Abstract
Sapoviruses (SaVs) are enteric caliciviruses that have been detected in multiple mammalian species, including humans, pigs, mink, dogs, sea lions, chimpanzees, and rats. They show a high level of diversity. A SaV genome commonly encodes seven nonstructural proteins (NSs), including the RNA polymerase protein NS7, and two structural proteins (VP1 and VP2). We classified human and animal SaVs into 15 genogroups (G) based on available VP1 sequences, including three newly characterized genomes from this study. We sequenced the full length genomes of one new genogroup V (GV), one GVII and one GVIII porcine SaV using long range RT-PCR including newly designed forward primers located in the conserved motifs of the putative NS3, and also 5' RACE methods. We also determined the 5’- and 3’-ends of sea lion GV SaV and canine GXIII SaV. Although the complete genomic sequences of GIX-GXII, and GXV SaVs are unavailable, common features of SaV genomes include: 1) “GTG” at the 5′-end of the genome, and a short (9~14 nt) 5′-untranslated region; and 2) the first five amino acids (M [A/V] S [K/R] P) of the putative NS1 and the five amino acids (FEMEG) surrounding the putative cleavage site between NS7 and VP1 were conserved among the chimpanzee, two of five genogroups of pig (GV and GVIII), sea lion, canine, and human SaVs. In contrast, these two amino acid motifs were clearly different in three genogroups of porcine (GIII, GVI and GVII), and bat SaVs. Our results suggest that several animal SaVs have genetic similarities to human SaVs. However, the ability of SaVs to be transmitted between humans and animals is uncertain.
Introduction
Sapoviruses (SaVs), members of the Sapovirus genus within the Caliciviridae family, have been detected from humans [1] and animals (pigs [2, 3], mink [4], dogs [5], sea lions [6], bats [7], chimpanzees [8], and rats [9], listed in the order of their discovery). The SaV genome is a linear, positive sense, single stranded RNA of 7.1–7.7 kb in size with a poly(A) tail at the 3’-end. It is predicted to encode viral nonstructural proteins (NSs) (NS1, NS2, putative NTPase [NS3], NS4, VPg [NS5], and fused protease–RNA-dependent RNA polymerase [RdRp] [NS6-NS7]), and structural proteins VP1 and VP2 [1]. SaVs are genetically highly diverse and currently classified into 14 genogroups (G) based on the complete VP1 amino acid sequences [10]. Typical conserved amino acid sequences were identified in the NS3 (GAPGIGKT), NS5 (KGKTK and DDEYDE), NS6 (GDCG), NS7 (WKGL, KDEL, DYSKWDST, GLPSG, and YGDD), and VP1 (PPG and GWS) among the GI-GV SaVs [1].
SaVs have been detected from pigs (GIII, GV-GXI), mink (GXII), and bats (GXIV), using primers targeting the NS7 region [2, 4, 7, 10–15]. In addition, SaV sequences have been detected from dogs (GXIII) and sea lions (GV) by next generation sequencing [5, 6]. A variety of SaVs were also detected from humans using primers targeting the NS7 or VP1 region, or by next generation sequencing [1, 16]. Current animal and human SaVs were classified into 11 genogroups (GIII, and GV–GXIV) and four genogroups (GI, GII, GIV, and GV), respectively, based on the complete VP1 sequences [10]. GV SaVs are detected from both animals and humans [1].
At the beginning of this study, there were 28 complete sapovirus genomes available in the literature or Genbank database: GI (n = 9), GII (n = 5), GIV (n = 4), and GV (n = 3) from humans; GIII (n = 3), GVI (n = 2), and GVII (n = 1) from swine; and GXIV (n = 1) from bats. In addition, the nearly full length genomic sequences, excluding the 5’- and 3’-ends, of GV sea lion and GXIII canine SaVs were reported [5, 6].
During our investigation, new SaVs were detected from chimpanzees [8] and rats [9] by next generation sequencing. The nearly full length genomic sequences, excluding the 5’- and 3’-ends, of the chimpanzee SaVs have been determined and are classified as GI based on the complete VP1 sequences. The VP1 sequences of rat SaVs have been determined, but they are not yet classified [9]. Furthermore, one more complete genome sequence of GIII porcine SaV (CH430 strain) has been determined [17].
Available complete genome sequences for animal SaVs are still limited compared to human SaVs. Therefore, the aims of this study were to determine additional complete genomic sequences of animal SaVs, to identify the common genetic characteristics of SaVs, and to examine the genetic relatedness among human and animal SaVs. We also proposed 15 genogroups for human and animal SaVs based on the complete VP1 sequences.
Materials and Methods
New forward primers designed for the amplification of swine and mink SaVs by RT-PCR
The genomic sequences of seven animal SaVs [3 GIII (Genbank accession no. AF182760, FJ387164, and JX678943), 2 GVI (AY974192 and KJ508818), 1 GVII (AB221130) and 1 GXIV (JN899075)] were aligned using ClustalW version 2.1 (http://clustalw.ddbj.nig.ac.jp/top-j.html). New primers, 1550F [5′- CCBTDMCCAYTGRAYTGTGA-3′] and 1571F [5′- CCCTWMCCAYTGAATTGTGA-3′] targeting the conserved “PL (N / D) CD” sequence, and 1578F [5′- TGGGACGAGTTTGACAC -3′] targeting “WDEFD” sequence, were designed and synthesized at Integrated DNA Technologies, Inc.
Fecal specimens
Swine, mink, dog, and sea lion fecal samples from previous studies [4–6, 10, 18–20] were stored at -80°C and were used in this study for further sequence analyses. SaVs of swine fecal origin used in this study (GVII WGP247 [KC309421]; GVIII WG194D [KC309416], WG197C [KC309417], and WG214D [KC309419]; GIX WG214C [KC309418]) had been determined previously for the 3’-end ~ 3kb fragment, covering from the partial NS7 to the 3’-end of the genome. Mink SaV sequence had been determined for only partial NS7 sequence using mixed samples of five feces [4]. The nearly complete genomic sequences of sea lion SaV (GV/CSL9775 [Genbank accession no. JN420370]) and dog SaV (GXII/AN210D [JN387134]) fecal origin had been de novo assembled using next generation sequencing data [5, 6]. We included them in this study because the 5′ and 3′ ends of the sea lion SaV, and the 5′ end of dog SaV were lacking based on comparisons to other SaV full genomes using The Basic Local Alignment Search Tool (BLAST http://www.ncbi.nlm.nih.gov/BLAST).
RNA extraction, cDNA synthesis, and PCR
Viral RNA was extracted from 200 μL of fecal suspensions using RNeasy Mini kit (Qiagen) with slight modification. Briefly, 200 μL of fecal suspensions was mixed with 350 μL of RLT buffer and incubate for 5 min, then 296 μL of ethanol was added and mixed well. The mixture was applied onto RNeasy mini column and then washed and eluted according to the manufacture’s instructions. Purified RNA was eluted in 40 μL UltraPure DNase/RNase-Free Distilled Water (Invitrogen) and used freshly or stored at -80°C.
cDNA was synthesized as follows: 8.5 μL of viral RNA solution was mixed with 0.5 μL of the 10 pmol/μl reverse primer [TX30SXN (5′- GACTAGTTCTAGATCGCGAGCGGCCGC CCT30−3′) [21] that was complementary to the 3’-end including partial polyA tail or the gene-specific reverse primer targeting the NS7 and VP1 junction region], and 1 μL of 10 mM dNTP. The mixtures were incubated at 65°C for 5 min, cooled on ice, and then mixed with 2 μL of 10 × First strand buffer (Invitrogen), 2 μL of 100 mM DTT (Invitrogen), 4 μL of 25 mM MgCl2 (Invitrogen), 1 μL of RNase OUT ribonuclease inhibitor (40 U/μl) (Invitrogen), and 1 μL of SuperScript III reverse transcriptase (200 U/μl) (Invitrogen). This mixture was incubated first at 25°C for 5 min, then at 50°C for 50 min, and finally was inactivated at 95°C for 5 min.Next, 1 μL of RNase H (2 U/μl) (Invitrogen) was added and incubated at 37°C for 20min.
The SaV genomic sequences spanning the putative NS3 to NS7 region or the putative NS3 to the end of the genome were amplified by RT-PCR with one of the newly designed forward primers (1550F, 1571F, or 1578F) and reverse primer TX30SXN or the strain-specific reverse primer targeting the NS7 -VP1 junction region, using high-fidelity PCR enzyme, PrimeSTAR GXL DNA polymerase (TaKaRa Mirus Bio). A final volume of 50 μ l of the PCR reaction mixture contained 2 μL of the cDNA or the first PCR products, 10 μ l of 5 × PrimeSTAR GXL DNA polymerase buffer, 4 μ l of 2.5 mM dNTPs, 2 μ l of forward primer (10 pmol/μ l), 2 μ l of reverse primers (10 pmol/μ l), and 1 μL of PrimeSTAR GXL DNA polymerase (1.25 U/μ l). PCR was performed at 98°C for 10 sec followed by 45 cycles of 98°C for 10 s, 55°C for 15 s, and 72°C for 2 min, and a final extension at 72°C for 10 min.
For the new porcine GV SaV, the sequence covering the partial NS3 to the 3’-end of the genome was amplified by semi-nested RT-PCR using gene-specific forward primers and TX30SXN primer, as described above.
5′ RACE
The 5′ terminal nucleotide sequences of the SaV genomes were determined using 5' Rapid Amplification of cDNA Ends (RACE), Version 2.0 (Invitrogen) with a slight modification from the original protocol. Briefly, cDNA was synthesized from RNA as follows: 8.5 μL of viral RNA was mixed with 0.5 μL of 10 pmol/μl strain-specific primer and 1 μL of 10 mM dNTPs. The mixture was incubated at 80°C for 3 min, cooled on ice, and then mixed with 2 μL of 10 × First strand buffer, 2 μL of 100 mM DTT, 4 μL of 25 mM MgCl2, 1 μL of RNase OUT ribonuclease inhibitor (40 U/μl), and 1 μL of SuperScript III reverse transcriptase (200 U/μl). This mixture was incubated first at 25°C for 5min, then at 50°C for ~3h, and finally at 85°C for 5 min. Afterwards, 1 μ l of RNase H or RNase T1 mixture (Invitrogen) was added to the mixture that was incubated at 37°C for 30 min. The cDNA was purified using the SNAP column in the 5' RACE System (Invitrogen) or the column in the QIAGEN PCR purification kit (Qiagen) according to the manufacture’s instructions. Finally, the purified cDNA was eluted in 50 μL of UltraPure DNase/RNase-Free Distilled Water. Homopolymeric (dC or dA) tailing was added on the purified cDNA as follows: 10 μL of the purified cDNA solution, 2.5 μL of 2.5mM dATP (promega) or dCTP (Invitrogen), 5 μL of 5 x tailing buffer, and 6.5 μL of water was mixed, incubated at 94°C for 3 min, and cooled on ice.Then 1 μL of Terminal deoxynucleotidyl transferase (20 U/μl) (Invitrogen) was added and the mixture was incubated first at 37°C for 10min and then at 65°C for 10 min to inactivate the enzyme. Nested PCRs were performed with the gene specific reverse primers and the abridged anchor primer AAP (5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′) for the primary PCR and the abridged universal primer AUAP (5′-GGCCACGCGTCGACTAGTAC-3′) (Invitrogen) for the secondary PCR for the poly (C)-tailed cDNA, and QT (5′- CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCT17-3′) and QO (5′- CCAGTGAGCAGAGTGACG-3′)for poly (A)-tailed cDNA, using high-fidelity PCR enzyme, PrimeSTAR GXL DNA polymerase or PrimeSTAR HS DNA polymerase (TaKaRa Mirus Bio). A final volume of 50 μ l of the reaction mixture contained 5 μL of the homopolymeric (dC or dA) tailing cDNA or the 0.2 μL of the first PCR reaction mixture, 10 μ l of 5 × PrimeSTAR DNA polymerase buffer, 4 μ l of 2.5 mM dNTPs, 2 μ l of forward primer (10 pmol/μ l), 2 μ l of reverse primer (10 pmol/μl), and 1 μL of PrimeSTAR HS DNA polymerase (2.5 U/μl) or PrimeSTAR GXL DNA polymerase (1.25 U/μ l). PCR was performed at 95°C for 5 min for initial denaturing, followed by 45 cycles of 94°C for 15 s, 60°C for 15 s, and 72°C for 2 min, and a final extension at 72°C for 10 min.
Amplification of the 3′ ends of the SaV genomes
The 3′ end of a SaV genome was amplified by RT-PCR with gene specific forward primer, and the reverse primer TX30SXN, using high-fidelity PCR enzyme. A final volume of 50 μl of the reaction mixture contained 2 μL of the cDNA synthesized with the TX30SXN primer, 10 μl of 5 × PrimeSTAR DNA polymerase buffer, 4 μ l of 2.5 mM dNTPs, 2 μ l of forward primer (10 pmol/μ l), 2 μ l of reverse primer (10 pmol/μ l), and 1 μL of PrimeSTAR HS DNA polymerase (2.5 U/μ l) or PrimeSTAR GXL DNA polymerase (1.25 U/μ l). PCR was performed at 95°C for 5 min for initial denaturing followed by 35 cycles of 98°C for 10 s, 55°C for 15 s, and 72°C for 1 min, and a final extension at 72°C for 10 min.
Cloning, sequencing, phylogenetic analyses, and genogrouping
The PCR products were separated by agarose gel electrophoresis, purified using a QIAquick Gel Extraction Kit (Qiagen), and sequenced directly or cloned into the pCR4Blunt-TOPO vector (Invitrogen) before sequencing by primer walking methods using a set of gene-specific primers. For cloned samples, at least three positive clones of each sample were selected for sequencing. Samples were sequenced using BigDye Terminator cycle chemistry and an automated ABI Prism3100xl sequencer (Applied Biosystems). Sequence editing and assembly were performed using the Sequencher program v4.10.1 (GeneCodes) and analyzed by Genetyx-Mac software v16.0.4 (Genetyx Corporation). The Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov) was employed to find homologous hits. Amino acid sequences were aligned using ClustalW version 2.1 (http://clustalw.ddbj.nig.ac.jp/top-j.html). The construction of Maximum-likelihood phylogenetic trees with 1,000 bootstrap replications, and the calculation of amino acid sequence pairwise distances were performed using MEGA6 software [22]. Identity = 1—distance.
Nucleotide sequence accession numbers
The SaV nucleotide sequences determined in this study have been deposited in GenBank under accession numbers KX000383 for GV/WG194D-1, KX000384 for GVII/RV0042, and KX000385 for GXII/WD1237. The nucleotide sequences of GV/CSL9775 (JN420370), GVII/WGP247 (KC309421), GVIII/WG214D (KC309419), GVIII/WG194D (KC309416), GVIII/WG197C (KC309417), GIX/214C (KC309418), and GXIII/AN210D (JN387134) SaVs have been updated in GenBank database.
Results and Discussion
Forward Primers targeting the putative NS3 region were designed to amplify SaVs from different animal species
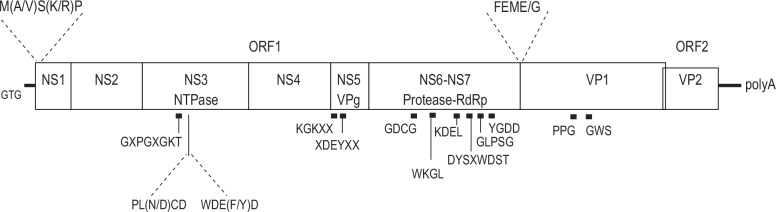
We found that the regions suitable for PCR primer design were in the putative NS3 region based on the full genomic sequence alignments of the seven animal SaVs. We designed three forward primers, 1550F, 1571F, and 1578F (see Materials and Methods). The primers 1550F and 1571F targeted the same PL (N / D) CD amino acid motif that was conserved among GIII, GVI, GVII, and GXIV SaVs, and the primer 1578F targeted the WDEFD amino acid motif of GXIV SaVs. This motif (WDEYD) differed slightly in GVI and GVII SaVs. These motifs were located downstream of the typical GXPGXGKT motif of the putative NS3 (Fig 1).
Fig 1. Sapovirus genomic organization: non-structural (NS) and structural proteins VP1 and VP2, and the conserved motifs.
Genomic organization of a common sapovirus, including two open reading frames (ORF1 and ORF2). ORF1 encodes the predicted viral NS proteins (NS1, NS2, NS3 [NTPase], NS4, NS5 [VPg], NS6-NS7 [protease-RNA-dependent RNA polymerase (RdRp)], and the major capsid protein (VP1). ORF2 encodes the minor capsid protein VP2. The following amino acid or nucleotide motifs are conserved among all available sequenced SaVs: GXPGXGKT, PL (N / D) CD, and WDE (F / Y) D of NS3, KGKXX and XDEYXX of NS5, GDCG of NS6, WKGL, KDEL, DYSXWDST, GLPSG, and YGDD of NS7, PPG and WGS of VP1, and the first three nucleotides (GTG) at the 5’–end. Also shown are the first five amino acids of NS1 (M [A / V] S [K / R] P) and around the NS7-VP1 cleavage site (FEME / G, the slash indicates the putative cleavage site by viral protease NS6) that are conserved among GI-GV, GVIII, and GXIII SaVs.
Successful amplification of GV, GVII, GVIII, and GXII SaVs, but not GIX SaVs, using the newly designed forward primers
We amplified the sequence fragments for two porcine GVII (RV0042 and WGP247), three porcine GVIII (WG214D, WG194D and WG197C), and one mink GXII (WD1237]) SaVs using the newly designed forward primers 1550F, 1571F, or 1578F and strain-specific reverse primers or the modified oligo dT primer, TX30SXN. The strain specific reverse primer for WD1237 was designed based on the partial sequence obtained by next generation sequence (NGS) as described previously (Li et al., 2011).
A long PCR product (approximately 6kb) was amplified for SaV strain RV0042 using primer set 1571F and TX30SXN. Approximately 3.5 kb-fragments were amplified for other samples using these three forward primer(s) and the corresponding strain-specific reverse primers targeting the NS7-VP1 junction region.
We could not amplify the fragment from GIX SaV (WG214C) using those forward primers. In most successful cases, only one of the three forward primers (1550F, 1571F, or 1578F) amplified a SaV strain, except for GVIII WG194D and WG197C samples. Two different SaVs, GVIII (WG194D) and GV SaVs (WG194D-1) were amplified from the same fecal sample WG194D using 1550F and 1578F primers, respectively. Identical sequences were amplified for WG197C strain using 1550F and 1571F forward primers.
The complete full length genomes of GV, GVII, GVIII, GXIII SaVs were determined using RACE methods
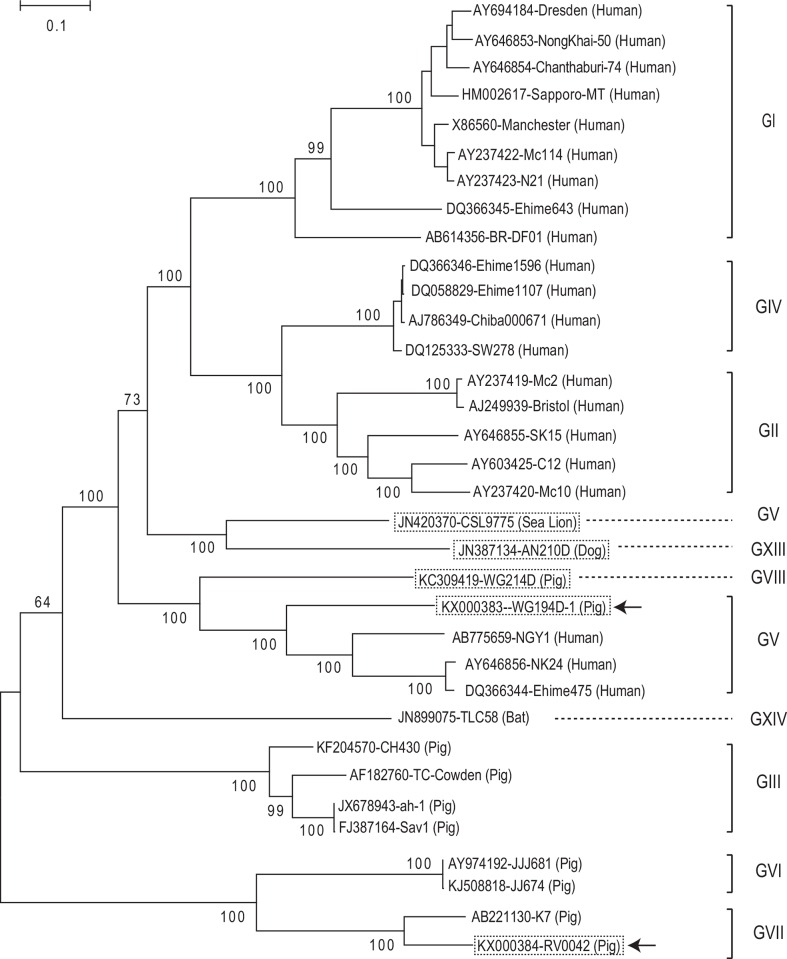
We further performed 5′-RACE for the porcine SaVs (GV, GVII, GVIII, and GIX) and mink SaV (GXII). Among the eight SaV strains, we successfully determined the complete genomic sequences of three strains (Po/SaV/GV/WG194D, Po/SaV/GVII/RV0042, and Po/SaV/GVIII/WG214D) (Table 1, Fig 2). The genome of GV/WG194D-1 was 7496 nt long, excluding the poly(A) tail, and shared 68% nt identity with other GV SaV genomes (Genbank accession numbers AB775659, AY646856, and DQ366344). The genome of GVII/RV0042 was 7150 nt long, and it shared 83% nt identity with the GVII/K7 strain (AB221130) detected in Japan. The genome of GVIII/214D strain was 7497 nt long and represented the first complete genome sequence for a porcine GVIII SaV. In this study, we also determined the 5’- and 3’-ends of sea lion/GV/CSL9775 strain (60 nt of 5′ end and 60 nt of 3′ end) and Canine/GXIII/AN210D strain (330 nt of 5′ end), and their genome sizes were 7497 and 7469 nt long, respectively (Table 1, Fig 2).
Table 1. Genomic characteristics of 50 representative sapovirus strains in 15 genogroups.
| Genogroup | Genbank accession no. | Strain name* | Species | Genome size (nt)** | The 5'-end 30 nt sequences*** | Length of 5-UTR (nt) | First five amino acid residues in the NS1 | Five amino acid residues at the putative NS7-VP1 cleavage site |
|---|---|---|---|---|---|---|---|---|
| GI | X86560 | Manchester | Human | 7431 | gtgattggtt agATGgtttc caagccattc | 12 | MVSKP | FEMEG |
| HM002617 | Sapporo-MT | Human | 7433 | gtgattggtt agATGgtttc caagccattc | 12 | MVSKP | FEMEG | |
| AY694184 | Dresden | Human | 7429 | gtgattggtt agATGgtttc caagccattc | 12 | MVSKP | FEMEG | |
| AY237422 | Mc114 | Human | 7429 | gtgattggtt agATGgcttc caagccattc | 12 | MASKP | FEMEG | |
| AY237423 | N21 | Human | 7429 | gtgattggtt agATGgcttc caagccattc | 12 | MASKP | FEMEG | |
| AY646853 | NongKhai-50 | Human | 7429 | gtgattggtt ggATGgcttc caagccattc | 12 | MASKP | FEMEG | |
| AY646854 | Chanthaburi-74 | Human | 7429 | gtgattggtt ggATGgcttc caagccattc | 12 | MASKP | FEMEG | |
| AB614356 | BR-DF01 | Human | 7476 | gtgattggtt agATGgtttc caagccatac | 12 | MVSKP | FEMEG | |
| DQ366345 | Ehime643 | Human | 7447 | gtgattggtt agATGgcttc taagccatat | 12 | MASKP | FEMEG | |
| KJ858686 | IJC04 | Chimpanzee | (7320) | ———-—ATGgcttc caagccattc | - | MASKP | FEMEG | |
| GII | AJ249939 | Bristol | Human | 7490 | gtgattggtt ggtATGgctt ctaagccatt | 13 | MASKP | FEMEG |
| AY237419 | Mc2 | Human | 7490 | gtgattggtt ggtATGgctt ctaagccatt | 13 | MASKP | FEMEG | |
| AY237420 | Mc10 | Human | 7458 | gtgattggtt agtATGgctt ccaagccatt | 13 | MASKP | FEMEG | |
| AY603425 | C12 | Human | 7476 | gtgattggtt ggATGgcttc caagccattc | 12 | MASKP | FEMEG | |
| AY646855 | SK15 | Human | 7459 | gtgattggtt agtATGgctt ccaagccatt | 13 | MASKP | FEMEG | |
| KJ950881 | NYC-A1 | Rat | (1650) | NA**** | - | NA | —MEG | |
| KJ950882 | NYC-B2 | Rat | (1650) | NA | - | NA | —MEG | |
| GIII | AF182760 | TC-PEC | Pig | 7320 | gtgatcgtgA TGgctaattg ccgtccgttg | 9 | MANCR | FVMEA |
| AY425671 | LL14 | Pig | (7291) | ———A TGgctaattg ccgtccgtta | - | MANCR | FVMEA | |
| FJ387164 | SaV1 | Pig | 7541 | gtgatcgtgA TGgctaattg ccgtccgtta | 9 | MANCR | FVMEA | |
| JX678943 | ah-1 | Pig | 7342 | gtgatcgtgA TGgctaattg ccgtccgtta | 9 | MANCR | FVMEA | |
| KF204570 | CH430 | Pig | 7341 | gtgatcgtgA TGcctaattg ccgtccgttg | 9 | MPNCR | FVMEA | |
| GIV | AJ786349 | Chiba00671 | Human | 7420 | gtgattggtt agtATGgctt ctaagccatt | 13 | MASKP | FEMEG |
| DQ058829 | Ehime1107 | Human | 7427 | gtgattggtt agtATGgctt ctaagccatt | 13 | MASKP | FEMEG | |
| DQ125333 | SW278 | Human | 7437 | gtgattggtt agtATGgctt ctaagccatt | 13 | MASKP | FEMEG | |
| DQ366346 | Ehime1596 | Human | 7428 | gtgattggtt agtATGgctt ctaagccatt | 13 | MASKP | FEMEG | |
| GV | AY646856 | NK24 | Human | 7500 | gtgatcacct tgggATGgct tccaagccac | 14 | MASKP | FEMEG |
| DQ366344 | Ehime475 | Human | 7500 | gtgatcacct tgggATGgct tccaagccac | 14 | MASKP | FEMEG | |
| AB775659 | NGY-1 | Human | 7521 | gtgatcacct tgggATGgct tccaagccat | 14 | MASKP | FEMEG | |
| AB521771 | TYMPo239 | Pig | (3949) | NA | - | NA | FEMEG | |
| AB521772 | TYMPo31 | Pig | (3949) | NA | - | NA | FEMEG | |
| JN420370 | CSL9775 | Sea lion | 7497 | gtgattggtt tgcgATGgcc tcaaagccat | 14 | MASKP | FEMEG | |
| KX000383 | WG194D-1 | Pig | 7496 | gtgatcactt tgagATGgct tcaaagccat | 14 | MASKP | FEMEG | |
| GVI | AY974192 | JJ681 | Pig | 7198 | gtgtatagtt ATGgcggcta cttgccgtca | 10 | MAATC | YTMEG |
| KJ508818 | JJ674 | Pig | 7198 | gtgtatagtt ATGgcggcta cttgccgtca | 10 | MAATC | YTMEG | |
| GVII | AB221130 | K7 | Pig | 7144 | gtgaacgttA TGgcggctac ttgccgtcat | 9 | MAATC | YKMEG |
| KX000384 | RV0042 | Pig | 7150 | gtgaacgttA TGgcggctgt ttgccgtcat | 9 | MAAVC | YKMEG | |
| KC309421 | WGP247 | Pig | (6052) | NA | - | NA | YVMEG | |
| GVIII | KC309419 | WG214D | Pig | 7497 | gtgatagctt tgATGgcctc ccggcctttc | 12 | MASRP | FEMEG |
| KC309416 | WG194D | Pig | (6654) | NA | - | NA | FEMEG | |
| KC309417 | WG197C | Pig | (6497) | NA | - | NA | FEMEG | |
| GIX | KC309418 | WG214C | Pig | (3695) | NA | - | NA | YVMEG |
| GX | AB242873 | K8 | Pig | (1617) | NA | - | NA | —MEG |
| GXI | DQ359100 | 2053P4 | Pig | (1635) | NA | - | NA | —MEG |
| GXII | KX000385 | WD1237 | Mink | (5816) | NA | - | NA | FEMEG |
| AY144337 | Canada 151A | Mink | (4556) | NA | - | NA | FEMEG | |
| GXIII | JN387134 | AN210D | Dog | 7469 | gtgatttgtt gttATGgctt ccaagccatt | 13 | MASKP | FEMEG |
| GXIV | JN899075 | TLC58 | Bat | 7695 | gtgatagtgA TGgcggcgtt aagccgtgtg | 9 | MAALS | FVMEG |
| GXV | KJ950878 | NYC-A19 | Rat | (1653) | not available | - | NA | —MEG |
| KJ950880 | NYC-E48 | Rat | (1653) | not available | - | NA | —MEG |
*The strains whose complete or partial genomic sequence were determined in this study are bolded.
** The complete genomic size does not exclude poly(A) tail, and the strains whose complete genomic sequence are not available yet are indicated by parentheses.
*** The predicted start codon is shown as capital letters.
****NA: Not available
Fig 2. Phylogenetic tree of the full length genomic sequences of 34 sapovirus strains using MEGA 6.
The five strains whose complete genomes were determined in this study are boxed with dotted lines. Among these, the two SaV strains that were newly identified in this study are also indicated with arrows. The number on each branch indicates the bootstrap value. The scale represents the amino acid substitutions per site. Each sapovirus strain is indicated in the following format: Genbank accession number-strain name (species).
The 5′-end of the newly determined GV, GVII, GVIII, and GXIII SaVs started with “GTG”, which was the same as for the other SaV strains (Table 1). The 5′- untranslated regions were 9 to 14 nt long, which shared the same size as other SaVs (Table 1).
We also determined the partial genomic sequences covering from the amino acid sequence GXPGXGKT in the putative NS3 region to the 3’-end of Po/GVII/WGP247 (6052nt), Po/GVIII/WG194D (6654nt) and Po/GVIII/WG197C (6497nt) strains, and covering amino acid sequence PLNCD in the putative NS3 region to the 3’- end of Mink/GXII/WD1237 (5816nt) strain, and covering from the amino acid sequence WKGL in the putative NS7 region to the 3’- end of Po/GIX/214C (3695nt) strain (Table 1 and Fig 1). These newly determined complete and partial SaV genomic sequences had typical conserved amino acid motifs for NS5 (KGKXX and XDEYXX), NS6 (GDCG), NS7 (WKGL, KDEL, DYSXWDST, GLPSG, and YGDD), and VP1 (PPG and WGS).
Genetic comparisons among animal and human SaVs
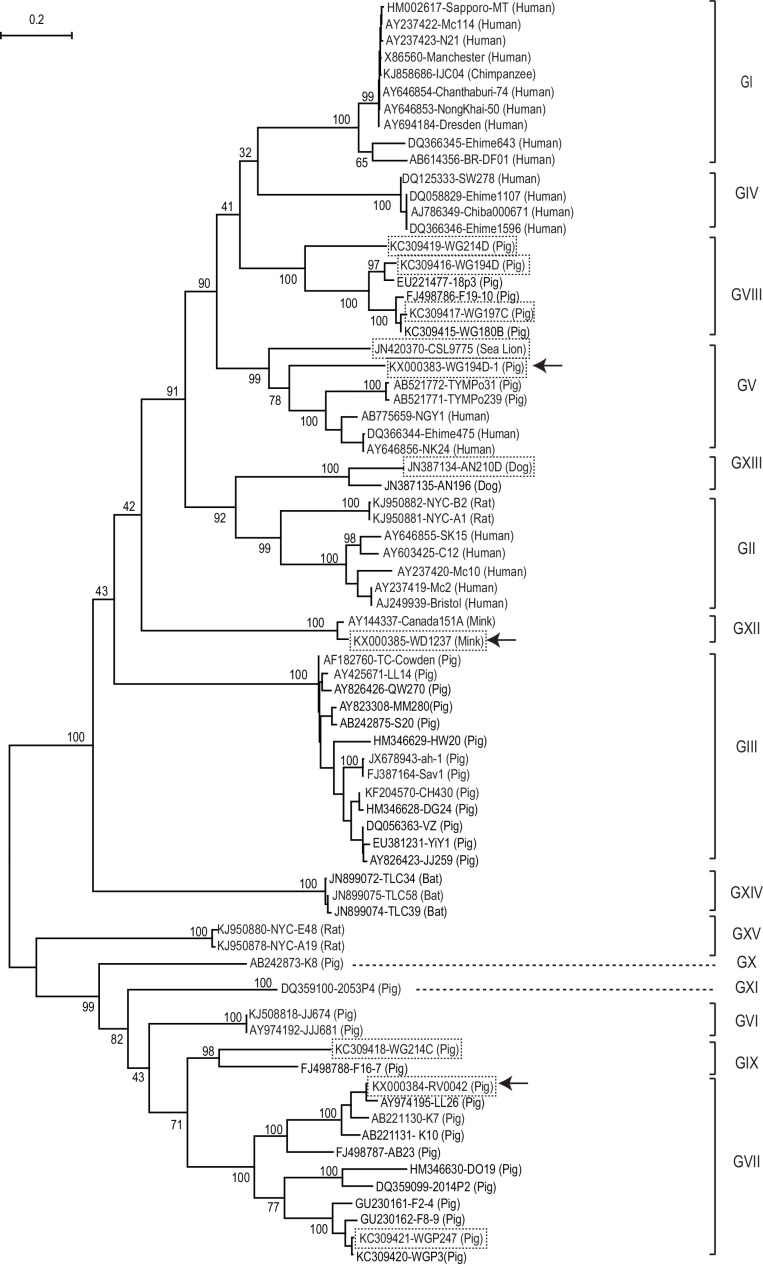
The two porcine SaVs (WG194D-1 and RV0042), and the mink SaV (WD1237) clustered with GV, GVII, and GXII SaV strains, respectively, based on phylogenetic analysis of the complete VP1 amino acid sequences (Fig 3).
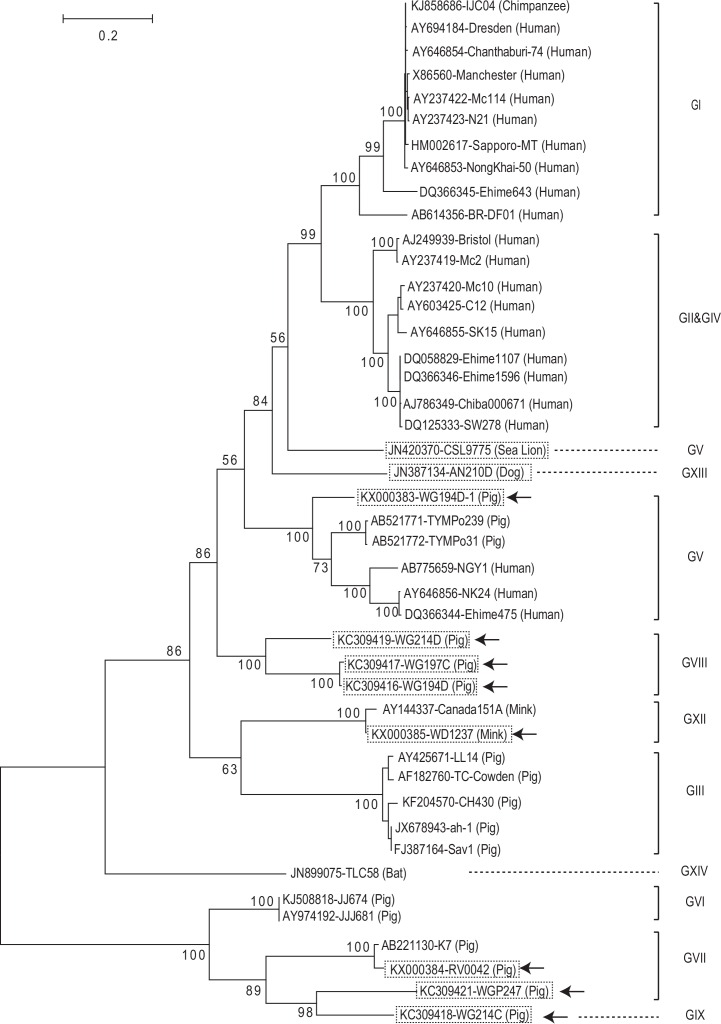
Fig 3. Phylogenetic tree based on the complete VP1 amino acid sequences of 74 sapovirus strains.
These strains represent all reported 14 genogroups (GI-GXIV) and the newly reported rat SaV strains using MEGA 6. The 10 animal SaV strains with additional sequences determined in this study are boxed with dotted lines. Among these, the three SaV strains that were newly identified in this study are also indicated with arrows. The number on each branch indicates the bootstrap value. The scale represents the amino acid substitutions per site. Each sapovirus strain is indicated in the following format: Genbank accession number-strain name (species).
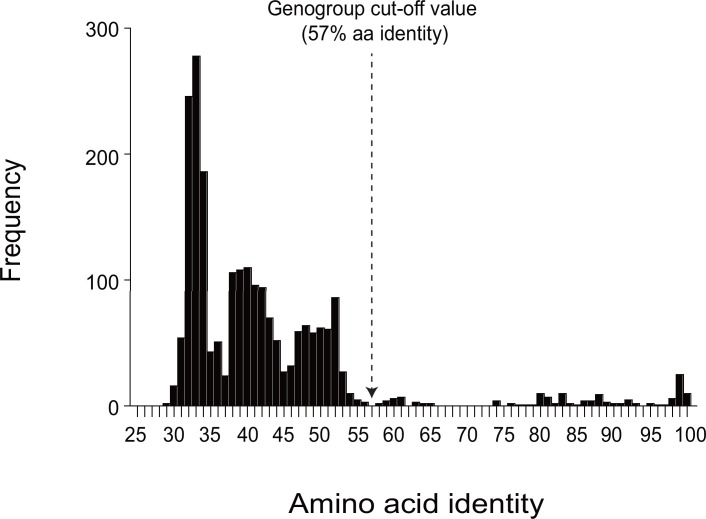
The SaVs detected from rat formed two distinct clusters (NYC-A19 and E48 cluster, and NYC-A1 and B2 cluster) based on the VP1 aa sequence similarity (Fig 3), as reported recently [9]. We proposed NYC-A19 and E48 as a new genogroup GXV, because they shared only 29.4–41.8% aa identity to other genogroups of SaVs. We also proposed NYC-A1 and B2 strains as GII, because they shared 59.6–61.1% aa identity with other human GII SaV strains. Compared with other genogroups, lower intra-genogroup aa identity was observed for GV (≥ 57.5%) and GVII (≥ 57.3%) SaV strains. We adjusted the previously proposed cut-off value of 60% VP1 aa identity for genogrouping [10] to a slightly lower value (57% identity or 43% distance) based on phylogenetic and amino acid sequence identity analysis with the newly available SaV sequence data (Figs 3 and 4).
Fig 4. Identity distribution of the VP1 amino acid sequences of 74 SaV strains.
The arrow indicates the genogrouping cut-off (≥ 57% aa identity) used in this study.The genogrouping of GV, GVII, GVIII, GIX, GXII, and GXIII SaVs sequenced in this study (as indicated as dotted box in Figs 3 and 5) based on NS7 and VP1 matched, except for the sea lion GV/CSL9775 and the porcine GVII/WGP247 strains. The Sea lion/GV/CSL9775 strain clustered together with other human and porcine GV SaV strains in the VP1 region, but it was separated from other GV strains in the NS7 region as recently discussed [1]. Similarly, GVII/ WGP247 strain clustered together with other GVII strains in the VP1 region, but it was closer to porcine GIX/WG214C in the NS7 region. The NS7 sequences of porcine GX and GXI SaVs and rat (GII and GXV) SaVs are not yet available (Table 1).
Fig 5. Phylogenetic tree of the putative NS7 amino acid sequences (approx. 500 aa) of sapovirus strains.
These 44 strains represent 12 genogroups (GI-GV, GVI, GVII, GVIII, GIX, GXII, GXIII, and GXIV) using MEGA 6. The amino acid sequences covering from “WKGL” sequence to the end of the putative NS7, “XXME”, were used. Only 44 of the 74 SaV strains in the VP1-tree (Fig 3) have sequences for this region. The 10 strains analyzed in this study are boxed with dotted lines. Among these, the eight SaV strains whose corresponding sequences were determined in this study are indicated with arrows. The NS7 sequences of porcine GIX and GX SaVs, and rat GII and GXV SaVs are not yet available. The number on each branch indicates the bootstrap value. The scale represents the amino acid substitutions per site. Each sapovirus strain is indicated in the following format: Genbank accession number-strain name (species).
From a different perspective, we noted that the first five amino acid sequences (M [A or V] S [K or R] P) of the putative NS1 and the five amino acid sequences surrounding the putative cleavage site between NS7 and VP1 (FEME/G: slash is the putative cleavage site) [23] of porcine GV and GVIII, sea lion GV and dog GXIII SaVs were similar to those of GI, GII, GIV, and GV human SaVs (M[A or V] S [K or R] P and FEME/G, respectively). In contrast, these sequences are clearly different in porcine GIII (M [A/ P] NCR and FVME/A), GVI (MAATC and YTMEG), GVII (MAA [V/T] C and Y [K/V] MEG), and bat GXIV SaVs (MAALS and FVMEG) (Table 1).
From these observations we can infer that some animal SaVs (porcine GIII, GVI and GVII and bat GXIV) evolved more distantly from human SaVs than others (porcine GV and GVIII, sea lion GV and dog GXIII). We compared such amino acid sequence characteristics partially for GIX and GXII SaVs. Only the putative cleavage sites between NS7 and VP1, YVMEG and FEMEG of GIX and GXII SaVs, respectively, are available. We could not do a similar analysis for porcine GX and GXI, and the rat GII and GXV SaVs, because the corresponding amino acid sequences are not available (Table 1).
Conclusions
Using the long RT-PCR strategy with our newly designed forward primer targeting the conserved region of the putative NS3 and the 5′ RACE methods, we identified and/or determined additional complete genomic sequences for GV, GVII, and GVIII SaVs. We also characterized the genomic extremes of sea lion GV and canine GXIII SaVs. Further determination of animal SaV genome sequences, including those SaVs (porcine GIX, GX and GXI, mink GXII, and rat GII and GXV), whose complete genomes have not been determined, may enable the design of more universal SaV primers to detect SaVs from both animals and humans. In future studies it will also be interesting to evaluate the potential interspecies transmission of closely related animal and human SaVs, such as GI, GII and GV SaVs, using experimental animals.
Acknowledgments
Nucleotide sequencing was performed at the Molecular and Cellular Imaging Center, the Ohio Agricultural Research and Development Center (OARDC), The Ohio State University. We thank Xiaohong Wang and Susan Sommer-Wagner for their technical assistance.
Data Availability
All sequence data are available from GenBank database (accession number(s) KX000383, KX000384, and KX000385, JN420370, KC309421, KC309419, KC309416, KC309417, KC309418, and JN387134).
Funding Statement
This research was supported by The Ohio State University (www.osu.edu) through start-up funding for Q. Wang. This research was also supported by the USDA, National Institute of Food and Agriculture, grant 2011-68003-30395 (to L.J. Saif and Q. Wang; subaward co-PIs)(https://nifa.usda.gov/). E.L. Delwart (PI) and T. Phan were supported by Blood Systems Research Institute and NIH (R01-HL-105770) (http://www.nih.gov/). Salaries and research support were provided by state and federal funds provided to the Ohio Agricultural Research and Development Center (OARDC), The Ohio State University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Oka T, Wang Q, Katayama K, Saif LJ. Comprehensive review of human sapoviruses. Clin Microbiol Rev. 2015;28(1):32–53. Epub 2015/01/09. 10.1128/CMR.00011-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo M, Chang KO, Hardy ME, Zhang Q, Parwani AV, Saif LJ. Molecular characterization of a porcine enteric calicivirus genetically related to Sapporo-like human caliciviruses. J Virol. 1999;73(11):9625–31. Epub 1999/10/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saif LJ, Bohl EH, Theil KW, Cross RF, House JA. Rotavirus-like, calicivirus-like, and 23-nm virus-like particles associated with diarrhea in young pigs. J Clin Microbiol. 1980;12(1):105–11. Epub 1980/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo M, Evermann JF, Saif LJ. Detection and molecular characterization of cultivable caliciviruses from clinically normal mink and enteric caliciviruses associated with diarrhea in mink. Arch Virol. 2001;146(3):479–93. Epub 2001/05/08. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L, Pesavento PA, Shan T, Leutenegger CM, Wang C, Delwart E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J Gen Virol. 2011;92(Pt 11):2534–41. Epub 2011/07/22. doi: vir.0.034611–0 [pii] 10.1099/vir.0.034611-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L, Shan T, Wang C, Cote C, Kolman J, Onions D, et al. The fecal viral flora of California sea lions. Journal of virology. 2011;85(19):9909–17. Epub 2011/07/29. 10.1128/JVI.05026-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tse H, Chan WM, Li KS, Lau SK, Woo PC, Yuen KY. Discovery and genomic characterization of a novel bat sapovirus with unusual genomic features and phylogenetic position. PLoS One. 2012;7(4):e34987 Epub 2012/04/20. 10.1371/journal.pone.0034987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mombo IM, Berthet N, Bouchier C, Fair JN, Schneider BS, Renaud F, et al. Characterization of a genogroup I sapovirus isolated from chimpanzees in the republic of congo. Genome Announc. 2014;2(4). Epub 2014/07/19. 10.1128/genomeA.00680-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P, et al. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. MBio. 2014;5(5):e01933–14. Epub 2014/10/16. 10.1128/mBio.01933-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheuer KA, Oka T, Hoet AE, Gebreyes WA, Molla BZ, Saif LJ, et al. Prevalence of porcine noroviruses, molecular characterization of emerging porcine sapoviruses from finisher swine in the United States, and unified classification scheme for sapoviruses. J Clin Microbiol. 2013;51(7):2344–53. Epub 2013/05/17. 10.1128/JCM.00865-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura K, Saga Y, Iwai M, Obara M, Horimoto E, Hasegawa S, et al. Frequent detection of noroviruses and sapoviruses in swine and high genetic diversity of porcine sapovirus in Japan during Fiscal Year 2008. J Clin Microbiol. 2010;48(4):1215–22. Epub 2010/02/19. doi: JCM.02130-09 [pii] 10.1128/JCM.02130-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang QH, Chang KO, Han MG, Sreevatsan S, Saif LJ. Development of a new microwell hybridization assay and an internal control RNA for the detection of porcine noroviruses and sapoviruses by reverse transcription-PCR. Journal of virological methods. 2006;132(1–2):135–45. Epub 2005/11/09. 10.1016/j.jviromet.2005.10.003 . [DOI] [PubMed] [Google Scholar]
- 13.Wang QH, Costantini V, Saif LJ. Porcine enteric caliciviruses: genetic and antigenic relatedness to human caliciviruses, diagnosis and epidemiology. Vaccine. 2007;25(30):5453–66. Epub 2007/01/20. 10.1016/j.vaccine.2006.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang QH, Han MG, Funk JA, Bowman G, Janies DA, Saif LJ. Genetic diversity and recombination of porcine sapoviruses. J Clin Microbiol. 2005;43(12):5963–72. Epub 2005/12/08. doi: 43/12/5963 [pii] 10.1128/JCM.43.12.5963-5972.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martella V, Lorusso E, Banyai K, Decaro N, Corrente M, Elia G, et al. Identification of a porcine calicivirus related genetically to human sapoviruses. J Clin Microbiol. 2008;46(6):1907–13. Epub 2008/04/18. doi: JCM.00341-08 [pii] 10.1128/JCM.00341-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibata S, Sekizuka T, Kodaira A, Kuroda M, Haga K, Doan YH, et al. Complete Genome Sequence of a Novel GV.2 Sapovirus Strain, NGY-1, Detected from a Suspected Foodborne Gastroenteritis Outbreak. Genome Announc. 2015;3(1). Epub 2015/02/14. 10.1128/genomeA.01553-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W, Yang B, Wang E, Liu J, Lan X. Complete sequence and phylogenetic analysis of a porcine sapovirus strain isolated from western China. Virus Genes. 2014;49(1):100–5. Epub 2014/05/06. 10.1007/s11262-014-1078-4 . [DOI] [PubMed] [Google Scholar]
- 18.Amimo JO, Vlasova AN, Saif LJ. Prevalence and genetic heterogeneity of porcine group C rotaviruses in nursing and weaned piglets in Ohio, USA and identification of a potential new VP4 genotype. Vet Microbiol. 2013;164(1–2):27–38. Epub 2013/02/23. 10.1016/j.vetmic.2013.01.039 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amimo JO, Vlasova AN, Saif LJ. Detection and genetic diversity of porcine group A rotaviruses in historic (2004) and recent (2011 and 2012) swine fecal samples in Ohio: predominance of the G9P[13] genotype in nursing piglets. J Clin Microbiol. 2013;51(4):1142–51. Epub 2013/02/01. 10.1128/JCM.03193-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sisay Z, Wang Q, Oka T, Saif L. Prevalence and molecular characterization of porcine enteric caliciviruses and first detection of porcine kobuviruses in US swine. Archives of virology. 2013;158(7):1583–8. Epub 2013/03/05. 10.1007/s00705-013-1619-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oka T, Takagi H, Saif LJ, Wang Q. Complete genome sequence of the feline calicivirus 2280 strain from the american tissue culture collection. Genome Announc. 2013;1(3). Epub 2013/06/22. 10.1128/genomeA.00349-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. Epub 2013/10/18. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oka T, Yamamoto M, Katayama K, Hansman GS, Ogawa S, Miyamura T, et al. Identification of the cleavage sites of sapovirus open reading frame 1 polyprotein. J Gen Virol. 2006;87(Pt 11):3329–38. Epub 2006/10/13. doi: 87/11/3329 [pii] 10.1099/vir.0.81799-0 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequence data are available from GenBank database (accession number(s) KX000383, KX000384, and KX000385, JN420370, KC309421, KC309419, KC309416, KC309417, KC309418, and JN387134).