver RNA were applied to slots as GH mRNA controls. 1 ng of rGH gene plasmid, also in the presence of 30 μg of control liver RNA, was applied as a base-resistant control.
If all processing signals in the fusion gene are correctly recognized, a fusion mRNA 63 nucleotides larger than bona fide rat GH mRNA (~1 kb) would be generated10,13. Denaturing gel electrophoresis and RNA Northern blot analysis of the liver RNA from MGH-21 show that its size is similar, as expected, to that of authentic mouse and rat pituitary GH mRNA (Fig. 5a). Liver RNA from a control mouse shows no GH-reactive sequences (Fig. 5a, lane 1). Because the GH cDNA probe used for the RNA blot analysis recognizes both rat and mouse GH mRNAs, it is necessary to establish that the hybridizing species in the liver is actually the product of the fusion gene and not mouse GH mRNA due to an unexpected activation of the endogenous mouse GH gene. The use of a XhoI linker for the construction of the fusion gene generates a sequence that will be uniquely present in MGH mRNA. Thus, the XhoI site of pMGH was labelled using [γ- 32P]ATP and polynucleotide kinase, followed by cleavage with SstI (Fig. 1c). This 221-nucleotide fragment was gel-purified, denatured, and used as hybridization probe in a single-strand-specific nuclease protection assay. Hybridization to MGH mRNA should generate a 74-base nuclease-resistant fragment while mouse GH mRNA or metallothionein mRNA will be unable to protect the kinased end and should therefore be negative. Figure 5b, lane 6 shows that the predicted 74-base fragment is present in liver RNA of mouse MGH-21, but not in normal mouse pituitary RNA (lane 5), control mouse liver RNA (lane 4) or in the liver RNA of MGH-3 (lane 3), an animal showing no accelerated growth (Fig. 3). Thus, it seems that transcription is initiating properly at the MT-I promoter and continuing through the putative termination site of the GH gene, the four GH intervening sequences are being properly spliced and the MGH mRNA is polyadenylated.
Fig. 5.
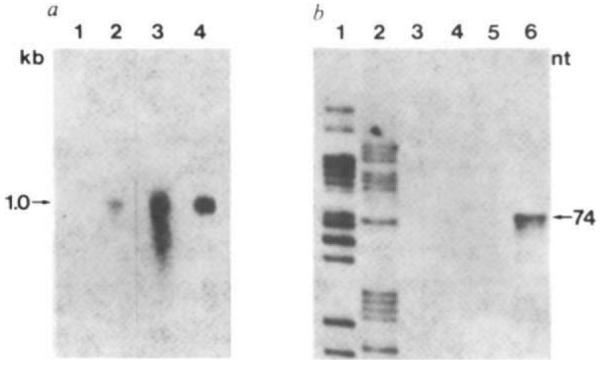
Structure of MGH mRNAs in liver of transgenic mice. a, Northern blot analysis. RNAs were resuspended 10 mM NaH2PO4, pH 7.4/50% formamide/2.2 M formaldehyde, heated at 65 °C for 5 min, and subjected to electrophoresis on a slab gel composed of 1.5% agarose in 10 mM NaH2PO4, pH 7.4/0.55 mM EDTA/1.1 M formaldehyde. The running buffer was 10 mM NaH2PO4, pH 7.4/0.5 M formaldehyde. The gel was stained with acridine orange to identify rRNA markers, photographed, incubated for 90 min with 50 mM NaOH, and then neutralized with two washes of 0.2 M NaOAc, pH 4.3. The RNA was then transferred to diazotized paper and prehybridized as described elsewhere32. Lane 1, total liver RNA (15 μg) from a control littermate; lane 2, total liver RNA (15 μg) from MGH-21; 3, total mouse pituitary RNA; 4, 40 ng of rat pituitary mRNA, poly(A)+. Lanes 1 and 2 exposed for 48 h; lanes 3 and 4 exposed for 5 h. b, Single-strand-specific nuclease protection assay. 20 μg of RNA were hybridized at 47 °C for 5 h in 40 μl of a solution containing 40 mM PIPES pH 6.4, 0.4 M NaCl, 80% formamide and 50,000 c.p.m. gel-purified 221-bp SstI–XhoI fragment of pMGH end-labelled at the XhoI site with 32P (see Fig. 1c). The samples were then diluted with 0.3 ml of 280 mM NaCl, 30 mM NaOAc pH 4.4, 4.5 mM ZnSO4, 20 μg ml−1 salmon sperm DNA and 150 units of mung bean single-strand-specific nuclease (Collaborative Research) and incubated at 47 °C for 1 h. Samples were ethanol-precipitated, resuspended in 90% formamide containing bromophenol blue and Xylene Cyanol FF, loaded on to an 8% acrylamide–urea sequencing gel, electrophoresed for 1.5 h at 2,000 V, dried and autoradiographed for 7 days at −70 °C with an intensifying screen. Lanes 1 and 2, sequencing ladder used for size standards; 3, MGH-3 RNA; 4, control liver RNA; 5, mouse pituitary RNA; 6, MGH-21 RNA.
Increased growth hormone in sera of MGH mice
The correlation between MGH mRNA levels and growth of the mice sugggests that expression of the MGH gene accounts for the observed biological consequences; this was supported by the three independent RNA analyses above. These results suggest that the circulating levels of GH should be increased. Blood was taken from the transgenic mice and from littermates and assayed for GH by radioimmunoassay (RIA). The GH levels in four of the transgenic mice were 100–800 times greater than levels in control littermates; one mouse had 112 μg ml−1 of GH in its serum (Table 1). Two of the transgenic mice showing a slow but significantly elevated growth rate had serum GH levels at the high end of the normal range. The lack of physiological regulation and the ectopic production of GH in these animals probably account for their accelerated growth.
Discussion
These data strongly suggest that the altered phenotype of the mice is a direct result of the integration and expression of the metallothionein–growth hormone fusion gene. The elevated level of GH present in some of these mice corresponds to a high level of MGH mRNA in the liver (up to 3,000 molecules per cell). The accumulation of MGH mRNA in the liver is comparable to the endogenous level of MT-I mRNA14, but is ~100-fold higher than that obtained from metallothionein–TK fusion genes studied previously2. This difference is probably the consequence of the intrinsic stability of the GH mRNA relative to TK mRNA; however, differences in transcription rates or processing efficiency due to variations in fusion gene construction are also possible. The high level of MGH gene expression in transgenic mice will greatly facilitate direct comparison of MGH and MT-I mRNA production in different tissues, thus allowing us to determine whether chromosomal location has an important effect on tissue-specific expression of the MT-I promoter.
Growth hormone levels in some of the transgenic mice were as much as 800-fold higher than in normal mice, resulting in animals nearly twice the weight of their unaffected littermates. This greater than normal accumulation of GH undoubtedly reflects both the lack of normal feedback mechanisms and expression of this gene in many large organs including liver, kidney and intestine. The effect of chronic exposure to high levels of GH is well documented15, resulting in the clinical condition referred to as gigantism. This condition in humans is usually associated with pituitary adenomas15 and more rarely with ectopic expression of GH by lung carcinomas16,17.
Some of the diverse effects of GH are mediated directly by the hormone18–20. However, it is generally believed that the major effect of GH is stimulation of somatomedin production in the liver18,19. Somatomedins are insulin-like growth factors that promote proliferation of mesodermal tissues such as muscle, cartilage and bone18,19. The involvement of somatomedins in the GH response provides an explanation for the growth of animals such as MGH-10, 14 and 16 in which the circulating level of GH was only slightly higher than normal. In these animals, GH produced in the liver may be sufficient to stimulate somatomedin production because the local GH concentration is relatively high. Thus, in these animals, GH may mimic the local paracrine function of some hormones21. However, the reason for lack of growth of MGH-3 is unclear.
The implicit possibility is to use this technology to stimulate rapid growth of commercially valuable animals. Benefit would presumably accrue from a shorter production time and possibly from increased efficiency of food utilization. The effects of continuously elevated GH levels on the quality of meat will need to be evaluated. By choosing the appropriate GH gene, milk production may be disproportionately increased considering that GH is homologous to prolactin, that GH of some species binds to prolactin receptors, and that exogenously administered GH has been shown to increase milk yield22,23. Having a regulatable promoter may be particularly advantageous for timely expression of GH. Applying these techniques to large animals will be more difficult. Nevertheless, when genes for desired traits can be isolated, this approach should provide a valuable adjunct to traditional breeding methods. Optimizing the conditions for integration and expression of foreign genes in mice should facilitate the eventual application of these techniques to other animals.
Another possibility is the use of this technology either to correct or to mimic certain genetic diseases. There are several inbred dwarf strains of mice, one of which, little, lacks GH and is about half normal size when homozygous24. Thus, introduction of the fusion gene described here or a natural GH gene might restore normal growth to these animals. On the other hand, the experiments described above show that gigantism can be created. Once these mice are inbred to give homozygous stocks, they will provide a valuable model system for biochemical studies on the consequences of excess GH production.
Finally, the exceptionally high levels of GH found in the sera of some of these mice raises the possibility of extending this technology to the production of other important polypeptides in farm animals. The concentrations of GH in MGH-21 serum was 10–100-fold higher than that reported for bacterial or mammalian cell cultures that were genetically engineered for GH production10,25–27. This genetic farming concept is comparable to the practice of raising valuable antisera in animals, except that a single injection of a gene into a fertilized egg would substitute for multiple somatic injections; moreover, the expression of that gene is likely to be heritable9. This approach may be particularly applicable in those cases where the protein of interest requires special covalent modifications (for example, proteolytic cleavage, glycosylation, or γ-carboxylation) for activity or stability.
Clearly the ability to introduce into mice, and by extrapolation into other animals, functional genes of selected construction offers wide ranging experimental as well as practical opportunities.
Acknowledgments
We thank Joan Vaughan for performing GH radioimmunoassays; Wylie Vale and Marcia Barinaga for valuable discussions; our colleagues for helpful comments; and Abby Dudley for secretarial assistance in the preparation of the manuscript. N.C.B. was supported by a NCI training grant (CA09370 HHS). The work was supported by research grants from the NIH (HD-09172, HD-15477, GM-26444, AM-21567) and NSF (PCM-8107172).
References
- 1.Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Proc. natn. Acad. Sci. U.S.A. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinster RL, et al. Cell. 1981;27:223–231. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Constantini F, Lacy E. Nature. 1981;294:92–94. doi: 10.1038/294092a0. [DOI] [PubMed] [Google Scholar]
- 4.Harbers K, Jähner D, Jaenisch R. Nature. 1981;293:540–542. doi: 10.1038/293540a0. [DOI] [PubMed] [Google Scholar]
- 5.Rusconi S, Schaffner W. Proc. natn. Acad. Sci. U.S.A. 1981;78:5051–5055. doi: 10.1073/pnas.78.8.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner E, Stewart T, Mintz B. Proc. natn. Acad. Sci. U.S.A. 1981;78:5016–5020. doi: 10.1073/pnas.78.8.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner TE, et al. Proc. natn. Acad. Sci. U.S.A. 1981;78:6376–6380. doi: 10.1073/pnas.78.10.6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon JW, Ruddle FH. Science. 1981;214:1244–1246. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- 9.Palmiter RD, Chen HY, Brinster RL. Cell. 1982;29:701–710. doi: 10.1016/0092-8674(82)90186-6. [DOI] [PubMed] [Google Scholar]
- 10.Doehmer J, et al. Proc. natn. Acad. Sci. U.S.A. 1982;79:2268–2272. doi: 10.1073/pnas.79.7.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinster RL, Chen HY, Warren R, Sarthy A, Palmiter RD. Nature. 1982;296:39–42. doi: 10.1038/296039a0. [DOI] [PubMed] [Google Scholar]
- 12.Mayo KE, Warren R, Palmiter RD. Cell. 1982;29:99–108. doi: 10.1016/0092-8674(82)90094-0. [DOI] [PubMed] [Google Scholar]
- 13.Evans RM, Birnberg NC, Rosenfeld MG. Proc. natn. Acad. Sci. U.S.A. in the press. [Google Scholar]
- 14.Durnam DM, Palmiter RD. J. biol. Chem. 1981;256:5712–5716. [PubMed] [Google Scholar]
- 15.Richmond IL, Wilson CB. J. Neurosurg. 1978;49:163–167. doi: 10.3171/jns.1978.49.2.0163. [DOI] [PubMed] [Google Scholar]
- 16.Steiner H, Dahlback O, Waldenstarm J. Lancet. 1968;i:783–785. doi: 10.1016/s0140-6736(68)92229-0. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg PB, Beck C, Martin TJ, Burger HG. Lancet. 1972;i:350–352. doi: 10.1016/s0140-6736(72)92843-7. [DOI] [PubMed] [Google Scholar]
- 18.Daughaday WH, Herington AC, Phillips LS. A. Rev. Physiol. 1975;37:211–244. doi: 10.1146/annurev.ph.37.030175.001235. [DOI] [PubMed] [Google Scholar]
- 19.Kostyo JL, Isaksson O. Int. Rev. Physiol. 1977;13:255–274. [PubMed] [Google Scholar]
- 20.Morikawa M, Nixon T, Green H. Cell. 1982;29:783–789. doi: 10.1016/0092-8674(82)90440-8. [DOI] [PubMed] [Google Scholar]
- 21.Krieger DT, Martin JB. New Engl. J. Med. 1981;304:876–885. doi: 10.1056/NEJM198104093041505. [DOI] [PubMed] [Google Scholar]
- 22.Shiu RPC, Friesen HG. J. biol. Chem. 1974;249:7902–7911. [PubMed] [Google Scholar]
- 23.Peel CJ, Bauman DE, Gorewit RC, Sniffen CJ. J. Nutr. 1981;111:1662–1671. doi: 10.1093/jn/111.9.1662. [DOI] [PubMed] [Google Scholar]
- 24.Beamer WG, Eicher EM. J. Endocr. 1976;71:37–45. doi: 10.1677/joe.0.0710037. [DOI] [PubMed] [Google Scholar]
- 25.Goeddel DV, et al. Nature. 1979;281:544–548. doi: 10.1038/281544a0. [DOI] [PubMed] [Google Scholar]
- 26.Pavlakis GN, Hizuka N, Gorden P, Seeburg P, Hamer DH. Proc. natn. Acad. Sci. U.S.A. 1981;78:7398–7402. doi: 10.1073/pnas.78.12.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robins DM, Paek I, Seeburg PH, Axel R. Cell. 1982;29:623–631. doi: 10.1016/0092-8674(82)90178-7. [DOI] [PubMed] [Google Scholar]
- 28.Glanville N, Durnam DM, Palmiter RD. Nature. 1981;292:267–269. doi: 10.1038/292267a0. [DOI] [PubMed] [Google Scholar]
- 29.Chen CW, Thomas CA., Jr Analyt. Biochem. 1980;101:339–341. doi: 10.1016/0003-2697(80)90197-9. [DOI] [PubMed] [Google Scholar]
- 30.Barta A, Richards RI, Baxter JD, Shine J. Proc. natn. Acad. Sci. U.S.A. 1981;78:4867–4871. doi: 10.1073/pnas.78.8.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tlsty T, Brown PC, Johnston R, Schimke RT. In: Gene Amplification. Schimke RT, editor. Cold Spring Harbor Laboratory; New York: 1982. pp. 231–238. [Google Scholar]
- 32.Potter E, Nicolaisen K, Ong EO, Evans RM, Rosenfeld MG. Proc. natn. Acad. Sci. U.S.A. 1981;78:6662–6666. doi: 10.1073/pnas.78.11.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]


