Abstract
Auxin has been shown to modulate the fruit ripening process. However, the molecular mechanisms underlying auxin regulation of fruit ripening are still not clear. Illumina RNA sequencing was performed on mature green cherry tomato fruit 1 and 7 days after auxin treatment, with untreated fruit as a control. The results showed that exogenous auxin maintained system 1 ethylene synthesis and delayed the onset of system 2 ethylene synthesis and the ripening process. At the molecular level, genes associated with stress resistance were significantly up-regulated, but genes related to carotenoid metabolism, cell degradation and energy metabolism were strongly down-regulated by exogenous auxin. Furthermore, genes encoding DNA demethylases were inhibited by auxin, whereas genes encoding cytosine-5 DNA methyltransferases were induced, which contributed to the maintenance of high methylation levels in the nucleus and thus inhibited the ripening process. Additionally, exogenous auxin altered the expression patterns of ethylene and auxin signaling-related genes that were induced or repressed in the normal ripening process, suggesting significant crosstalk between these two hormones during tomato ripening. The present work is the first comprehensive transcriptome analysis of auxin-treated tomato fruit during ripening. Our results provide comprehensive insights into the effects of auxin on the tomato ripening process and the mechanism of crosstalk between auxin and ethylene.
Introduction
Fruit ripening is a complex and highly coordinated process, which includes rapid changes in color, texture and flavor. Ethylene and auxin are two important classes of phytohormones that have been reported to modulate fruit ripening [1–3]. Ethylene is the most important phytohormone in the climacteric fruit ripening process, and its function has been well documented [4–6]. Recent reports have indicated that the interaction between ethylene and auxin may be crucial for fruit ripening [7–9].
In some fruits, such as strawberry, grape and tomato, application of auxin in the pre-ripening stage delayed ripening [9–11]. However, in pear and apple, applying auxin to the whole tree instead of the fruits before the onset of ripening increased ethylene production and induced earlier softening, leading to the acceleration of ripening [12–14]. In tomato and grape berry, low levels of auxin were essential for triggering the onset of ripening, whereas elevated auxin coincided with the burst of ethylene during the ripening period [8, 15]. These results suggest that auxin not only acts as an inhibitor but also plays complex roles in modulating fruit ripening.
Auxin and ethylene have been shown to interact with each other to regulate many physiological processes [16, 17]. The transcriptional accumulation of many auxin response factors (ARF) and auxin/indole-3-acetic acid (Aux/IAA) genes in tomato is controlled by both ethylene and auxin in tomato seedlings [18–20]. Similarly, the expression of ethylene response factor (ERF) genes can also be regulated by auxin [21].
The genome sequencing of tomato (Solanum lycopersicum), a good model plant for investigating fruit development, has provided powerful insights into the molecular changes in fruit ripening [22]. However, the role of auxin in the fruit ripening process has not been fully elucidated, although the involvement of auxin response genes in the regulation of ripening has been demonstrated [23–25]. Moreover, the molecular mechanism and details of the crosstalk between auxin and ethylene are also ambiguous. To date, few comprehensive transcriptome studies have been conducted on auxin-treated tomato fruit during ripening. Thus, we employed Illumina RNA sequencing to analyze cherry tomato fruit 1 and 7 days after exogenous auxin application to elucidate the molecular regulatory mechanisms of auxin and its interplay with ethylene during the ripening period. The alteration in phytohormone signal transduction and ripening-related metabolic pathways were investigated, and the short-term and long-term effects of auxin on fruit ripening were determined. The results provide comprehensive insights into the regulatory mechanisms of exogenous auxin on tomato ripening.
Materials and Methods
Plant materials and treatments
Mature green cherry tomato fruit (Solanum lycopersicum cv. Xin Taiyang) with a uniform shape and size was collected from a standard greenhouse (20–25°C, 70%-85% RH) from Transfar Agribio Co., Ltd. in Xiaoshan County, Zhejiang Province, China. Fruits were sterilized by dipping them in 0.5% (v/v) sodium hypochlorite aqueous solution for 5 min, washing twice with sterile water and air-drying at room temperature. After removing pedicels, the fruits were randomly divided into two groups, then immersed into 0.45 mM 2, 4-dichlorophenoxyacetic acid or sterile water and infiltrated under a vacuum (35 kPa, 3 min). After treatment, the fruits were stored in the dark at 20 ± 2°C with 90 ± 5% relativity humidity (RH) for 25 days. Samples were taken at 0, 1, 4, 7, 10, 13, 15, 18, 21, 25 days after treatment (DAT). Pericarps of the sampled fruit were cut into pieces, frozen in liquid nitrogen and kept at -70°C for subsequent use.
Fruit color and texture
Five fruits from each group were used to measure fruit color and texture. Fruit color was expressed using the l*, a*, b* color space coordinates and measured with a Chroma meter (Konica Minolta, CR-400, Japan) at four symmetrical locations around the equator, as previously described [26]. Each color value was obtained from five fruits. Hue angle (H°) was calculated as H° = arctangent (b*/a*) × (180/π) to evaluate the color change. The hue angles near 120° and 0° represent pale green and red, respectively. Fruit firmness was measured by pushing a probe (5 mm diameter) into the pericarp at a speed of 1 mm·s-1 to a depth of 12 mm using a Texture Analyzer (TA-XT2i, Stable Microsystems Texture Technologies Inc., UK). The assessment was performed at two opposite locations along the fruit equator after peeling off the epidermis. The maximum force was recorded to represent fruit firmness.
Ethylene measurement
Ethylene was assayed as previously described [27]. Twenty fruits from the control or auxin groups were sealed in a 2000 mL jar and kept at 20°C in the dark for 2 h. Ethylene concentration was measured by injecting l mL headspace gas into a gas chromatograph (model SP 6800, Lunan Chemical Engineering Instrument Co., China) equipped with a GDX-502 column (JieDao TECH, China) and a flame ionization detector (Shimadzu GC-2014C, Shimadzu Corporation, Japan). Measurements were performed in triplicate.
ACO (1-aminocyclopropane-1-carboxylate oxidase) activity assay
ACO activity was measured using the method of Mei Zhang et al. [28] with slight modifications. Pericarp samples (1 g) were homogenized with 3 mL pre-chilled extract solution [100 mM Tris-HCl, pH 7.5, 10% (w/v) glycerin, 5% (w/v) polyvinylpyrrolidone, 5 mM DDT, 30 mM sodium ascorbic acid, 0.1 mM FeSO4] and centrifuged at 12,000 × g (4°C) for 10 min. The supernatant (0.5 mL) was added to 1.5 mL of the reaction solution (10% glycerin, 30 mM sodium ascorbic acid, 2 mM 1-aminocyclopropane-1-carboxylic acid, 0.1 mM FeSO4) in a 10 mL tube, sealed with a rubber plug and incubated at 30°C for 60 min. Ethylene in the head space was measured by gas chromatography. ACO activity was expressed as nmol C2H4 h-1 g-1 FW.
Library preparation and Illumina sequencing
Fruits from each group were sampled and denoted CK1d (samples from the control group at 1 DAT), CK7d (samples from the control group at 7 DAT), AX1d (samples from the auxin group at 1 DAT), or AX7d (samples from the auxin group at 7 DAT). Two biological replicates, each containing pooled pericarps from eight fruits, were prepared for the RNA-Seq assays. Library preparation and transcriptome sequencing were performed by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). Total RNA was isolated with TRIzol® LS Reagent as described by the manufacturer’s protocol. RNA concentration was measured using a Qubit® RNA Assay Kit with a Qubit® 2.0 Fluorometer (Life Technologies, CA, USA), and integrity was assessed using the RNA Nano 6000 Assay Kit with a Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
A total of 3 μg of RNA per sample was used as the input material for the RNA sample preparations. Sequencing libraries were generated using a NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina, USA) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq 2000 platform, and 100 bp paired-end reads were generated.
Transcriptome analysis
Clean data (clean reads), which were obtained by removing low quality reads from the raw data, were mapped to the tomato genome assembly SL2.50 using TopHat v2.0.2 [29]. Reads per kilobase of exon model per million mapped reads (RPKM) were calculated using HTSeq v0.6.1 to estimate gene expression levels [30].
Differential expression analysis of two groups was performed using the DESeq R package. P-values were adjusted by Q-values using the Benjamini-Hochberg method [31] for controlling the false discovery rate. Genes with Q-values < 0.05 were defined as differentially expressed.
Gene Ontology (GO) enrichment analysis of differentially expressed genes was performed using the GOseq R package as described by Young, M. D., et al. [32], in which gene length bias was corrected. GO terms with corrected P-values less than 0.05 were considered significantly enriched by differentially expressed genes. KOBAS software was used to test the statistical enrichment of differentially expressed genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways as previously described [33].
Quantitative real-time PCR assay
Total RNA was extracted using RNAiso plus (TaKaRa, Japan) according to the manufacturer’s protocol. cDNA was obtained from 1 μg of total RNA using a PrimeScript RT kit (TaKaRa, Japan). Quantitative real-time PCR assay was performed using SYBR® Premix Ex Taq (TaKaRa, Japan) on an ABI StepOne Real-Time PCR System (Applied Biosystems, USA) as previously described [27]. The tomato β-actin gene was used as the reference gene to calculate relative expression levels based on the 2-ΔΔCt method [34]. Primers are listed in S1 Table.
Correlation network analysis and statistics
Correlation network analysis was performed as previously described [35]. The network was visualized with Cytoscape version 3.2.1 (www.cytoscape.org) [36]. For biological and biochemical data, significant differences between samples were determined with a Student's t-test (independent samples) using SPSS version 20.0 (IBM Corp, Armonk, USA).
Results
Effects of auxin treatment on postharvest fruit color, firmness and ethylene synthesis
A significant inhibition in ripening was observed in auxin-treated fruit during storage (Fig 1E). The control fruit began to turn orange at 7 DAT coupled with a rapid softening, and the fruit turned red at 15 DAT. Changes in the fruit color and texture were delayed by auxin treatment. The auxin-treated fruit remained green and hard until 10 DAT (Fig 1A and 1B).
Fig 1. Physiological changes induced by exogenous auxin application in tomato fruit.
The changes in (A) hue angle, (B) firmness, (C) ACO activity, (D) and ethylene and the pictures of tomato fruits after auxin treatment. Error bars indicate the standard error of three replicates. Asterisks (*) represent significant differences between the control and auxin treatments (Student's t-test, P < 0.05).
Ethylene production was lower in auxin-treated fruit than that in control fruit during storage (Fig 1D). Auxin treatment delayed the initiation of climacteric ethylene for approximately 3 days compared with the control. ACO activity was also inhibited by auxin during the first two weeks after treatment, although the maximum activity was approximately the same in the control and auxin-treated fruits (Fig 1C).
Summary of transcriptome sequencing data
To elucidate the short-term and long-term effects of auxin treatment, fruits in both the auxin-treated and control groups were sampled at 1 day and 7 days after treatment for RNA-Seq analysis. Raw data generated by sequencing ranged from 30.3 to 37.6 million reads per sample, and more than 95% of them had a quality score ≥ Q20. After filtering, 29.9 to 37.0 million clean reads were obtained, approximately 91% of which could be mapped to the tomato reference genome. In addition, more than 90% of the clean reads were uniquely mapped reads, while the proportion of multiple mapped reads was less than 0.8% (Table 1).
Table 1. Throughput and the quality of RNA-Seq data.
| Sample name | Raw reads | Clean reads | Q20 (%) | Mapped reads | Mapped percent | Uniquely mapped reads | Multiple mapped reads |
|---|---|---|---|---|---|---|---|
| CK1d_1 | 37652220 | 37030234 | 96.67 | 68154908 | (92.03%) | 67623746 (91.31%) | 531162 (0.72%) |
| CK1d_2 | 30654627 | 30272262 | 96.35 | 55865924 | (92.27%) | 55467758 (91.61%) | 398166 (0.66%) |
| AX1d_1 | 30329099 | 29900210 | 95.71 | 54636510 | (91.36%) | 54284505 (90.78%) | 352005 (0.59%) |
| AX1d_2 | 30862471 | 30397218 | 95.94 | 55769863 | (91.74%) | 55392789 (91.11%) | 377074 (0.62%) |
| CK7d_1 | 33350282 | 32878536 | 95.74 | 60241973 | (91.61%) | 59851967 (91.02%) | 390006 (0.59%) |
| CK7d_2 | 31573280 | 31090705 | 95.66 | 56743198 | (91.25%) | 56333250 (90.60%) | 409948 (0.66%) |
| AX7d_1 | 35514663 | 34990712 | 95.87 | 63657461 | (90.96%) | 63271623 (90.41%) | 385838 (0.55%) |
| AX7d_2 | 33332070 | 32802692 | 95.78 | 60215602 | (91.78%) | 59839662 (91.21%) | 375940 (0.57%) |
Q20 and Q30 represent the percentage of the sequencing data with an error rate less than 1% and 0.1%, respectively.
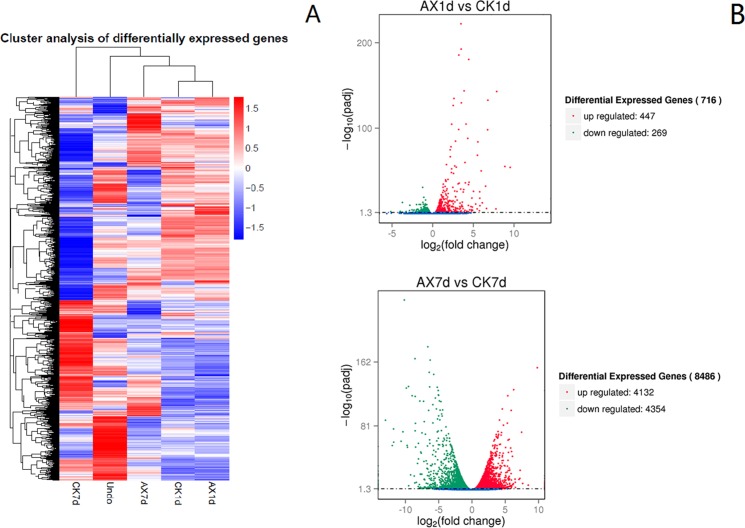
More than 34,000 genes with different abundances were detected, and approximately 50% of them had a RPKM ≥1 (S2 Table). Hierarchical cluster analysis was used to estimate the variance in differentially expressed genes (DEGs), and we obtained 16,157 DEGs in the samples (Fig 2A). Compared with the control, 447 and 4132 genes were up-regulated by auxin at 1 and 7 DAT, while 269 and 4354 genes were repressed, respectively (Fig 2B, S3 and S4 Tables).
Fig 2. Differentially expressed genes in the samples.
(A) Hierarchical clustering and heat map of differentially expressed genes based on the expression levels (RPKM). Genes in red and blue represent highly and lowly expressed genes, respectively. (B) The volcano plot shows the numbers of significantly differentially expressed genes in each comparison group.
GO enrichment and KEGG pathway analysis of DEGs
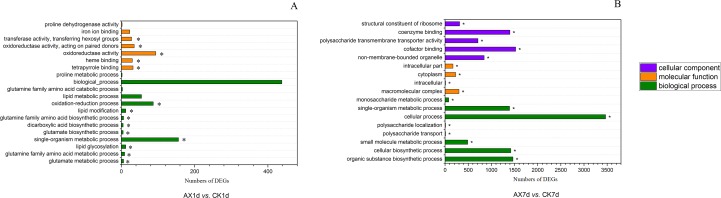
We performed GO enrichment analysis to investigate the distribution of DEGs in biological process (BP), cellular component (CC) and molecular function (MF). Fourteen GO terms (BP: 9 terms, MF: 5 terms) were significantly enriched in AX1d compared with CK1d, and glutamate metabolic process (GO: 0006536, 6 DEGs) and tetrapyrrole binding (GO: 0046906, 32 DEGs) were the most significantly enriched terms in BP and MF, respectively (Fig 3A). In addition, fifty-six GO terms (BP: 35 terms, MF: 6 terms, CC: 13 terms) were significantly enriched in AX7d compared with CK7d. The most significantly enriched terms in BP, MF and CC were cellular metabolic process (GO: 0044237, 2658 DEGs), cofactor binding (GO: 0048037, 301 DEGs) and macromolecular complex (GO: 0032991, 843 DEGs), respectively (Fig 3B). Detailed results of the GO enrichment analysis are shown in S5 and S6 Tables.
Fig 3. GO enrichment analysis of DEGs in all comparison groups.
The top twenty most enriched GO terms in the comparison groups (A) AX1d vs. CK1d (B) and AX7d vs. CK7d. Asterisks (*) indicate significantly (Q-value < 0.05) enriched GO terms.
To investigate the major pathways of the DEGs, we aligned all DEGs to KEGG pathways (S7 and S8 Tables). In the AX1d vs. CK1d group, DEGs were enriched in 84 KEGG metabolic pathways and “Metabolic pathways” (75 DEGs), “Biosynthesis of secondary metabolites” (49 DEGs) and “Plant hormone signal transduction” (28 DEGs) were the top three pathways containing the greatest number of DEGs. Among the 84 pathways, nine had P-values > 0.05, and “Glutathione metabolism”, “Plant hormone signal transduction”, and “Photosynthesis-antenna proteins” were the most significantly enriched pathways (Q-value < 0.05) (Fig 4). In the AX7d vs. CK7d group, one hundred and twenty-three KEGG metabolic pathways were identified, and the top three pathways containing the greatest number of DEGs were “Metabolic pathways” (861 DEGs), “Biosynthesis of secondary metabolites” (471 DEGs) and “Ribosome” (158 DEGs). Seven KEGG pathways had P-values < 0.05, and “Biosynthesis of amino acids” (144 DEGs), “Carbon metabolism” (151 DEGs), “Glycolysis/Gluconeogenesis” (77 DEGs) were the major enriched pathways (Fig 4).
Fig 4. KEGG enrichment analysis of DEGs in all comparison groups.
The top ten most enriched KEGG pathways in the comparison groups (A) AX1d vs. CK1d and (B) AX7d vs. CK7d. The bars in red, yellow and dark green represent the KEGG pathways with different enrichment levels (Q-value < 0.05, P-value < 0.05 but Q-value > 0.05, P-value > 0.05, respectively).
Expression of genes involved in ethylene synthesis and signal transduction
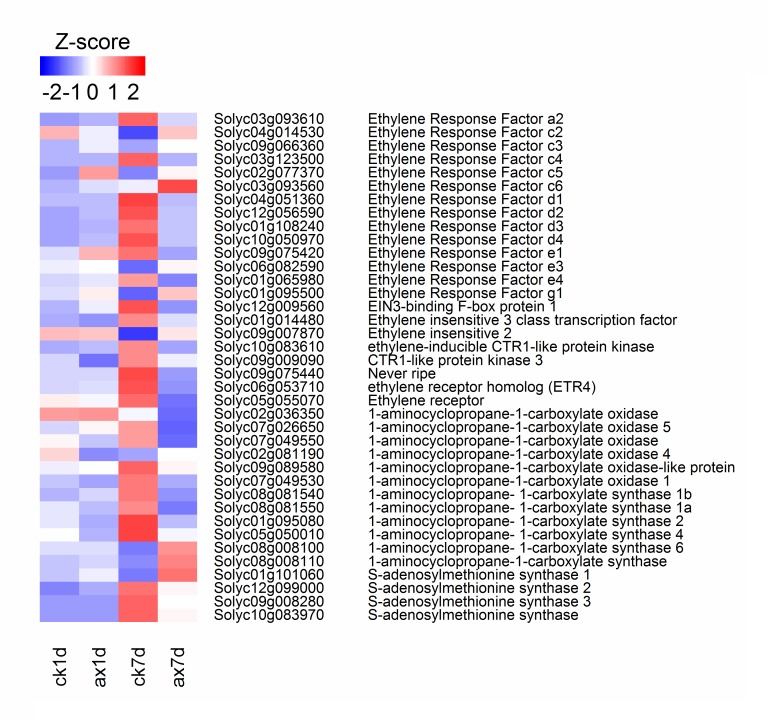
Thirty-nine genes related to ethylene synthesis and signal transduction were differentially expressed among the samples (Fig 5, Table 2). Compared with the control, the expression levels of SlACS4 (Solyc05g050010) and SlACO4 (Solyc02g081190) were reduced in auxin-treated samples at 1 DAT. However, at 7 DAT, one SAM (S-adenosylmethionine synthase) gene (Solyc01g101060) was induced by exogenous auxin, while the other three (Solyc10g083970, Solyc09g008280, Solyc12g099000) were down-regulated. ACS (1-aminocyclopropane-1-carboxylate synthase) and ACO (1-aminocyclopropane-1-carboxylate oxidase) are two important enzymes in ethylene biosynthesis. In this experiment, four ACS genes (Solyc05g050010, Solyc01g095080, Solyc08g081550, Solyc08g081540) and six ACO genes (Solyc07g049530, Solyc09g089580, Solyc02g081190, Solyc07g049550, Solyc07g026650, Solyc02g036350) were variously down-regulated in AX7d, but two ACS genes (Solyc08g008110, Solyc08g008100) were significantly up-regulated, although ethylene biosynthesis was repressed (Fig 1D).
Fig 5. Heat map of the expression levels of the DEGs involved in ethylene synthesis and signal transduction.
The RPKM values were normalized with log2 (RPKM+1) and converted to Z-scores to scale the expression levels of DEGs involved in ethylene synthesis and signal transduction. Red indicates high expression, while blue denotes low expression.
Table 2. DEGs involved in ethylene synthesis and signal transduction.
| Fold change (log2 ratio) | |||
|---|---|---|---|
| Gene ID | Annotation | AX1d vs.CK1d | AX1d vs.CK1d |
| Solyc10g083970 | S-adenosylmethionine synthase | - | -1.76 |
| Solyc09g008280 | S-adenosylmethionine synthase 3 | - | -2.74 |
| Solyc12g099000 | S-adenosylmethionine synthase 2 | - | -1.24 |
| Solyc01g101060 | S-adenosylmethionine synthase 1 | - | 1.47 |
| Solyc08g008110 | 1-aminocyclopropane-1-carboxylate synthase | - | 3.86 |
| Solyc08g008100 | 1-aminocyclopropane-1-carboxylate synthase 6 | - | 3.45 |
| Solyc05g050010 | 1-aminocyclopropane-1-carboxylate synthase 4 | -2.00 | -4.48 |
| Solyc01g095080 | 1-aminocyclopropane-1-carboxylate synthase 2 | - | -5.01 |
| Solyc08g081550 | 1-aminocyclopropane-1-carboxylate synthase 1a | - | -2.48 |
| Solyc08g081540 | 1-aminocyclopropane-1-carboxylate synthase 1b | - | -2.34 |
| Solyc07g049530 | 1-aminocyclopropane-1-carboxylate oxidase 1 | - | -3.77 |
| Solyc09g089580 | 1-aminocyclopropane-1-carboxylate oxidase-like protein | - | -2.25 |
| Solyc02g081190 | 1-aminocyclopropane-1-carboxylate oxidase 4 | -1.10 | - |
| Solyc07g049550 | 1-aminocyclopropane-1-carboxylate oxidase | - | -5.96 |
| Solyc07g026650 | 1-aminocyclopropane-1-carboxylate oxidase 5 | - | -2.24 |
| Solyc02g036350 | 1-aminocyclopropane-1-carboxylate oxidase | - | -0.87 |
| Solyc05g055070 | Ethylene receptor | - | -1.44 |
| Solyc06g053710 | Ethylene receptor homolog (ETR4) | - | -1.94 |
| Solyc09g075440 | Never ripe | - | -3.00 |
| Solyc09g009090 | CTR1-like protein kinase 3 | - | -0.46 |
| Solyc10g083610 | Ethylene-inducible CTR1-like protein kinase | - | -1.20 |
| Solyc09g007870 | Ethylene insensitive 2 | - | 0.66 |
| Solyc01g014480 | Ethylene insensitive 3 class transcription factor | - | -0.58 |
| Solyc07g008250 | EIN3-binding F-box protein | -2.13 | |
| Solyc12g009560 | EIN3-binding F-box protein 1 | - | -3.07 |
| Solyc01g095500 | Ethylene response factor g1 | - | 3.03 |
| Solyc01g065980 | Ethylene response factor e4 | - | -1.02 |
| Solyc06g082590 | Ethylene response factor e3 | - | 2.67 |
| Solyc09g075420 | Ethylene response factor e1 | - | -2.06 |
| Solyc10g050970 | Ethylene response factor d4 | - | -4.43 |
| Solyc01g108240 | Ethylene response factor d3 | - | -3.92 |
| Solyc12g056590 | Ethylene response factor d2 | - | -2.20 |
| Solyc04g051360 | Ethylene response factor d1 | - | -9.52 |
| Solyc03g093560 | Ethylene response factor c6 | - | 1.40 |
| Solyc02g077370 | Ethylene response factor c5 | 1.93 | 1.28 |
| Solyc03g123500 | Ethylene response factor c4 | - | -1.71 |
| Solyc09g066360 | Ethylene response factor c3 | - | 2.13 |
| Solyc04g014530 | Ethylene response factor c2 | -1.08 | 3.88 |
| Solyc03g093610 | Ethylene response factor a2 | - | -3.53 |
“-” represents no significant difference. The fold change value is represented by the log2 ratio.
Ethylene signal transduction plays crucial roles in mediating the biochemical changes caused by ethylene. At 1 DAT, the expressions of the most genes involved in ethylene signal transduction in auxin-treated samples showed no difference compared to the control except SlERF.c2 (Solyc04g014530) and SlERF.c5 (Solyc02g077370) (Fig 5, Table 2). However, at 7 DAT, expressions of Nr (never-ripe) (Solyc09g075440) and ETR4 (ethylene receptor) (Solyc06g053710), which showed higher levels (RPKM > 50) than the other ethylene receptors in the samples (S2 Table), were dramatically inhibited by auxin. The transcript levels of two EIN3 binding F-Box proteins-encoding genes (EBF) (Solyc07g008250, Solyc12g009560), which mediated ethylene signal transduction via the proteolysis of the transcription factor EIN3, were down-regulated in auxin-treated samples at 7 DAT. Moreover, Sl-ERF.c2 (Solyc04g014530) and Sl-ERF.d1 (Solyc04g051360) were the most significantly induced and repressed member of the Ethylene-responsive transcription factors (ERF) family in AX7d, respectively.
Expression of genes involved in indoleacetic acid (IAA) synthesis and signal transduction
Forty-five DGEs related to IAA biosynthesis and signal transduction were identified (Fig 6, Table 3). In the IAA biosynthesis pathway, of the two monooxygenases, the one with the highest abundance (Solyc06g008050) (S2 Table) was down-regulated by auxin at both 1 and 7 DAT. Auxin response factors (ARF) regulate the expressions of auxin-responsive genes, which contain auxin response elements in the promoter regions. At 1 DAT, most ARF genes were stably expressed in the auxin group fruit, except for one (Solyc11g069190), which changed slightly. At 7 DAT, among ten differentially expressed ARF genes, five (Solyc12g042070, Solyc07g016180, Solyc02g037530, Solyc09g007810, Solyc05g056040) were up-regulated, and the others (Solyc03g118290, Solyc11g069190, Solyc04g081240, Solyc05g047460, Solyc01g096070) were down-regulated in auxin-treated fruit. Three early auxin response gene families, auxin/indole-3-acetic acid (Aux/IAA), Gretchen Hagen-3 (GH3), and small auxin-up RNA (SAUR), were directly regulated by ARFs. Aux/IAA proteins are negative regulators of auxin signal transduction that bind to ARFs. Fifteen Aux/IAA genes were significantly up-regulated by auxin and maintained high expression levels at 7 DAT. For the GH3 gene family, four GH3 genes (Solyc02g092820, Solyc02g064830, Solyc01g107400, Solyc01g107390) were up-regulated in AX1d vs. CK1d, and three of them showed the same pattern in AX7d vs. CK7d except one (Solyc01g107400). Moreover, the expression levels of two GH3 genes (Solyc01g107400, Solyc01g107390) were higher in CK7d than those in CK1d. For the SAUR gene family, no differences were observed in most SAUR genes in AX1d vs. CK1d except SlSAUR58 (Solyc06g053260).
Fig 6. Heat map of expression levels of the DEGs involved in IAA synthesis and signal transduction.
The RPKM values were normalized with log2 (RPKM+1) and converted to Z-scores to scale the expression levels of DEGs involved in ethylene synthesis and signal transduction. Red indicates high expression, while blue indicates low expression.
Table 3. DEGs involved in IAA synthesis and signal transduction.
| Fold change (log2 ratio) | |||
|---|---|---|---|
| Gene ID | Annotation | AX1d vs. CK1d | AX1d vs. CK1d |
| Solyc06g065630 | Monooxygenase | - | -5.57 |
| Solyc06g008050 | Monooxygenase | -1.45 | -1.14 |
| Solyc09g074520 | Auxin F-box protein 5 | - | 1.22 |
| Solyc06g007840 | Auxin signaling F-box 2 | - | 1.58 |
| Solyc03g118290 | Auxin response factor 2a | - | -1.18 |
| Solyc12g042070 | Auxin response factor 2b | - | 0.50 |
| Solyc11g069190 | Auxin response factor 4 | 0.59 | -1.25 |
| Solyc04g081240 | Auxin response factor 5 | - | -0.95 |
| Solyc07g016180 | Auxin response factor 7a | - | 1.71 |
| Solyc05g047460 | Auxin response factor 7b | - | -0.79 |
| Solyc02g037530 | Auxin response factor 8b | - | 0.83 |
| Solyc09g007810 | Auxin response factor 16a | - | 0.57 |
| Solyc01g096070 | Auxin response factor 18 | - | -1.27 |
| Solyc05g056040 | Auxin response factor 24 | - | 1.11 |
| Solyc03g120390 | AUX/IAA protein 15 | 2.34 | 3.26 |
| Solyc06g053830 | AUX/IAA protein 7 | - | 2.48 |
| Solyc03g120500 | AUX/IAA protein 6 | -0.42 | 1.23 |
| Solyc06g066020 | AUX/IAA protein 36 | 8.87 | 4.38 |
| Solyc09g065850 | AUX/IAA protein 3 | 4.02 | -0.47 |
| Solyc12g007230 | AUX/IAA protein 8 | - | 1.71 |
| Solyc09g090910 | AUX/IAA protein 13 | 3.26 | 3.60 |
| Solyc09g083280 | AUX/IAA protein 1 | 3.67 | 3.14 |
| Solyc09g064530 | AUX/IAA protein 22 | 4.11 | 3.52 |
| Solyc06g084070 | AUX/IAA protein 2 | 3.74 | - |
| Solyc07g008020 | AUX/IAA protein 35 | 6.79 | 5.94 |
| Solyc01g097290 | AUX/IAA protein 9 | 3.97 | 4.17 |
| Solyc03g121060 | AUX/IAA protein 26 | - | 2.67 |
| Solyc06g008590 | AUX/IAA protein 10 | 2.45 | 7.20 |
| Solyc06g053840 | AUX/IAA protein 4 | 0.88 | 2.59 |
| Solyc09g008170 | SAUR protein 69 | - | -9.54 |
| Solyc07g014620 | SAUR protein 63 | - | 2.89 |
| Solyc06g072650 | SAUR protein 61 | - | -1.71 |
| Solyc06g053260 | SAUR protein 58 | 3.32 | - |
| Solyc04g081270 | SAUR protein 52 | - | 3.57 |
| Solyc04g081250 | SAUR protein 51 | - | 2.42 |
| Solyc03g082530 | SAUR protein 37 | - | -4.44 |
| Solyc02g084010 | SAUR protein 33 | - | 2.41 |
| Solyc01g110560 | SAUR protein 3 | - | 2.67 |
| Solyc01g096340 | SAUR protein 2 | - | -1.63 |
| Solyc01g110680 | SAUR protein 12 | - | 2.17 |
| Solyc01g091030 | SAUR protein 1 | - | 4.23 |
| Solyc02g092820 | IAA-amido synthetase 3–4 | 13.60 | 9.81 |
| Solyc02g064830 | IAA-amido synthetase 3–3 | 6.75 | 4.51 |
| Solyc01g107400 | IAA-amido synthetase | 1.04 | -5.50 |
| Solyc01g107390 | Auxin-responsive GH3 product | 3.52 | 1.98 |
Expression of genes involved in carotenoid metabolism
The color transition from green to red, with the accumulation of carotenoid and lycopene, is a major change during ripening of tomato fruit. We observed a significant inhibition of color change in auxin-treated fruit (Fig 1E). Therefore, we investigated the expressions of the genes encoding the enzymes associated with carotenoid metabolism. Most genes upstream of lycopene were down-regulated, with log2 fold changes ranging from -3.35 to -0.98 in AX7d compared with CK7d, except SlPSY2 (phytoene synthase 2) (Solyc02g081330) (Table 4). In the downstream carotenoid metabolic pathway, β-carotene is generated from lycopene and finally converted to neoxanthin by various enzymes. Two genes encoding lycopene beta-cyclase (Solyc10g079480, Solyc04g040190) and one gene encoding zeaxanthin epoxidase (SlZEP, Solyc10g083790) were up-regulated whereas the genes encoding β-carotene hydroxylase (Solyc03g007960) and carotenoid isomerase (Solyc10g081650) were down-regulated in AX7d vs. CK7d.
Table 4. DEGs associated with carotenoid metabolism.
| Fold change (log2 ratio) | |||
|---|---|---|---|
| Gene ID | Annotation | AX1d vs. CK1d | AX1d vs. CK1d |
| Solyc02g090890 | Zeaxanthin epoxidase | - | 2.65 |
| Solyc01g097810 | Zeta-carotene desaturase | - | -1.00 |
| Solyc02g081330 | Phytoene synthase 2 | - | 1.46 |
| Solyc03g031860 | Phytoene synthase 1 | - | -3.36 |
| Solyc10g079480 | Beta-lycopene cyclase | - | 2.27 |
| Solyc04g040190 | Lycopene beta-cyclase | - | 1.06 |
| Solyc06g074240 | Lycopene beta cyclase | - | 0.00 |
| Solyc10g083790 | Cytochrome P450 | - | 0.69 |
| Solyc10g081650 | Carotenoid isomerase | - | -0.98 |
| Solyc03g007960 | Beta-carotene hydroxylase-2 | - | -3.77 |
Expression of genes involved in cell wall degradation
In the present work, fruit softening was observed to be delayed by auxin treatment (Fig 1B). Polygalacturonase (PG), pectinesterase (PME), β-xylosidase, pectate lyase and expansin (EXP) play crucial roles in cell wall degradation. At 1 DAT, one (Solyc10g080210) of the genes encoding a polygalacturonase precursor was found to be repressed by exogenous auxin. At 7 DAT, most of the genes encoding these enzymes were significantly down-regulated in AX7d vs. CK7d (Table 5). In contrast to the other members of the expansin-coding genes, SlEXP1 (Solyc06g051800) was greatly reduced in AX7d vs. CK7d.
Table 5. DEGs associated with cell wall degradation.
| Fold change (log2 ratio) | |||
|---|---|---|---|
| Gene ID | Annotation | AX1d vs. CK1d | AX1d vs. CK1d |
| Solyc10g080210 | Polygalacturonase-2 precursor | -1.67 | -10.17 |
| Solyc06g060170 | Probable polygalacturonase-like | - | -1.88 |
| Solyc05g005170 | Polygalacturonase | - | -8.58 |
| Solyc12g098340 | Probable pectinesterase 29-like | - | 0.65 |
| Solyc03g083360 | Probable pectinesterase | - | -1.17 |
| Solyc03g078090 | Probable pectinesterase | - | -6.10 |
| Solyc07g017600 | Pectinesterase | - | -2.31 |
| Solyc08g081620 | Endo-1,4-beta-glucanase precursor | - | -2.63 |
| Solyc09g010210 | Endo-1,4-beta-glucanase precursor | - | -2.77 |
| Solyc02g091680 | Probable beta-D-xylosidase 6-like | - | -1.12 |
| Solyc01g104950 | Beta-xylosidase | - | -2.29 |
| Solyc10g047030 | Beta-D-xylosidase 1 precursor | - | -6.44 |
| Solyc09g005850 | Probable pectate lyase 4-like | - | -1.22 |
| Solyc03g111690 | Probable pectate lyase 18-like | - | -4.67 |
| Solyc09g091430 | Probable pectate lyase 15-like | - | -6.87 |
| Solyc03g031840 | Expansin precursor | - | 1.58 |
| Solyc06g051800 | Expansin 1 | - | -1.28 |
| Solyc10g086520 | Expansin precursor 6 | - | 2.31 |
| Solyc02g088100 | Expansin precursor 5 | - | 1.87 |
Expression of genes involved in energy metabolism
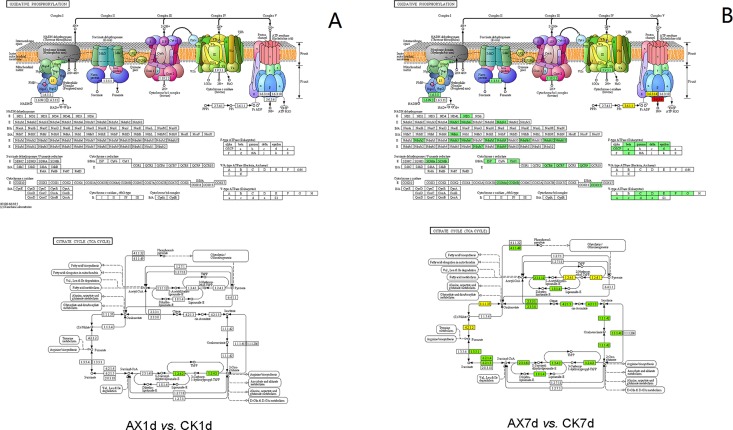
Fruit ripening is a highly complex process that requires a sufficient energy supply for the synthesis of a large number of mRNAs, proteins, flavor compounds, and other molecules. In the comparison group AX1d vs. CK1d, only one gene (Solyc04g011350) associated with the citrate cycle (TCA cycle) was slightly depressed, and no DEGs were enriched in the oxidative phosphorylation pathway (Fig 7A). However, most of the genes involved in the TCA cycle and oxidative phosphorylation pathway were significantly down-regulated in AX7d vs. CK7d (Fig 7B). These results indicate that auxin may inhibit fruit respiration rate and reduce fruit vitality during ripening.
Fig 7. The expression of DEGs in oxidative phosphorylation and the citrate cycle.
The expression patterns of DEGs involved in oxidative phosphorylation and the citrate cycle in the comparison groups (A) AX1d vs. CK1d and (B) AX7d vs. CK7d. Red and green boxes represent the genes that are up-regulated and down-regulated, respectively. The yellow box represents the genes that are both up- and down-regulated.
Validation of RNA-Seq data by RT-qPCR
The expression patterns of twelve genes that were randomly selected from the RNA-Seq data were validated by RT-qPCR. The results of linear regression analysis indicated a high correlation (r2 = 0.94) between the data of RT-qPCR and RNA-Seq (S1 Fig).
Discussion
The short-term and long-term effects of exogenous auxin on tomato fruit
Auxin regulates many plant physiological processes by modulating the expressions of auxin-response genes. Three gene families, AUX/IAA, GH3 and SAUR, exhibit rapid changes in expression in response to exogenous auxin, indicating the necessity for a quick response to auxin signaling in the plant [37]. However, many auxin-induced physiological or molecular changes did not appear in a short-term period, suggesting the importance of the long-term effects of auxin [8–10]. Here, we analyzed the short-term (1 DAT) and long-term (7 DAT) effects of auxin on the biology and molecular processes in postharvest tomato fruit.
For short-term effects, we found that glutathione metabolism was the most enriched KEGG pathway in AX1d vs. CK1d (Fig 4). Thirteen genes encoding glutathione S-transferases (GSTs) were strongly up-regulated by auxin treatment. GSTs catalyze the reaction converting glutathione (GSH) to R-S-glutathione and are essential enzymes in plant stress resistance that protect the tissues against oxidative stress, toxins, herbicide stress, and other stressors [38–40]. The improvement of stress tolerance and resistance by overexpression of GST-encoding genes has been reported in other studies [41–43]. Additionally, the expression of GSTs can also be induced by auxin [44, 45] and many other phytohormones [46, 47], indicating the involvement of GSTs in the hormone signal response. In addition to GSTs, twelve and four members of the AUX/IAA and GH3 gene family were up-regulated with various log2 fold changes in auxin-treated fruit, respectively (Table 3). Most of these auxin response genes decreased at 7 DAT (Table 3), suggesting that they may mediate the short-time effects of auxin. With regard to TFs, NAC (NAM, ATAF, and CUC) family genes were the major regulated TF genes in AX1d vs. CK1d (S2 Fig, S9 Table). One NAC family member, SlNAM3 (Solyc06g069710), was the most highly induced TF gene in auxin-treated fruit (S9 Table). NAC TFs participate in various biological processes, including stress resistance [48]. Overexpression of NAC TFs can enhance plant drought resistance and salt tolerance [49–51]. Moreover, high levels of auxin can also result in stress in the plant, which involves inducing defense- and detoxication-related genes to maintain auxin homeostasis. Based on these reports, we hypothesize that the improvement of defense and stress resistance functions may be one of the major short-term effects of auxin in postharvest tomato fruit.
For the long-term effects, we observed that most of the genes associated with carotenoid metabolism, cell wall degradation and energy metabolism were down-regulated in auxin-treated fruit. In tomato, SlPSY2 (Solyc02g081330), which has been reported to be responsible for carotenoid accumulation in root and green tissues [52], was up-regulated by auxin. In contrast, SlPSY1 (Solyc03g031860) is functional during fruit ripening, and its level are higher than that of SlPSY2 in fruit [52]. The expression of SlPSY1 was strongly repressed by auxin at 7 DAT, suggesting that auxin has a longer inhibitory effect than that described in the previous report [9]. Regarding the effects on fruit softening, auxin may play contradictory roles in different fruits and development stages. Auxin appears to promote softening of peach [53] and apple [54, 55], but it maintains the firmness of strawberry [56] and citrus [57]. In the present study, we confirmed that applying auxin to tomato fruit before ripening repressed the genes associated with cell wall degradation. In addition, another significant long-term effect of auxin was the repression of fruit energy metabolism. The auxin-mediated restriction on respiration has been observed in apple [58] and loquat fruitlets [59], but reports examining the direct effects of auxin on energy metabolism are still lacking. Based on our transcriptome sequencing results, the inhibition of energy metabolism by auxin was also a long-term effect. The expressions of genes associated with the TCA cycle and oxidative phosphorylation in auxin-treated fruit showed a marked decrease at 7 DAT but almost no difference at 1 DAT compared with the control fruit (Fig 7). Notably, compared with the fruit at AX1d, most of oxidative phosphorylation-related genes were down-regulated at AX7d, suggesting a homeostatic regulation of the energy supply corresponding to the lower energy demand due to delayed ripening.
Auxin may maintain system 1 ethylene synthesis and prevent the initiation of system 2 ethylene
The transition from system 1 to system 2 ethylene production is essential for climacteric fruit ripening. In tomato, at least nine ACS genes (SlACS1a, SlACS1b, SlACS2-8) and five ACO genes (SLACO1-5) have been identified, and their expression patterns have been studied [60–64]. SlACS1a and SlACS6 are responsible for system 1 ethylene production [60, 65]. The transcript levels of these two genes declined in mature green tomato fruit after treatment with exogenous ethylene, demonstrating the negative effect of ethylene on the regulation of these two genes [65]. Moreover, SlACS1a also plays an important role in the transition from system 1 to system 2 ethylene [65]. Two other members of the ACS gene family, SlACS2 and SlACS4, which are repressed in the pre-climacteric period but greatly induced at the ripening stage, are responsible for system 2 ethylene synthesis [60, 65, 66]. In addition, SlACO1, SlACO3 and SlACO4 are expressed at low levels before the climacteric period but are increased at the onset of the ripening stage [67]. In the present study, we found that the system 2 ethylene initiated at 7 DAT and the expressions of SlACS1a (Solyc08g081550), SlACS1b (Solyc08g081540), SlACS2 (Solyc01g095080), SlACS4 (Solyc05g050010), SlACO1 (Solyc07g049530), SlACO3 (Solyc09g089580), ACO4 (Solyc07g049550) and SlACO5 (Solyc07g026650) were increased. However, in auxin-treated fruit, the expressions of both ACS and ACO, which are associated with system 2 ethylene synthesis, were still at low levels (Table 2), resulting in low ethylene production (Fig 1E). These results suggest that auxin application in the pre-climacteric period may delay the onset of system 2 ethylene biosynthesis. We also observed that SlACS6 (Solyc08g008100), a crucial ACS gene related to system 1 ethylene synthesis, was greatly induced in auxin-treated fruit at 7 DAT (Table 2, Fig 5), indicating that auxin may maintain system 1 ethylene synthesis.
Previous studies have shown that epigenetic remodeling plays a crucial role in the transition from ethylene system 1 to ethylene system 2 [68, 69]. The methylation level of promoters affects the binding affinity of transcription factors, leading to the modulation of gene expression. The promoter region of the ACS and ACO genes is hypermethylated at the developing stage and demethylated during ripening [69]. In addition, the expressions of many DNA methyltransferase-coding genes are also modulated by 1-methylcyclopropene (1-MCP), an ethylene inhibitor, demonstrating the complex interaction between ethylene and methylation [68]. In control fruit, from 1 DAT to 7 DAT, we observed a significant increase in three DNA demethylase-coding genes (Solyc09g009080, Solyc10g083630, Solyc11g007580), along with a decrease in five cytosine-5 DNA methyltransferase-coding genes (Solyc11g030600, Solyc04g005250, Solyc01g006100, Solyc12g100330, Solyc02g062740) (Table 6). Compared with the control, most of the cytosine-5 DNA methyltransferase-encoding genes and DNA demethylase-encoding genes were induced and repressed in auxin-treated fruit, respectively (Table 6). This may contribute to the maintenance of high methylation levels in the nucleus, inhibiting the demethylation of system 2 ethylene-related genes and delaying the onset of climacteric ethylene.
Table 6. DEGs associated with DNA methylation.
| Fold change (log2 ratio) | ||||
|---|---|---|---|---|
| Gene ID | Annotation | AX1d vs. CK1d | AX1d vs. CK1d | AX1d vs. CK1d |
| Solyc09g009080 | DNA demethylase1 | - | - | 0.87 |
| Solyc10g083630 | DNA demethylase 2 | - | -1.43 | 1.01 |
| Solyc11g007580 | DNA demethylase 3 | -0.49 | -1.84 | 1.76 |
| Solyc11g030600 | Cytosine-5 DNA methyltransferase | - | 0.89 | -1.36 |
| Solyc04g005250 | Cytosine-5 DNA methyltransferase 1 | - | 1.25 | -1.22 |
| Solyc01g006100 | Cytosine-5 DNA methyltransferase 1L | - | - | -6.36 |
| Solyc12g100330 | Cytosine-5 DNA methyltransferase 3L | - | -0.49 | -0.69 |
| Solyc02g062740 | Cytosine-5 DNA methyltransferase 5 | - | - | -0.43 |
The crosstalk between auxin and ethylene in the fruit ripening process
The crosstalk between auxin and ethylene may occur through the expression of genes that contain both auxin response elements (AuxRE) and ethylene response motifs (ERELEE4) in their promoter or respond to both of the phytohormones [17]. Many ARF and ERF promoters contain several AuxRE and ERELEE4 motifs, allowing these transcription factors to modulate the expression of each other [20, 70]. In Arabidopsis, AtARF7 and AtARF19 have been reported to be regulators involved in the ethylene response [71]. CpARF7, a homolog of AtARF7 in papaya (Carica papaya L.), was also shown to be involved in fruit ripening via the regulation of ethylene signaling [72]. In addition to the regulation by ethylene, several ERF genes are also induced by auxin [7, 21, 73]. The existence of ethylene-auxin interactions during fruit ripening has been demonstrated in a previous study [7]. According to our results, eight ERF genes, especially SlERF.a2 (Solyc03g093610), SlERF.d1 (Solyc04g051360), SlERF.d3 (Solyc01g108240) and SlERF.d4 (Solyc10g050970), were dramatically repressed by auxin (Fig 5). However, of the five auxin-induced ERF genes, the expression of SlERFc2 (Solyc04g014530) and SlERFe3 (Solyc06g082590) showed the highest increase in the AX7d vs. CK7d comparison. These results indicate that auxin participates in the ethylene response by modulating ERF gene expression. With regard to ARF genes, five (SlARF2a, Solyc03g118290; SlARF4, Solyc11g069190; SlARF5, Solyc04g081240; SlARF7b, Solyc05g047460; SlARF18, Solyc01g096070) were up-regulated and three (SlARF7a, Solyc07g016180; SlARF8b, Solyc02g037530; SlARF24, Solyc05g056040) were down-regulated during ripening (Fig 6). These ARF genes may play contrary roles in ripening. For example, SlARF4 plays an important role in the control of sugar and chlorophyll metabolism. Down-regulation of SlARF4 results in dark green fruit and blocks ripening [23, 24]. The ARF genes SlARF2a and SlARF2b are involved in the regulation of many key TF genes, such as RIN, CNR and NOR, and are down-regulated, resulting in the inhibition of ripening [74]. Most of the ARF gene expression patterns were consistent with a previous study [20], except SlARF4. This slight difference may be due to the tomato cultivar. In addition to the ARF and ERF genes, the TF genes associated with ripening were differentially regulated by auxin and ethylene. Three crucial ripening-related TF genes, RIN (Solyc05g012020), CNR (Solyc02g077920) and TAGL1 (Solyc07g055920), were all repressed by auxin (RIN, log2 ratio: -2.89; CNR, log2 ratio: -2.16; TAGL1, log2 ratio: -0.64) (S4 Table). These findings suggest that auxin may regulate the expression of ripening-related TF genes, leading to changes in carotenoid biosynthesis, cell wall degradation and energy metabolism, finally delaying the ripening process.
We also performed correlation network analysis to assess the potential interactions between auxin and ethylene as previously described [35]. Only correlations between auxin- and ethylene-related genes are shown in this network (Fig 8). Five auxin-related nodes with high correlation values (node strength > 0.6) (S10 Table), SlSAUR63, SlSAUR52, SlIAA8, SlARF7a, and SlARF8b, predominantly showed negative correlations with ethylene-related genes, whereas SlSAUR69, SlSAUR2 and SlARF2a exhibited positive correlations. Although the auxin response factor gene SlARF2a has been shown to modulate tomato ripening [74], the functions of the other genes showing strong correlations to ethylene-related genes should be investigated further.
Fig 8. Correlation network of auxin- and ethylene-related genes.
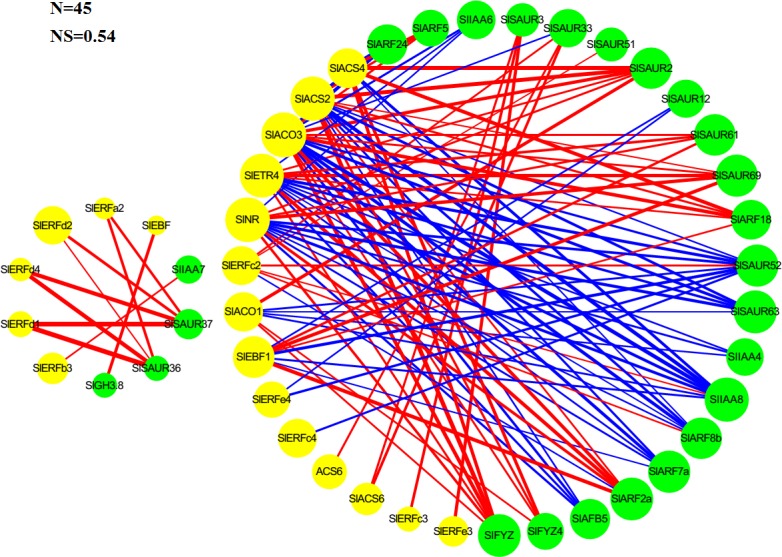
“NS” and “N” indicate network strength and the number of nodes, respectively. Node strength and network strength were calculated as previously described [35] and are listed in S10 Table. Each node represents an auxin-related gene (green) or an ethylene-related gene (yellow). Node size is proportional to node strength. Red and blue lines represent the positive and negative correlation between two genes, respectively. Line thickness is proportional to the absolute value of the correlation coefficient (r). Only high-level correlations (∣r∣< 0.75) between auxin- and ethylene-related genes are shown in the network.
Based on our study, we established a model to explore the effects of ethylene-auxin crosstalk on fruit ripening (Fig 9). During normal ripening, the methylation level in the nucleus is deceased, leading to the activation of TFs (RIN, CNR, TAGL1) and the transition to system 2 ethylene. The genes involved in ethylene (SlERF.a2, SlERF.b3, and others) or auxin (SlIAA3, SlARF7a, and others) signal transduction are differentially expressed to modulate the ripening process (Fig 9A). After treatment of exogenous auxin, demethylation is inhibited, leading to a block of the function of the TFs (RIN, CNR, TAGL1). The synthesis of system 2 ethylene (auto stimulatory) is inhibited, whereas system 1 ethylene (auto inhibitory) is maintained. Meanwhile, the expression of the auxin- or ethylene-related genes that are up- or down-regulated in normal ripening are disturbed by exogenous auxin, leading to a final delay in ripening (Fig 9B).
Fig 9. Model for the regulation of exogenous auxin on fruit ripening cooperating with ethylene.
(A) Normal ripening process with the regulation of ethylene and ripening-related TFs (RIN, CNR, TAGL1, and others). (B) Exogenous auxin inhibits ripening by modulating epigenomic remodeling, ethylene synthesis and the expression of auxin response genes.
Conclusions
Auxin has been shown to delay the fruit ripening process. Our present work revealed possible mechanisms for the modulation of ripening by auxin-ethylene interactions. Auxin treatment up-regulated the expression of auxin response genes, GST-encoding genes and stress tolerance-related TF genes and induced the recruitment of a large number of defense-associated genes to improve stress resistance and maintain auxin homeostasis. During the subsequent period, auxin enhanced the transcript levels of the genes involved in system 1 ethylene synthesis and maintained the high methylation level in the nucleus to repress the expression of system 2 ethylene synthesis-related genes. Finally, the expressions of the genes associated with carotenoid metabolism, cell wall degradation and energy metabolism were strongly repressed, and the ripening process was retarded. Additionally, the expression patterns of the genes involved in ethylene biosynthesis and signal transduction were markedly disturbed by exogenous auxin. The results of correlation network analysis revealed a strong correlation between auxin- and ethylene-related genes, suggesting significant crosstalk between auxin and ethylene during tomato ripening.
Supporting Information
(TIF)
Differentially expressed TF genes in response to exogenous auxin at (A) 1 DAT and (B) 7 DAT.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We would like to express our deep gratitude to all the members of our lab for their kindness and assistance with the experiments and with writing the manuscript.
Data Availability
All relevant data are within the paper and its supporting information files.
Funding Statement
This work was supported by grants from National Basic Research Program of China (2013CB127101). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Srivastava A, Handa AK. Hormonal regulation of tomato fruit development: A molecular perspective. J Plant Growth Regul. 2005;24(2):67–82. doi: 10.1007/s00344-005-0015-0. [Google Scholar]
- 2.Seymour GB, Chapman NH, Chew BL, Rose JK. Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnol J. 2013;11(3):269–78. 10.1111/j.1467-7652.2012.00738.x [DOI] [PubMed] [Google Scholar]
- 3.Seymour GB, Ostergaard L, Chapman NH, Knapp S, Martin C. Fruit development and ripening. Annu Rev Plant Biol. 2013;64:219–41. 10.1146/annurev-arplant-050312-120057 [DOI] [PubMed] [Google Scholar]
- 4.Cara B, Giovannoni JJ. Molecular biology of ethylene during tomato fruit development and maturation. Plant Sci. 2008;175(1–2):106–13. 10.1016/j.plantsci.2008.03.021 [DOI] [Google Scholar]
- 5.Barry CS, Giovannoni JJ. Ethylene and fruit ripening. J Plant Growth Regul. 2007;26(2):143–59. 10.1007/s00344-007-9002-y [DOI] [Google Scholar]
- 6.Alexander L, Grierson D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot. 2002;53(377):2039–55. 10.1093/jxb/erf072 [DOI] [PubMed] [Google Scholar]
- 7.Trainotti L, Tadiello A, Casadoro G. The involvement of auxin in the ripening of climacteric fruits comes of age: the hormone plays a role of its own and has an intense interplay with ethylene in ripening peaches. J Exp Bot. 2007;58(12):3299–308. 10.1093/jxb/erm178 [DOI] [PubMed] [Google Scholar]
- 8.Bottcher C, Burbidge CA, Boss PK, Davies C. Interactions between ethylene and auxin are crucial to the control of grape (Vitis vinifera L.) berry ripening. BMC Plant Biol. 2013;13(1):222 10.1186/1471-2229-13-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, et al. Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol. 2015;15(1):114 10.1186/s12870-015-0495-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottcher C, Boss PK, Davies C. Delaying Riesling grape berry ripening with a synthetic auxin affects malic acid metabolism and sugar accumulation, and alters wine sensory characters. Funct Plant Biol. 2012;39(9):745–53. 10.1071/FP12132 [DOI] [PubMed] [Google Scholar]
- 11.Symons GM, Chua YJ, Ross JJ, Quittenden LJ, Davies NW, Reid JB. Hormonal changes during non-climacteric ripening in strawberry. J Exp Bot. 2012;63(13):4741–50. 10.1093/jxb/ers147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Yuan R. NAA and ethylene regulate expression of genes related to ethylene biosynthesis, perception, and cell wall degradation during fruit abscission and ripening in ‘Delicious’ apples. J Plant Growth Regul. 2008;27(3):283–95. 10.1007/s00344-008-9055-6 [DOI] [Google Scholar]
- 13.Kondo S, Takano Y. Cell Wall Metabolism and Induction of Ripening Capacity inLa France'Pear as Influenced by 2, 4-DP. J Am Soc Hortic Sci. 2000;125(2):242–7. [Google Scholar]
- 14.Yuan R, Carbaugh DH. Effects of NAA, AVG, and 1-MCP on ethylene biosynthesis, preharvest fruit drop, fruit maturity, and quality of ‘Golden Supreme’and ‘Golden Delicious’ apples. HortScience. 2007;42(1):101–5. [Google Scholar]
- 15.Bottcher C, Keyzers RA, Boss PK, Davies C. Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J Exp Bot. 2010;61(13):3615–25. 10.1093/jxb/erq174 [DOI] [PubMed] [Google Scholar]
- 16.Liu KD, Kang BC, Jiang H, Moore SL, Li HX, Watkins CB, et al. A GH3-like gene, CcGH3, isolated from Capsicum chinense L. fruit is regulated by auxin and ethylene. Plant Mol Biol. 2005;58(4):447–64. 10.1007/s11103-005-6505-4 [DOI] [PubMed] [Google Scholar]
- 17.Muday GK, Rahman A, Binder BM. Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 2012;17(4):181–95. 10.1016/j.tplants.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 18.Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol. 2012;53(4):659–72. 10.1093/pcp/pcs022 [DOI] [PubMed] [Google Scholar]
- 19.Kumar R, Tyagi AK, Sharma AK. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol Genet Genomics. 2011;285(3):245–60. 10.1007/s00438-011-0602-7 [DOI] [PubMed] [Google Scholar]
- 20.Zouine M, Fu Y, Chateigner-Boutin AL, Mila I, Frasse P, Wang H, et al. Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLoS One. 2014;9(1):e84203 10.1371/journal.pone.0084203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pirrello J, Prasad BC, Zhang W, Chen K, Mila I, Zouine M, et al. Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol. 2012;12(1):190 10.1186/1471-2229-12-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium TG. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485(7400):635–41. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones B, Frasse P, Olmos E, Zegzouti H, Li ZG, Latche A, et al. Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J. 2002;32(4):603–13. 10.1046/j.1365-313X.2002.01450.x [DOI] [PubMed] [Google Scholar]
- 24.Sagar M, Chervin C, Mila I, Hao Y, Roustan JP, Benichou M, et al. SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 2013;161(3):1362–74. 10.1104/pp.113.213843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaabouni S, Jones B, Delalande C, Wang H, Li ZG, Mila I, et al. Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J Exp Bot. 2009;60(4):1349–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fagundes C, Moraes K, Perez-Gago MB, Palou L, Maraschin M, Monteiro AR. Effect of active modified atmosphere and cold storage on the postharvest quality of cherry tomatoes. Postharvest Biol Technol. 2015;109:73–81. 10.1016/j.postharvbio.2015.05.017 [DOI] [Google Scholar]
- 27.Bu JW, Yu YC, Aisikaer G, Ying TJ. Postharvest UV-C irradiation inhibits the production of ethylene and the activity of cell wall-degrading enzymes during softening of tomato (Lycopersicon esculentum L.) fruit. Postharvest Biol Technol. 2013;86:337–45. 10.1016/j.postharvbio.2013.07.026 [DOI] [Google Scholar]
- 28.Zhang M, Yuan B, Leng P. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot. 2009;60(6):1579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate—a Practical And Powerful Approach To Multiple Testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
- 32.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Method Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21(19):3787–93. 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 35.Diretto G, Al-Babili S, Tavazza R, Scossa F, Papacchioli V, Migliore M, et al. Transcriptional-metabolic networks in beta-carotene-enriched potato tubers: the long and winding road to the Golden phenotype. Plant Physiol. 2010;154(2):899–912. 10.1104/pp.110.159368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quint M, Gray WM. Auxin signaling. Curr Opin Plant Biol. 2006;9(5):448–53. 10.1016/j.pbi.2006.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cummins I, Dixon DP, Freitag-Pohl S, Skipsey M, Edwards R. Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metab Rev. 2011;43(2):266–80. 10.3109/03602532.2011.552910 [DOI] [PubMed] [Google Scholar]
- 39.Dixon DP, Lapthorn A, Edwards R. Plant glutathione transferases. Genome Biol. 2002;3(3):REVIEWS3004 10.1186/gb-2002-3-3-reviews3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frova C. The plant glutathione transferase gene family: genomic structure, functions, expression and evolution. Physiol Plant. 2003;119(4):469–79. 10.1046/j.1399-3054.2003.00183.x [DOI] [Google Scholar]
- 41.Yu T, Li YS, Chen XF, Hu J, Chang X, Zhu YG. Transgenic tobacco plants overexpressing cotton glutathione S-transferase (GST) show enhanced resistance to methyl viologen. J Plant Physiol. 2003;160(11):1305–11. 10.1078/0176-1617-01205 [DOI] [PubMed] [Google Scholar]
- 42.Karavangeli M, Labrou NE, Clonis YD, Tsaftaris A. Development of transgenic tobacco plants overexpressing maize glutathione S-transferase I for chloroacetanilide herbicides phytoremediation. Biomol Eng. 2005;22(4):121–8. 10.1016/j.bioeng.2005.03.001 [DOI] [PubMed] [Google Scholar]
- 43.Sharma R, Sahoo A, Devendran R, Jain M. Over-expression of a rice tau class glutathione s-transferase gene improves tolerance to salinity and oxidative stresses in Arabidopsis. PLoS One. 2014;9(3):e92900 10.1371/journal.pone.0092900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.George S, Venkataraman G, Parida A. A chloroplast-localized and auxin-induced glutathione S-transferase from phreatophyte Prosopis juliflora confer drought tolerance on tobacco. J Plant Physiol. 2010;167(4):311–8. 10.1016/j.jplph.2009.09.004 [DOI] [PubMed] [Google Scholar]
- 45.Shi HY, Li ZH, Zhang YX, Chen L, Xiang DY, Zhang YF. Two pear glutathione S-transferases genes are regulated during fruit development and involved in response to salicylic acid, auxin, and glucose signaling. PLoS One. 2014;9(2):e89926 10.1371/journal.pone.0089926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou JM, Goldsbrough PB. An Arabidopsis Gene with Homology To Glutathione S-Transferases Is Regulated by Ethylene. Plant Mol Biol. 1993;22(3):517–23. 10.1007/Bf00015980 [DOI] [PubMed] [Google Scholar]
- 47.Xiang C, Miao ZH, Lam E. Coordinated activation of as-1-type elements and a tobacco glutathione S-transferase gene by auxins, salicylic acid, methyl-jasmonate and hydrogen peroxide. Plant Mol Biol. 1996;32(3):415–26. 10.1007/BF00019093 [DOI] [PubMed] [Google Scholar]
- 48.Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10(2):79–87. 10.1016/j.tplants.2004.12.010 [DOI] [PubMed] [Google Scholar]
- 49.Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, et al. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A. 2006;103(35):12987–92. 10.1073/pnas.0604882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng X, Chen B, Lu G, Han B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun. 2009;379(4):985–9. 10.1016/j.bbrc.2008.12.163 [DOI] [PubMed] [Google Scholar]
- 51.Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16(9):2481–98. 10.1105/tpc.104.022699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giorio G, Stigliani AL, D'Ambrosio C. Phytoene synthase genes in tomato (Solanumlycopersicum L.)—new data on the structures, the deduced amino acid sequences and the expression patterns. FEBS J. 2008;275(3):527–35. 10.1111/j.1742-4658.2007.06219.x [DOI] [PubMed] [Google Scholar]
- 53.Tatsuki M, Nakajima N, Fujii H, Shimada T, Nakano M, Hayashi K, et al. Increased levels of IAA are required for system 2 ethylene synthesis causing fruit softening in peach (Prunus persica L. Batsch). J Exp Bot. 2013;64(4):1049–59. 10.1093/jxb/ers381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang CY, Steffens GL. Postharvest Responses Of Spartan Apples To Preharvest Paclobutrazol Treatment. Hortscience. 1987;22(2):276–8. [Google Scholar]
- 55.Curry EA, Greene DW. Cppu Influences Fruit-Quality, Fruit-Set, Return Bloom, And Preharvest Drop Of Apples. Hortscience. 1993;28(2):115–9. [Google Scholar]
- 56.Chen J, Mao L, Lu W, Ying T, Luo Z. Transcriptome profiling of postharvest strawberry fruit in response to exogenous auxin and abscisic acid. Planta. 2016;243(1):183–97. 10.1007/s00425-015-2402-5 [DOI] [PubMed] [Google Scholar]
- 57.Ma QL, Ding YD, Chang JW, Sun XH, Zhang L, Wei QJ, et al. Comprehensive insights on how 2,4-dichlorophenoxyacetic acid retards senescence in post-harvest citrus fruits using transcriptomic and proteomic approaches. J Exp Bot. 2014;65(1):61–74. 10.1093/jxb/ert344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stopar M, Resnik M, Pongrac VZ. Non-structural carbohydrate status and CO2 exchange rate of apple fruitlets at the time of abscission influenced by shade, NAA or BA. Sci Hortic. 2001;87(1–2):65–76. 10.1016/S0304-4238(00)00169-2 [DOI] [Google Scholar]
- 59.Amorós A, Zapata P, Pretel M, Botella M, Almansa M, Serrano M. Role of naphthalene acetic acid and phenothiol treatments on increasing fruit size and advancing fruit maturity in loquat. Sci Hortic. 2004;101(4):387–98. 10.1016/j.scienta.2003.11.010 [DOI] [Google Scholar]
- 60.Nakatsuka A, Murachi S, Okunishi H, Shiomi S, Nakano R, Kubo Y, et al. Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 1998;118(4):1295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oetiker JH, Olson DC, Shiu OY, Yang SF. Differential induction of seven 1-aminocyclopropane-1-carboxylate synthase genes by elicitor in suspension cultures of tomato (Lycopersicon esculentum). Plant Mol Biol. 1997;34(2):275–86. 10.1023/A:1005800511372 [DOI] [PubMed] [Google Scholar]
- 62.Zhang Z, Zhang H, Quan R, Wang XC, Huang R. Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol. 2009;150(1):365–77. 10.1104/pp.109.135830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jafari Z, Haddad R, Hosseini R, Garoosi G. Cloning, identification and expression analysis of ACC oxidase gene involved in ethylene production pathway. Mol Biol Rep. 2013;40(2):1341–50. 10.1007/s11033-012-2178-7 [DOI] [PubMed] [Google Scholar]
- 64.Sell S, Hehl R. A fifth member of the tomato 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase gene family harbours a leucine zipper and is anaerobically induced. DNA Seq. 2005;16(1):80–2. 10.1080/10425170500050817 [DOI] [PubMed] [Google Scholar]
- 65.Barry CS, Llop-Tous MI, Grierson D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 2000;123(3):979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lincoln JE, Campbell AD, Oetiker J, Rottmann WH, Oeller PW, Shen NF, et al. LE-ACS4, a fruit ripening and wound-induced 1-aminocyclopropane-1-carboxylate synthase gene of tomato (Lycopersicon esculentum). Expression in Escherichia coli, structural characterization, expression characteristics, and phylogenetic analysis. J Biol Chem. 1993;268(26):19422–30. [PubMed] [Google Scholar]
- 67.Barry CS, Blume B, Bouzayen M, Cooper W, Hamilton AJ, Grierson D. Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 1996;9(4):525–35. 10.1046/j.1365-313X.1996.09040525.x [DOI] [PubMed] [Google Scholar]
- 68.Cao D, Ju Z, Gao C, Mei X, Fu D, Zhu H, et al. Genome-wide identification of cytosine-5 DNA methyltransferases and demethylases in Solanum lycopersicum. Gene. 2014;550(2):230–7. 10.1016/j.gene.2014.08.034 [DOI] [PubMed] [Google Scholar]
- 69.Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol. 2013;31(2):154–9. 10.1038/nbt.2462 [DOI] [PubMed] [Google Scholar]
- 70.Mantiri FR, Kurdyukov S, Lohar DP, Sharopova N, Saeed NA, Wang XD, et al. The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiol. 2008;146(4):1622–36. 10.1104/pp.107.110379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J, Dai X, Zhao Y. A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol. 2006;140(3):899–908. 10.1104/pp.105.070987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu K, Yuan C, Li H, Lin W, Yang Y, Shen C, et al. Genome-wide identification and characterization of auxin response factor (ARF) family genes related to flower and fruit development in papaya (Carica papaya L.). BMC Genomics. 2015;16(1):901 10.1186/s12864-015-2182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol. 2004;7(4):465–71. 10.1016/j.pbi.2004.04.007 [DOI] [PubMed] [Google Scholar]
- 74.Hao Y. Auxin-mediated fruit development and ripening: new insight on the role of ARFs and their action mechanism in tomato (S. lycopersicum). PhD. Thesis, The University of Toulouse, 2014. Available: http://oatao.univ-toulouse.fr/13598.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Differentially expressed TF genes in response to exogenous auxin at (A) 1 DAT and (B) 7 DAT.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its supporting information files.