Abstract
The MPT transports Pi to synthesize ATP. PsMPT, a chilling-induced gene, was previously reported to promote energy metabolism during bud dormancy release in tree peony. In this study, the regulatory elements of PsMPT promoter involved in chilling response were further analyzed. The PsMPT transcript was detected in different tree peony tissues and was highly expressed in the flower organs, including petal, stigma and stamen. An 1174 bp of the PsMPT promoter was isolated by TAIL-PCR, and the PsMPT promoter::GUS transgenic Arabidopsis was generated and analyzed. GUS staining and qPCR showed that the promoter was active in mainly the flower stigma and stamen. Moreover, it was found that the promoter activity was enhanced by chilling, NaCl, GA, ACC and NAA, but inhibited by ABA, mannitol and PEG. In transgenic plants harboring 421 bp of the PsMPT promoter, the GUS gene expression and the activity were significantly increased by chilling treatment. When the fragment from -421 to -408 containing a MYC cis-element was deleted, the chilling response could not be observed. Further mutation analysis confirmed that the MYC element was one of the key motifs responding to chilling in the PsMPT promoter. The present study provides useful information for further investigation of the regulatory mechanism of PsMPT during the endo-dormancy release.
Introduction
The mitochondrial phosphate transporter (MPT) shuttles inorganic phosphate (Pi) into the mitochondrial matrix, where Pi is utilized for oxidative phosphorylation to synthesize ATP from ADP. MPT encoding genes have been cloned from mammals [1–3], yeast [4], and wood frogs [5] with most studies focusing mainly on the structure and catalytic function of the transporters.
Recently, cloning and characterization of MPT were reported in several plants [6–12]. Plant MPT genes were identified to be involved in abiotic stress responses, and their expression patterns showed tissue preferences. Birch Mpt1 was ozone-inducible and highly expressed in the tissue of dividing cells, such as root tips, shoot apices and developing root nodules [6]. AtMPTs play an important role in response to salt stress in Arabidopsis. Furthermore, with different expression profiles in various tissues and conditions, transcription of AtMPTs has been detected in all tissue except siliques [12]. The sequences and structures of 26 potential PT family genes in rice were analyzed, and six MPTs also showed tissue preferential expression profiles, among which OsPT17 and OsPT19 were differently regulated under hormone treatment conditions. In addition, six putative cis-elements were found in all of the OsPT genes including ARR1AT, CAATBOX1, CACTFTPPCAL, GATABOX, GT1CONSENSUS and GTGANTG10. Specifically, GATABOX and GT1CONSENSUS are light-responsive cis-elements, and CACTFTPPCAL is necessary for carbon metabolism [11]. Current knowledge of MPT regulation and the molecular mechanisms mediating its biological functions in plants is still incomplete.
Tree peony (Paeonia suffruticosa Andrews) is one of the most well-known horticultural and medicinal plants in the world. One of the main production mechanisms in the tree peony industry, especially for the Spring Festival flower market in China, is forcing culture. Dormancy is a major obstacle for the forced culture of tree peony in winter, and sufficient chilling is an efficient way to break dormancy. Therefore, it is important to determine how chilling induces dormancy release in tree peony. PsMPT was previously isolated from the tree peony subtractive cDNA library of burst buds and strongly induced by chilling treatment to promote ATP production during the release of bud dormancy. In addition, ectopic-expression of PsMPT in Arabidopsis showed that PsMPT enhanced ATP synthesis and affected plant growth and development [10]. These results suggested that PsMPT plays an important role in energy production during bud dormancy release in tree peony [13]. However, the expression characteristics of PsMPT and its regulatory mechanisms are unclear.
In this study, we isolated the promoter of PsMPT and constructed PsMPT promoter::GUS engineered Arabidopsis. We investigated: 1) the temporal and spatial characteristics of the PsMPT promoter in Arabidopsis and PsMPT expression in tree peony; 2) how plant hormones and abiotic stresses, including chilling, affects the activity of the PsMPT promoter; 3) which one of cis-elements among the PsMPT promoter is involved in the chilling response.
Materials and Methods
Plant materials
Four-year-old tree peonies (Paeonia suffruticosa ‘Luhehong’) were obtained from the Tree Peony Research Center of Heze (Shandong, China). According to the method of Huang et al.[10], plants were treated in cold conditions (0–4°C) for 21 days to break bud dormancy, as the daily mean temperature was under 10°C in Qingdao, Shandong, China. The plants were then transferred to a greenhouse (18–22°C, 8-h-light/16-h-dark cycle) to resume growth. Tissues (root, stem, leaf, calyx, petal, stamen and carpel at the early stage of flowering) were collected and stored at -80°C until use. One hundred μmol·L-1 ABA and 50 μmol·L-1 GA3 were applied to non-chilling buds with double-distilled water as the control, and buds were collected after 0, 1, 6, 12, 24 and 48 h. Three replicates (3 plants/replicate) were performed for all treatments.
Isolation of the PsMPT promoter
Genomic DNA was extracted from tree peony buds using the cetyltrimethylammonium bromide (CTAB) extraction method as previously described [14]. DNA samples were qualified photometrically, then checked on agarose gel, and stored at -20°C for use. Based on the cDNA sequence of PsMPT (Genbank accession No.: EU072922), three gene-specific primers, SP1, SP2 and SP3, in nested positions were designed with primer premier 5.0 and synthesized (Table 1). The PsMPT promoter was amplified using gene-specific primers and four short arbitrary primers (AP) with the Genome Walking Kit (TaKaRa). The PCR products were ligated into the pMD18-T vector (TaKaRa), and sequenced at BGI-Beijing, China.
Table 1. The primers used in this paper.
| Primers | Sequences (5’-3’) | purpose |
|---|---|---|
| SP1 | CTGATGTTAGGGTTCTACTTTCCTCTTTCTCTC’ | Promoter isolation |
| SP2 | ATGTCTGCGTTACCCAAGGTCGTCCC | Promoter isolation |
| SP3 | ATAGGGCATTCCCAGAAACGATTGTCC | Promoter isolation |
| FP1 | GGGAAGCTTGGGACCCAGTGTGT (Hind III) | Promoter analysis |
| FP2 | GGGAAGCTTTGGGGACTCAATTGT (Hind III) | Promoter analysis |
| FP3 | GCGAAGCTTGATACAATGGGAGAGGAG (Hind III) | Promoter analysis |
| FP4 | GGGAAGCTTGGTCGCATTCGTCG (Hind III) | Promoter analysis |
| FP5 | GGGAAGCTTAGAACAAGAATCGTGGAG (Hind III) | Promoter analysis |
| FP6 | GGGAAGCTTAGCTCGGCATTCAGTG (Hind III) | Promoter analysis |
| RP | CGAGGATCCCATATCTGATGTTAGG (BamH I) | Promoter analysis |
| FDP | GGGAAGCTTCATATTATGTCAAATTGG (Hind III) | Motif identification |
| RM1 | TCCAATTTGACATAATATGTTACAAGGT | Motif identification |
| FM1 | TTATGTCAAATTGGAGACTTGATT | Motif identification |
| GUSFW | AGTGGCAGTGAAGGGCGAACAGT | qPCR of GUS |
| GUSRV | TCAGCGTAAGGGTAATGCGAGGT | qPCR of GUS |
| ActinFW | GAGAGATTCCGTTGCCCTGA | qPCR of β-actin |
| ActinRV | CTCAGGAGGAGCAACCACC | qPCR of β-actin |
| MPTFW | GCTGGAGGAATAATGAGTTGTGG | qPCR of PsMPT |
| MPTRV | GCACCTTGAGCACTGTAACCA | qPCR of PsMPT |
Bioinformatics analysis of the promoter sequence
Regulatory elements in the promoter were analyzed using the online program PLACE (http://www.dna.affrc.go.jp/PLACE/) [15, 16] and Plantcare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) [17].
Construction of the promoter-reporter plasmids
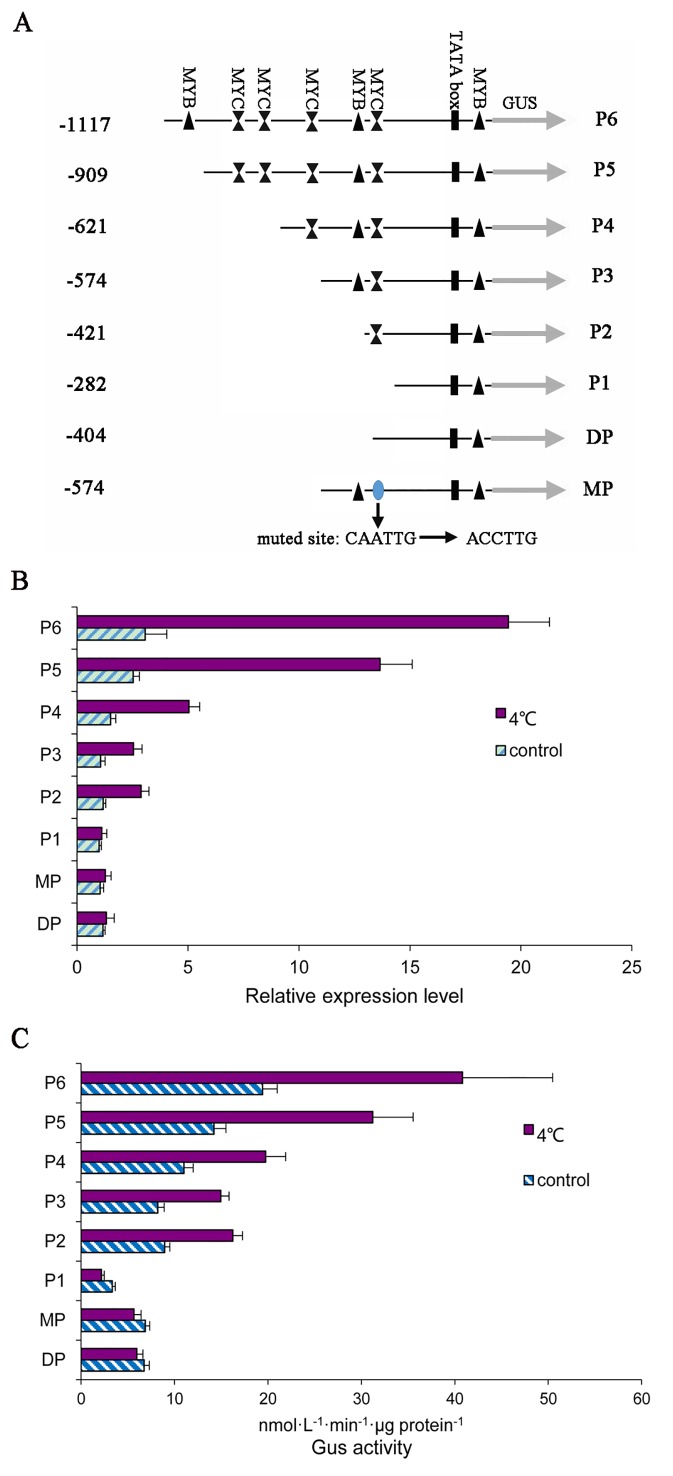
To construct the binary vector consisting of the β-glucuronidase (GUS) coding sequence driven by the PsMPT promoter, a fragment from -1117 to -1 relative to the translation initiation codon was obtained using the high fidelity DNA polymerase and promoter specific primers (FP6 and RP, Table 1). The serial deletions of the PsMPT promoter (-909, -621, -574, -421, and -282 to -1) were amplified by PCR with the corresponding forward (FP1, FP2, FP3, FP4, FP5) and reverse primers (RP) that contained a Hind III and BamH I site at the 5' end of each primer, respectively. For chilling response element identification, the MYC element -413 relative to the translation initiation site was subsequently deleted from the P2 construct with the FDP primer, and mutated to form the MP construct by recombination PCR with the FM1 and RM1 primers (Table 1). PCR products were retrieved and cloned into a pMD18-T simple vector (TaKaRa) followed by sequencing conformation at BGI Beijing, China. The Hind III/BamH I digested DNA fragments were inserted into the corresponding sites of pBI121, in place of the deleted CaMV35S promoter upstream of the GUS coding sequence. The pBI121 vector with the CaMV35S::GUS was used as the positive control. The resulting constructs were named P1 (-282), P2 (-421), P3 (-574), P4 (-621), P5 (-909), P6 (-1117), DP (-404), and MP (-574), respectively.
Arabidopsis transformation
Arabidopsis thaliana wild-type (Col-0) plants were transformed using the Agrobacterium tumefaciens strain EHA105 carrying the above constructs according to the floral dip method [18]. The transformants were screened on MS medium containing 50 mg L-1 kanamycin, and positive plants were identified by PCR amplification using GUS specific primers (GUSFW and GUSRV, Table 1). The corresponding T1 transgenic seedlings that segregated at a ratio of 3:1 (resistant: sensitive) were selected to propagate T2 individuals for further analysis. Five to ten transgenic lines were obtained for every construct.
Hormone and abiotic stress treatments
The transformed Arabidopsis plants grew at 18°C in a controlled growth chamber (16-h-light/8-h-dark cycle), and 5-week-old Arabidopsis plants were used to analyze the response of the PsMPT promoter to hormone and abiotic stresses. For chilling treatment, plants were cultured under 4°C for 24 h, and those at 18°C were used as controls. Moreover, the inflorescence of 5-week-old Arabidopsis plants were sprayed by GA3 (50 μmol L-1), NAA (100 μmol L-1), ABA (100 μmol L-1), ACC (250 mmol L-1), NaCl (200 mmol L-1), mannitol (40 μmol L-1) and PEG (100 mmol L-1) treatments for 24 h, respectively. Double-distilled water was used as a control. Samples were collected after 0, 1, 3, 6, 12 and 24 h treatments and stored at -80°C.
GUS histochemical assay and quantitative analysis of GUS activity
GUS activity was determined in tissue extracts using a standard protocol [19]. GUS fluorescence was measured with a Microplate Spectrofluorometer, the data were obtained by subtracting the background 4-methyiumbelliferyl glucuronide of the PPsMPT::GUS transgenic plants. The average GUS activity was obtained from at least five independent transformants, and each assay was repeated three times. GUS histochemical staining was performed using identified homozygous transgenic plants by a modified Jefferson’s method [19]. In brief, plant tissues were incubated in a 100 mmol L-1 sodium phosphate buffer (pH 7.0) containing 0.1% Triton X-100, 10 mmol L-1 EDTA, 1 mmol L-1 X-gluc and 0.5 mmol L-1 potassium ferricyanide at 37°C overnight. The stained tissues were then washed several times with 70% ethanol to bleach the chlorophyll.
Quantitative real-time PCR (qPCR)
Total RNA was extracted from the inflorescence of Arabidopsis and buds of tree peony as previously described [20], and then treated with DNase I (TaKaRa) according to the manufacturer’s instructions. First strand cDNA was synthesized from 2 μg of total RNA using the PrimerScript™ RT reagent Kit (TaKaRa). PCR reactions were performed in a 25 μL system including 12.5 μL 2×SYBR Green Master mix (TaKaRa), 300 nmol L-1 each primer (Table 1) and 2 μL 10-fold diluted cDNA template. PCR reactions were run in a Roche LightCycler® 480 (Roche, Germany) using the following program: 95°C for 2 min and 45 cycles of 95°C for 5 s, 57°C for 30 s and 72°C for 30 s. The reactions were run in triplicate. The expression was normalized to beta-actin. Quantification of the relative gene expressions was performed using the 2-ΔΔCt method [21]. Statistical analyses were performed using SPSS 13.0 (SPSS, USA).
Results
Expression characteristics of PsMPT in tree peony
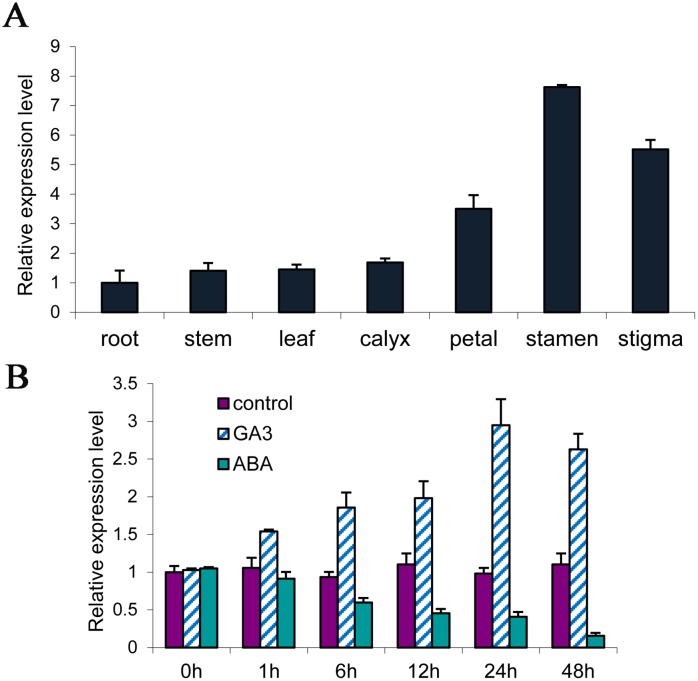
The expression of PsMPT was previously reported during dormancy release in tree peony [13]. In this study, the temporal and spatial expression of PsMPT was further detected at the early stage of flowering in tree peony. The results of qPCR indicated that the transcription of PsMPT was detected in all tree peony tissues; however, the PsMPT transcript was very low in root, stem, leaf and calyx, but high in flower organs, including petal, stamen and stigma (Fig 1A). The PsMPT transcripts in the stamen were expressed 6-fold as compared to that of the root. The results indicated that PsMPT was expressed preferentially in flower organs of tree peony.
Fig 1. Tissue-specific expression of PsMPT in germinated buds (A) and transcriptional levels of PsMPT in response of GA3 and ABA of tree peony by qPCR (B).
100 μmol L-1 ABA and 50 μmol L-1 GA3 was sprayed to the dormant buds in green house (18–22°C, 8-h-light/16-h-dark cycle). Values are means ±SD of three replicates.
The response of PsMPT in dormant buds to Gibberellic Acid (GA) and Abscisic Acid (ABA) were analyzed by qPCR. When GA was applied, the PsMPT transcript was quickly promoted and peaked at 24 h, then declined slightly. Conversely, ABA application dramatically decreased the expression of PsMPT (Fig 1B).
Isolation of the PsMPT promoter and the putative cis-acting element
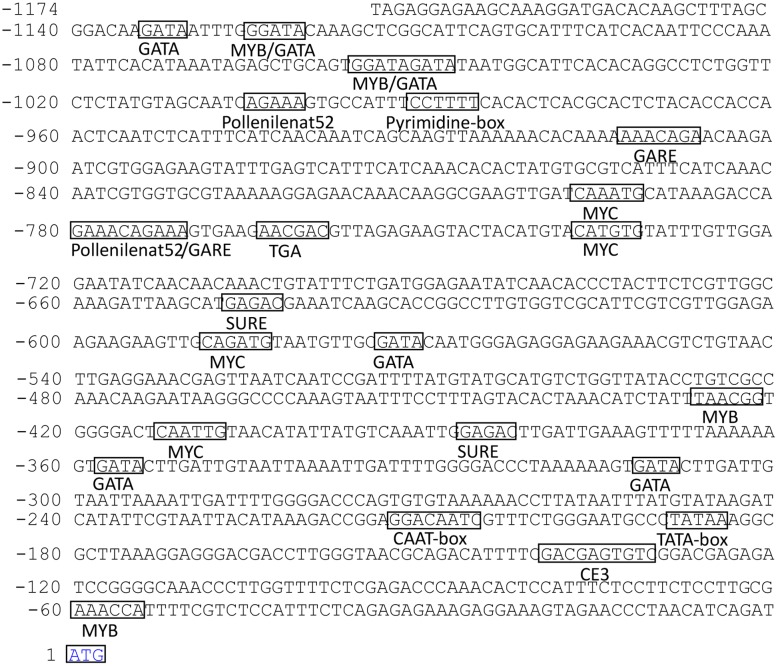
In this study, the 1174 bp upstream genomic DNA sequence of PsMPT was isolated by TAIL-PCR (Fig 2). The adenosine of the translation initiation codon (ATG) of the PsMPT gene was defined as +1 (Fig 2). A motif search was carried out using PLACE and PlantCare to analyze the putative cis-elements. As shown in Fig 2, the putative TATA box was found at position -189/-185 and the CAAT box at position -210/-207. In addition, a number of regulatory motifs potentially related to environmental signals were found, which included auxin-, GA- and dehydration-responsive and tissue-specific elements. Among the cis-elements, one putative pyrimidine box and two GA-responsive elements (GAREs) were located at -991/-986, -913/-907 and -780/-771, respectively, which were related to GA response and sugar repression [22, 23]. Four putative Myelocytomatosis viral oncogene homolog (MYC) (5’-CANNTG -3’) motifs and four Myeloblastosis viral oncogene homolog (MYB) (5’-WAACCA -3’, 5’-YAACKG -3’ or 5’-GGATA-3’) motifs were located at positions -797/-792, -737/-732, -589/-584, -412/-408 and -1127/-1122, -1056/-1051, -427/-422, -40/-35, respectively. These motifs had previously been identified in response to dehydration in Arabidopsis [24–26]. Several MYB and MYC motifs were thought to respond to chilling or freezing in Arabidopsis [24–26]. There were two putative sulfur-responsive element (SURE) motifs at positions -647/-643 and -387/-382, and one putative TGA element (auxin responsive element) at position -764/-759. SURE contains the auxin response factor binding sequence [27]. Therefore, PsMPT might be regulated by chilling and auxin. Interestingly, eight putative GATA boxes were present in the sequence, which were thought to be involved in tissue-specific expression and light response [28, 29]. Moreover, three putative pollen1lelat52 elements, related to the pollen specific expression [2, 30], were found at -775/-771, -782/-777, and -1005/-1001, respectively.
Fig 2. PsMPT promoter sequence of the 5’ region upstream from the start codon (ATG) and putative cis-elements predicted in the promoter region.
Numbers indicate the positions relative to the translation start codon starting from the adenosine (+1). The putative TATA box at position -189, CAAT box at position -210, and the ATG start codon is framed in box and denoted with blue color. The other important putative cis-elements are framed and labeled below.
Temporal and spatial expression of the PsMPT promoter in Arabidopsis
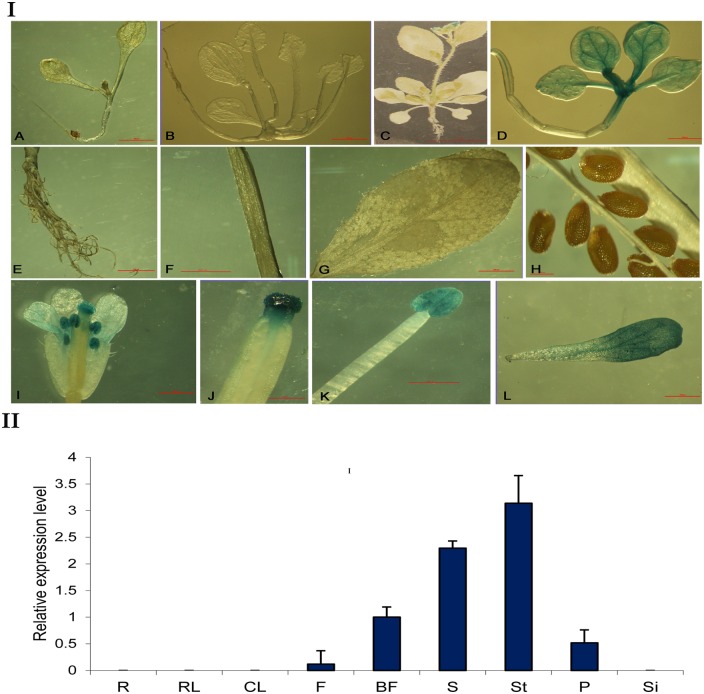
To identify the expression patterns of the PsMPT promoter, the promoter::GUS chimeric construct (PPsMPT::GUS) was transformed into Arabidopsis, and histochemical GUS staining was carried out in various organs throughout plant development (Fig 3I). These results showed that GUS activity was not detected in the seedling tissues (Fig 3IA and 3IB) but were observed in the flower organs (Fig 3IC, 3IE–3IG). GUS activity in transgenic plants was more pronounced in the stigma and stamen compared with the sepals (Fig 3II–3IL). GUS staining was not observed in the silique and seeds (Fig 3IH). These results suggested that PsMPT is preferentially expressed in the flower tissues.
Fig 3. Histochemical localization (I) and tissue-specific expression of GUS (II) in transgenic Arabidopsis thaliana carrying the PsMPT promoter::GUS construct.
(I)Histochemical localization of GUS expression by staining with X-gluc in transgenic Arabidopsis thaliana carrying the PsMPT promoter::GUS construct. Arrow bar shows GUS staining in flower. A 7-day-plants from seeding; B 21-day-plants from seeding; C 28-day-plants from seeding; D Positive control (Ca35S promoter driven); E roots; F: stem; G leaf; H: mature silique; I flower; J stigma; K stamen; L petal. (II)Total RNA was isolated from roots (R), rosette leaf (RL), cauline leaf (CL), flower bud (F), bloomed flower (BF), stamen (S), stigma (St), petal (P) and siliques (Si) of 35-day-old transgenic plants from seeding. The transcriptional levels were analyzed by qPCR using GUS gene-specific PCR primers, which were normalized with beta-actin. Values are means ± SD of three replicates.
qPCR was performed to evaluate the spatial expression of the PsMPT promoter. The results showed that the GUS transcript was only detected in the flower (Fig 3II). No transcript of GUS was detected in the roots, rosette leaves, cauline leaves and silique. GUS transcript levels in bloomed flowers were approximately 10-fold higher compared with flower buds. In bloomed flowers, the most abundant expression was found in stigma, followed by in stamen, with the lowest amount in the petals. In summary, the GUS staining results are in accordance with that of qPCR. The GUS reporter did not appear in the immature flower buds possibly due to the low abundance of the transcripts. The flower-specific expression characteristics of the promoter implied that PsMPT might participate in plant anthesis and gametophyte development.
Responses of the PsMPT promoter to hormones and abiotic stresses
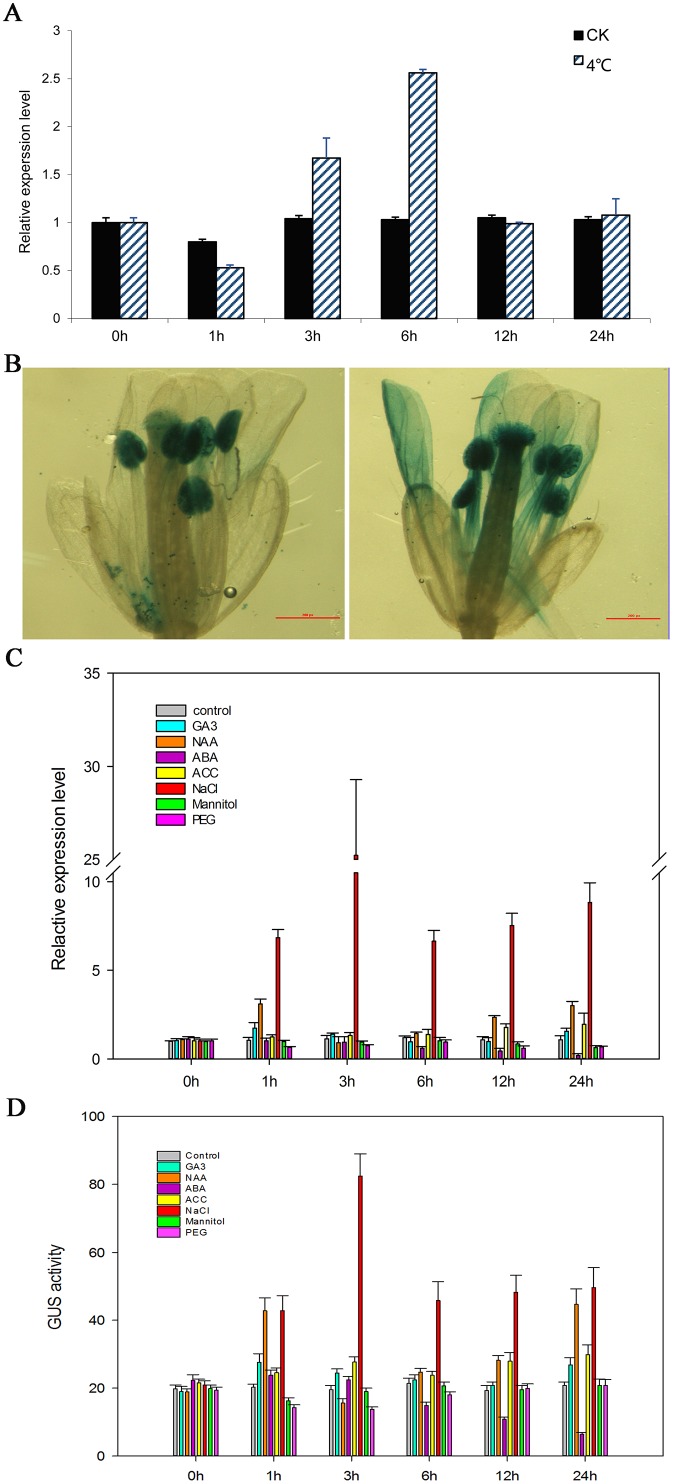
The transgenic plants carrying the PPsMPT::GUS cassette were treated with hormones and abiotic stresses, and the transcription of GUS was evaluated by qPCR, respectively. Overall, GUS activity changed rapidly and fluctuated during the entire period for all of the treatments (Fig 4). Chilling increased the GUS transcript during 3 h to 6 h after treatment, with a peak approximately 2.5-fold higher at 6 h (Fig 4A), which was also verified by GUS staining (Fig 4B). The results of qPCR showed that GA3 and NAA treatments enhanced GUS expression at 1 h, then decreased, followed by another peak at 24 h. ACC accelerated the transcript of GUS during the entire process. Conversely, ABA inhibited GUS expression throughout the process. In addition, the GUS transcript was continuously decreased by mannitol and PEG until 3 h, followed by a slight increase at 12 h and 24 h; however, the GUS transcript levels were lower than that of control. Notably, NaCl dramatically enhanced GUS activity, and it reached a peak at 3 h with an approximate 25-fold increase (Fig 4C). The results of GUS activity were consistent with GUS expressions of qPCR (Fig 4D).
Fig 4. Relative expression levels of PsMPT promoter in response to hormone and various abiotic stresses treatments in transgenic Arabidopsis plants.
(A) GUS expressions when exposed to 4°C temperature. Plants were transferred to a cold chamber maintained at 4°C, and the control grew at 18°C. Error bars represent ±SD. (B) GUS staining of transgenic Arabidopsis grew at 18°C (left) and at 4°C for 6 h (right). (C) GUS expressions were measured by qPCR using 35-day-old plants from seeding. 50 μmol L-1 GA3, 100 μmol L-1 NAA, 100 μmol L-1 ABA, 250 mmol L-1 ACC, 200 mmol L-1 NaCl, 40 μmol L-1 mannitol and 100 mmol L-1 PEG was sprayed to the inflorescence at 18°C, and double-distilled water treatment was used as control. (D) GUS fluorescence (nmol L-1 min-1 μg protein-1) were measured by a Microplate Spectrofluorometer using 35-day-old plants from seeding. 50 μmol L-1 GA3, 100 μmol L-1 NAA, 100 μmol L-1 ABA, 250 mmol L-1 ACC, 200 mmol L-1 NaCl, 40 μmol L-1 mannitol and 100 mmol L-1 PEG was sprayed to the inflorescence at 18°C, and double-distilled water treatment was used as control.
Identification of chilling response elements
As reported previously, PsMPT was chilling inducible. Furthermore, PsMPT was involved in chilling induced dormancy release in tree peony. To confirm the PsMPT promoter region involved in the chilling response, a number of truncated promoter fragments (P1 (-282), P2 (-421), P3 (-574), P4 (-621), P5 (-909) and P6 (-1117)) were isolated and fused to the GUS reporter gene into pBI121 vector (Fig 5A). Transgenic Arabidopsis plants with each promoter-GUS construct were generated. The transcription levels of GUS were detected by qPCR (Fig 5B). In the transgenic plants, the highest level of GUS expression was detected in the engineered Arabidopsis with the P6 construct, which contained the full-length PsMPT promoter (−1117/−1). GUS transcript decreased in order from the P6 to P1 construct, and P2 and P3 had similar abundance. These results suggested that the transcription enhancer might exist in the upstream of the PsMPT promoter.
Fig 5. Assays for GUS expression driven by the series PsMPT promoter.
(A) Schematic diagram of the PsMPT promoter deletions and mutation that were used to analyze the activity of different fragments of the PsMPT promoter. All fragmented promoter were fused to a GUS reporter gene. (B) Quantitative analyses of GUS expression in transgenic plants driven by deletion or muted constructs of PsMPT promoter in response to chilling. (C) GUS activity in transgenic Arabidopsis plants. The inflorescence of 5-week-old Arabidopsis plants was used as material, and five independent lines for every treatment.
We also detected GUS expression of the successive deletions at 4°C for 6 h compared to 18°C (Fig 5B). After exposure to 4°C, the GUS transcripts were up-regulated in the P2 to P6 constructs. The largest increase in GUS abundance was observed in plants with the P6 construct (6.34-fold). The next-largest increase was observed in the P5 construct (-909/-1, 5.40-fold). Similar enhancement was observed in the P2 and P3 constructs with 2.45 and 2.41-fold, respectively. However, the P1 construct (-282/-1), which only contained basic transcriptional elements, could not induce GUS expression at 4°C. These results indicated that the −421/−282 region of the PsMPT promoter could lead to efficient chilling induction. Bioinformatics analyses showed that a MYC cis-element existed in this region, and several MYCs were found within the promoter. We speculated thus that the MYC elements in the promoter might be involved in the chilling response.
To verify our hypothesis, another deletion construct (DP) (-404) and one mutated construct (MP) (-574) were constructed (Fig 5A). In the DP construct, the MYC element located at -412 in the promoter was deleted. When exposed to 4°C, no significant difference was observed as compared to that of 18°C treatment, which indicated that there was a loss of function for chilling induction with the MYC element deleted in the PsMPT promoter (Fig 5B). In the MP construct where the MYC element (-412) was mutated from CAATTG to ACCTTG, a complete elimination of the chilling response was observed (Fig 5B).
To confirm the results obtained by qPCR, a quantitative measurement of GUS activity was performed using the series constructs, P1-P6, DP and MP. Consistent with the results obtained by qPCR, the GUS activity was observed to increase from the P2 to P6 after exposed to 4°C, and no difference was observed in the P1, DP and MP. In summary, we concluded that the MYC element involved in chilling treatment responses (Fig 5C).
Discussion
MPT can shuttle inorganic phosphate (Pi) into the mitochondrial matrix, where ATP is synthesized. Thus, MPT plays a key role in cellular ATP regeneration. ATP is essential for almost all biological processes in the cell, and MPTs have been reported to be involved in abiotic responses [6, 12], bud dormancy release, growth and development [13]. Although several MPTs have been cloned and functionally annotated, their characteristics and regulatory mechanisms are poorly understood.
We previously cloned PsMPT, a chilling induced gene, in tree peony, which accelerated ATP synthesis and dormancy release [13]. In this study, we isolated the PsMPT promoter and detected its activity using GUS as the reporter in transgenic Arabidopsis. GUS-staining and qPCR of the transgenic plants revealed that the PsMPT promoter was preferentially expressed in the flower, mainly in stamen and stigma. Therefore, the tissue-specific expression may be related to the putative GATA boxes and pollen1lelat52 elements founded in the promoter [30, 31]. The results suggested that PsMPT might play an important role during gametophyte development, pollination and fertilization, and Pi might be transported to reproductive organs during the reproductive development stage. Differing from ectopic expression of the PsMPT promoter, PsMPT mRNA was detected in all tissues of tree peony by qPCR. This discrepancy has also been reported in mice [32, 33] and is believed to be related to different biological species or incomplete isolation of promoter sequences.
The temporal and spatial expression of MPT has been reported in several plants, and all of the results showed tissue-specific expression. Birch MPT1 was highly expressed in tissues containing dividing cells [6]. There were six MPT members in rice, and a microarray analysis also revealed tissue-specific expression [11]. In Arabidopsis, the transcription of the AtMPT1 was pronounced in the stamens of flowers, and AtMPT2 mRNA was abundant in rosette leaves; whereas, AtMPT3 was strongly expressed in leaves and weakly expressed in the roots and flowers [12]. Overall, the spatial pattern of PsMPT was similar to AtMPT1. In contrast, PsMPT showed only 51% sequence identity with AtMPT1, and 79% sequence similarity with AtMPT3 (S1 Table). The organization of promoters between PsMPT and AtMPTs was also compared, and large differences were observed (S1 Fig). Similarly discrepant MPT sequences and expression patterns between tree peony and Arabidopsis were also found in grape and Arabidopsis [11]. Taylor et al. investigated the relationship between the promoter and coding sequence selective constraint and suggested that they were generally uncorrelated [34], which implied partially independent evolution of promoters and their coding sequences between species. Therefore, we speculated that the discrepancy might be due to species-specific independent evolution of MPTs and their promoters.
The response of the PsMPT promoter to abiotic stress and hormones was analyzed by qPCR. GUS transcript increased during chilling treatment in the transgenic plants driven by the PsMPT promoter. In addition, the PsMPT promoter was also induced by salt, GA3, ACC and NAA, while ABA, mannitol and PEG suppressed its activity. In rice, OsPT19 was suppressed by five hormone treatments including ABA, 2, 4-D, GA3, KT, and NAA, whereas OsPT17 was induced by the hormone treatments with NAA or GA3 [11]. These responses might be due to the cis-elements in the promoter, for instance, the putative pyrimidine box and two GARE motifs to GA response, putative TGA and SURE elements to NAA response, and MYB and MYC to abiotic stress response, such as chilling. Interestingly, NaCl treatment could significantly increase the expression of the reporter gene with a maximum of approximately 25-fold. Similar results were reported for Arabidopsis [12].
Among all of the factors influencing promoter activity, chilling and GA3 treatments are of interest because they effectively accelerate the dormancy release in winter [35–38]. Huang et al. found that PsMPT was induced by chilling [13]. Interestingly, it was reported that the chilling-induced expression of PsMPT was not maintained after being transferred to a greenhouse (18–22°C) when less than 21 days of chilling were applied. On the other hand, the levels of PsMPT transcripts remained high with a 21 d or longer chilling duration after returning to growth temperature [13]. In this study, ectopic expression analyses provided more evidence that the PsMPT promoter could be induced by chilling. We speculated that the early increase of the PsMPT transcript might be induced by chilling, and GA production might be a downstream effect of chilling, as proposed for dormant seeds [39, 40]. Meanwhile, chilling temperature was reported to enhance the accumulation of endogenous GA, and exogenous GA could partially replace chilling to accelerate endo-dormancy release [41]. In this study, we found that exogenous GA could activate the expression of the PsMPT promoter. Therefore, the reactivation of the PsMPT transcripts might be due to the high endogenous GA induced by sufficient chilling when transferred to growth condition. Buds chilled for less than 21 days had relatively low GA levels that could not activate the PsMPT expression required for the recovery of plant growth ability.
Considering the central role of PsMPT in energy metabolism during dormancy release, it is important to elucidate how chilling accelerates PsMPT expression. It is well-known that the transcription of mRNA is mainly regulated through the cooperation of transcript factors and corresponding cis-elements. Several cis-elements have been identified to be involved in chilling or cold responses, such as ABRE (ABA responsive element), DRE/CRT (dehydration-responsive element/C-repeat element, A/GCCGAC), MYB, MYC, and the E-box [24, 26, 42, 43, 44, 45]. Bioinformatics analysis of the isolated PsMPT promoter showed that four MYB and four MYC elements were present upstream of the promoter, which might be responsible for the chilling response. Based on the location of the MYC and MYB elements, deletion experiments were conducted to identify the candidate chilling response elements in the promoter. GUS expression and activity revealed that the P2 construct containing one MYC element (-412/-408, CAATTG) effectively responds to chilling, and the addition of a MYB element (P3 construct) did not improve the chilling response ability. When MYC was deleted or mutated, the chilling response character abated. Alternatively, increase of MYC elements in the P4, P5 and P6 constructs enhanced the chilling response activity, indicating there was an additive effect of MYC elements in the chilling response. This study demonstrates that the MYC elements in the PsMPT promoter play a crucial role in the chilling responses.
In conclusion, we isolated the PsMPT promoter in tree peony and found that it is a floral-preferential promoter. The promoter of PsMPT responded to chilling, ACC, PEG, NaCl, mannitol, auxin and GA. Deletion and mutation analyses demonstrated that the MYC cis-element functioned in the chilling response. This work provides useful information for further investigation of the regulatory mechanisms of PsMPT promoter during endo-dormancy release.
Supporting Information
(DOCX)
(DOC)
Acknowledgments
We would like to thank Dr. Chunhai Dong for constructive suggestions in writing and revision of this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Natural Science Foundation of China (31071828, 31372104 and 31471908).
References
- 1.Runswick MJ, Powell SJ, Nyren P, Walker JE. Sequence of the bovine mitochondrial phosphate carrier protein: structural relationship to ADP/ATP translocase and the brown fat mitochondria uncoupling protein. EMBO J. 1987. May; 6 (5):1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira GC, Pratt RD, Pedersen PL. Energy-linked anion transport. Cloning, sequencing, and characterization of a full length cDNA encoding the rat liver mitochondrial proton/phosphate symporter. J Biol Chem. 1989; 264 (26):15628–15633. [PubMed] [Google Scholar]
- 3.Dolce V, Fiermonte G, Messina A, Palmieri F. Nucleotide sequence of a human heart cDNA encoding the mitochondrial phosphate carrier. DNA Seq. 1991; 2 (2):133–135. [DOI] [PubMed] [Google Scholar]
- 4.Phelps A, Schobert CT, Wohlrab H. Cloning and characterization of the mitochondrial phosphate transport protein gene from the yeast Saccharomyces cerevisiae. Biochem. 1991. January; 30 (1):248–252. [DOI] [PubMed] [Google Scholar]
- 5.De Croos JA, McNally JD, Palmieri F, Storey KB. Upregulation of the mitochondrial phosphate carrier during freezing in the wood frog Rana sylvatica: potential roles of transporters in freeze tolerance. J Bioenerg Biomembr. 2004. June; 36 (3):229–239. [DOI] [PubMed] [Google Scholar]
- 6.Kiiskinen M, Korhonen M, Kangasjärvi J. Isolation and characterization of cDNA for a plant mitochondrial phosphate translo2 cator (Mpt1): ozone stress induces Mpt1 mRNA accumulation in birch (Betula pendula Roth). Plant Mol Biol. 1997. October; 35(3):271–279. [DOI] [PubMed] [Google Scholar]
- 7.Takabatake R, Hata S, Taniguchi M, Kouchi H, Sugiyama T, Izui K. Isolation and characterization of cDNAs encoding mitochondrial phosphate transporters in soybean, maize, rice, and Arabidopsis. Plant Mol Biol. 1999. June; 40 (3): 479–486. [DOI] [PubMed] [Google Scholar]
- 8.Nakamori K, Takabatake R, Umehara Y, Kouchi H, Izui K, Hata S. Cloning, functional expression, and mutational analysis of a cDNA for Lotus japonicus mitochondrial phosphate transporter. Plant Cell Physiol. 2002. July 25; 43 (10):1250–1253. [DOI] [PubMed] [Google Scholar]
- 9.Hamel P, Saint Georges Y, De Pinto B, Lachacinski N, Altamura N, Dujardin G. Redundancy in the function of mitochondrial phosphate transport in Saccharomyces cerevisiae and Arabidopsis thaliana. Mol Microbiol. 2004. January; 51 (2):307–317. [DOI] [PubMed] [Google Scholar]
- 10.Huang X, Xue T, Dai S, Gai S, Zheng C, Zheng G. Genes associated with the release of dormant buds in tree peonies (Paeonia suffruticosa). Acta Physiol Plant. 2008a. November; 30:797–806. [Google Scholar]
- 11.Liu F, Chang X, Ye Y, Xie W, Wu P, Lian X. Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol plant. 2011. November; 4 (6):1105–1122. 10.1093/mp/ssr058 [DOI] [PubMed] [Google Scholar]
- 12.Zhu W, Miao Q, Sun D, Yang G, Wu C, Huang J, et al. The mitochondrial phosphate transporters modulate plant responses to salt stress via affecting ATP and gibberellin metabolism in Arabidopsis thaliana. PLoS ONE. 2012. August; 7:e43530 10.1371/journal.pone.0043530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang X, Zhu W, Dai S, Gai S, Zheng G, Zheng C. The involvement of mitochondrial phosphate transporter in accelerating bud dormancy release during chilling treatment of tree peony (Paeonia suffruticosa). Planta. 2008b. September; 228:545–552. [DOI] [PubMed] [Google Scholar]
- 14.Gai S, Zhang Y, Mu P, Liu C, Liu S, Dong L, et al. Transcriptome analysis of tree peony during chilling requirement fulfillment: assembling, annotation and markers discovering. Gene. 2012. April 15; 497 (2):256–262. 10.1016/j.gene.2011.12.013 [DOI] [PubMed] [Google Scholar]
- 15.Prestridge DS. SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci. 1991. April; 7 (2): 203–206. [DOI] [PubMed] [Google Scholar]
- 16.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999. January 1; 27 (1):297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002. January 1; 30 (1):325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998. December; 16 (6):735–743. [DOI] [PubMed] [Google Scholar]
- 19.Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987. December; 5 (4):387–405. [Google Scholar]
- 20.Gai S, Zhang Y, Liu C, Zhang Y, Zheng G. Transcript Profiling of Paoenia ostii during Artificial Chilling Induced Dormancy Release Identifies Activation of GA Pathway and Carbohydrate Metabolism. PLoS ONE. 2013. February 6; 8:e55297 10.1371/journal.pone.0055297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2− ΔΔCT Method. Methods. 2001. December; 25 (4):402–408. [DOI] [PubMed] [Google Scholar]
- 22.Morita A, Umemura T, Kuroyanagi M, Futsuhara Y, Perata P, Yamaguchi J. Functional dissection of a sugar-repressed alpha-amylase gene (RAmy1A) promoter in rice embryos. FEBS Lett. 1998; 423 (1):81–85. [DOI] [PubMed] [Google Scholar]
- 23.Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P. A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley Aleurone. Plant Physiol. 2002. August 8; 130: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003. January; 15:63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chinnusamy V, Ohta M, Kanrar S, Lee B, Hong X, Agarwal M, et al. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Gene Dev. 2003. April 2; 17 (8):1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal M, Hao Y, Kapoor A, Dong C, Fujii H, Zheng X, et al. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006. October 2; 281 (49):37636–37645. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama Nakashita A, Nakamura Y, Watanabe Takahashi A, Inoue E, Yamaya T, Takahashi H. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 2005. February 17; 42 (3):305–314. [DOI] [PubMed] [Google Scholar]
- 28.Lam E, Chua N. ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell. 1989. December; 1 (12):1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vieweg MF, Frühling M, Quandt H, Heim U, Bäumlein H, Pühler A, et al. The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and nonlegume plants. Mol Plant Microbe In. 2004. January; 17 (1): 62–69. [DOI] [PubMed] [Google Scholar]
- 30.Bate N, Twell D. Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol Biol. 1998. July; 37 (5):859–869. [DOI] [PubMed] [Google Scholar]
- 31.Filichkin SA, Leonard JM, Monteros A, Liu PP, Nonogaki H. A novel endo-beta-mannanase gene in tomato LeMan5 is associated with anther and pollen development. Plant Physiol. 2004. March; 134 (3):1080–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farini E, Whitelaw CBA. Ectopic expression of β-1actoglobulin transgenes. Mol Gen Genet. 1995. November; 246 (6): 734–738. [DOI] [PubMed] [Google Scholar]
- 33.Kristiansen MT, Rasmussen LM, Olsen N, Asa SL, Jørgensen JO. Ectopic ACTH Syndrome: Discrepancy between Somatostatin Receptor Status in vivo and ex vivo, and between Immunostaining and Gene Transcription for POMC and CRH. Horm Res. 2002; 57 (5–6): 200–204. [DOI] [PubMed] [Google Scholar]
- 34.Taylor MS, Kai C, Kawai J, Carninci P, Hayashizaki Y, Semple CAM. Heterotachy in mammalian promoter evolution. PLoS Genet. 2006. April; 2(4): e30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horvath DP, Anderson JV, Chao WS, Foley ME. Knowing when to grow: signals regulating bud dormancy. Trend Plant Sci. 2003. November; 8 (11):534–540. [DOI] [PubMed] [Google Scholar]
- 36.Schrader J, Moyle R, Bhalerao R, Hertzberg M, Lundeberg J, Nilsson P, et al. Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. Plant J. 2004. October; 40 (2):173–187. [DOI] [PubMed] [Google Scholar]
- 37.Rohde A, Ruttink T, Hostyn V, Sterck L, Van Driessche K, Boerjan W. Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. J Exp Bot. 2007. September 28; 58 (15–16): 4047–4060. [DOI] [PubMed] [Google Scholar]
- 38.Rentzsch S, Podzimska D, Voegele A, Imbeck M, Müller K, Linkies A, et al. Dose-and tissue-specific interaction of monoterpenes with the gibberellin-mediated release of potato tuber bud dormancy, sprout growth and induction of α-amylases and β-amylases. Planta. 2012. January; 235 (1): 137–151. 10.1007/s00425-011-1501-1 [DOI] [PubMed] [Google Scholar]
- 39.Hazebroek JP, Metzger JD, Mansager ER. Thermoinductive regulation of gibberellin metabolism in Thlaspi arvense L. (II. Cold induction of enzymes in gibberellin biosynthesis). Plant Physiol. 1993. June; 102 (2):547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell. 2004. February; 16 (2): 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng GS, Gai SP, Gai WL. Changes of endogenous hormones during dormancy release by chilling in tree peony. Scientia Silvae Sinicae. 2009; 45:48–52. (Chinese in English abstract) [Google Scholar]
- 42.Chinnusamy V, Schumaker K, Zhu JK. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot. 2004. December 12; 55(395):225–236. [DOI] [PubMed] [Google Scholar]
- 43.Kim JC, Lee SH, Cheong YH, Yoo CM, Lee SI, Chun HJ, et al. A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. 2001. February; 25(3): 247–259. [DOI] [PubMed] [Google Scholar]
- 44.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011. April; 66(1): 94–116. 10.1111/j.1365-313X.2010.04459.x [DOI] [PubMed] [Google Scholar]
- 45.Licausi F, Ohme-Takagi M, Perata P. APETALA2/Ethylene responsive factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013. August; 199(3): 639–649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.